Abstract
Bile acids are a group of molecular species of acidic steroids with peculiar physical-chemical and biological characteristics. At high concentrations they become toxic to mammalian cells, and their presence is pertinent in the pathogenesis of several liver diseases and colon cancer. Bile acid cytoxicity has been related to membrane damage, but also to nondetergent effects, such as oxidative stress and apoptosis. Strikingly, hydrophilic ursodeoxycholic acid (UDCA), and its taurine-conjugated form (TUDCA), show profound cytoprotective properties. Indeed, these molecules have been described as potent inhibitors of classic pathways of apoptosis, although their precise mode of action remains to be clarified. UDCA, originally used for cholesterol gallstone dissolution, is currently considered the first choice therapy for several forms of cholestatic syndromes. However, the beneficial effects of both UDCA and TUDCA have been tested in other experimental pathological conditions with deregulated levels of apoptosis, including neurological disorders, such as Alzheimer's, Parkinson's, and Huntington's diseases. Here, we review the role of bile acids in modulating the apoptosis process, emphasizing the anti-apoptotic effects of UDCA and TUDCA, as well as their potential use as novel and alternate therapeutic agents for the treatment of apoptosis-related diseases.
Keywords: Bcl-2 family, cell death, liver, neuroprotection, nuclear steroid receptors
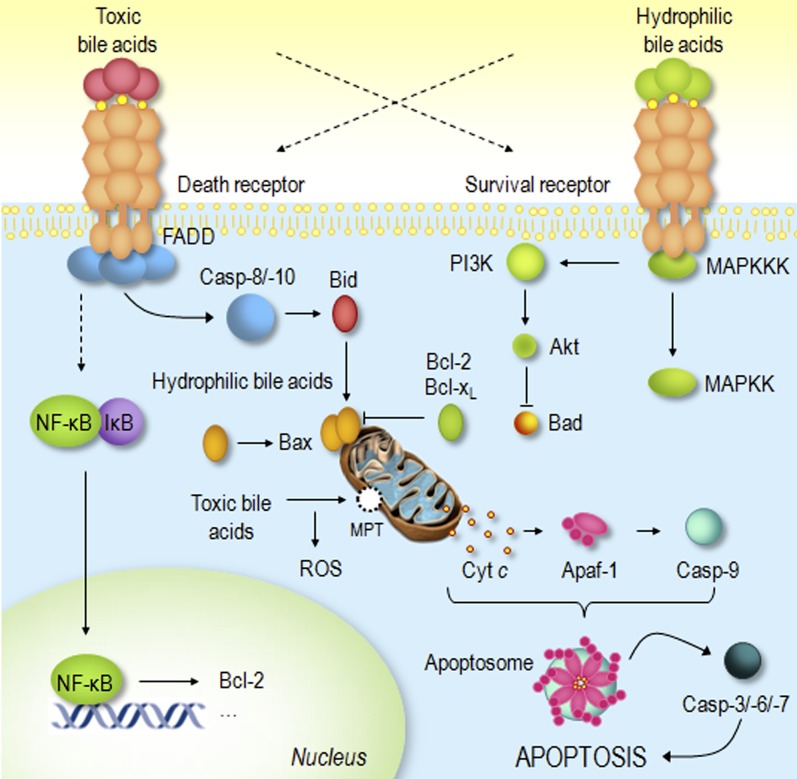
Bile acids, the major constituents of bile, are produced in the liver and secreted into the intestine, where they play crucial biological roles, such as solubilization of lipids in the intestinal lumen, among many others. Interest in bile acids has grown markedly over the last decades since discovering the role of these acidic steroids in several important physiological phenomena, including liver and intestinal pathology and pharmacology. Certain bile acids are cytotoxic molecules implicated in increased cell proliferation and cancer development in the intestinal tract (1) and/or cell death by necrosis and apoptosis. In fact, apoptosis has been described as a key event during hepatobiliary diseases (2). Curiously, not all bile acids are toxic, and previous studies suggest that this may be related to minor changes in their chemical structure (3). In this regard, accumulation of hydrophobic bile acids within the hepatocyte induces cell death of liver cells during cholestasis, while hydrophilic bile acids may be cytoprotective (Fig. 1).Ursodeoxycholic acid (UDCA), for instance, is an endogenous hydrophilic bile acid in clinical use for the treatment of certain liver diseases. There is now strong evidence that the cytoprotective effects of this molecule result, in part, from its ability to reduce the apoptotic threshold in several cell types through modulation of classical mitochondrial pathways. Interestingly, the use of UDCA as an agent to treat nonliver diseases associated with increased levels of apoptosis, such as neurodegenerative disorders, is now a major consideration. Moreover, in recent years, the use of bile acids in supramolecular chemistry, materials chemistry, and nanotechnology has also been the focus of intensive research (4).
Fig. 1.
Schematic representation of bile acid modulation of death and survival transduction pathways. Toxic bile acids induce apoptosis by activating both ligand-dependent and -independent death receptor oligomerization. The activation of death receptors by bile acids invariably signals the mitochondrial pathway of apoptosis in type II cells, such as hepatocytes. Moreover, toxic bile acids may directly target mitochondria, either through induction of the MPT and ROS production or activation of pro-apoptotic Bcl-2 family members. Finally, hydrophobic bile acids partially activate death-receptor-dependent survival pathways, such as the NF-κB. Hydrophilic bile acids do not induce apoptosis as they simultaneously activate survival signaling pathways, such as the MAPK and PI3K, antagonizing proapototic Bcl-2 family members and preventing mitochondrial dysfunction and apoptosis. See text for more complete description. Cyt c, cytochrome c; MAPKK, mitogen-activated protein kinase kinase; MAPKKK, mitogen-activated protein kinase kinase kinase; MPT, mitochondrial permeability transition.
BILE ACID METABOLISM AND PHYSIOLOGY
Bile acids are detergent molecules synthesized in the liver from neutral sterols by a complex series of chemical reactions (5). In humans and most animal species, bile acids are produced primarily from the cholesterol metabolic pathway (6). The complete synthesis of bile acids requires approximately 17 enzymes, in a process tightly regulated by nuclear hormone receptors and other transcription factors (7). Bile acids exert a negative feedback regulation on their own synthesis, in particular by inhibiting cholesterol 7α-hydroxylase (CYP7A1) and cholesterol 12α-hydrolase (CYP8B1) activities and expression, through mechanisms that are not entirely understood. The nuclear bile acid receptor farnesoid X receptor (FXR) is the primary mediator of this negative regulatory pathway. Two major FXR-dependent pathways for bile acid inhibition of CYP7A1 have been described and include the small heterodimer partner (SHP)/liver receptor homolog-1 (LRH-1) in liver and the fibroblast growth factor intestine 19 (FGF19)/tyrosine kinase FGF receptor 4 (FGFR4) in intestine. Nevertheless, other mechanisms independent of FXR signaling have also been identified (8, 9).
In the liver, activation of FXR by hydrophobic bile acids induces the expression of the SHP transcriptional repressor (10). SHP, in turn, negatively interacts with other transcription factors, such LRH-1, which has been shown to enhance cyp7a1 and cyp8b1 basal transcription (11). Because SHP binds efficiently to LRH-1 and inhibits its transcriptional activity, it was proposed that this interaction induces cyp7a1 and cyp8b1 repression (12, 13). Most notably, recent studies using mice models of LRH-1 tissue specific deficiency have changed the classic panorama of bile acid synthesis feedback inhibition (14, 15). Surprisingly, LRH-1 deficiency in mouse liver had no significant effect on either basal cyp7a1 expression or its repression by FXR. This was in agreement with previously published data showing that SHP gene knockout did not prevent bile acid inhibition of cyp7a1 mRNA expression in mice (16, 17), thereby suggesting an additional regulatory pathway. In contrast, cyp8b1 expression was significantly reduced and accompanied by predicted changes in the bile acid pool. LRH-1 also appears to induce the expression of other genes, including shp in liver and both shp and fgf15 in the ileum, as revealed by LRH-1 knockout in mouse liver and intestine, respectively (15).
Most studies on the feedback mechanisms involved in bile acid homeostasis have focused on liver with little attention paid to the intestine. Nevertheless, several reports reveal that the intestine also plays an active role in bile-acid-mediated suppression of bile acid synthesis in liver. For example, FXR induces FGF15, an ortholog of human FGF19, in mouse intestine where expression is inversely correlated to that of cyp7a1 mRNA levels in mouse liver (18). FGF19 was reported to activate FGFR4 signaling, thus inhibiting CYP7A1 mRNA expression levels in human hepatocytes (19). Moreover, specific liver and intestine FXR knockout confirmed that intestinal FXR is required for bile acid inhibition of CYP7A1 in liver (20). In fact, unlike inhibiting apical bile acid transport, blocking basolateral bile acid transport resulted in reduced hepatic bile acid synthesis with increased FGF15 ileal expression (21). FGF15 may function as an interface signal between the intestine and the liver, affecting CYP7A1 expression indirectly by modulating the activity of bile acids or other molecules that signal from intestine to liver. However, this model is hard to reconcile with the finding that induction of FGF15 expression in the ileum of bile duct ligated mice correlates with a significant reduction in CYP7A1 mRNA levels in liver. Further, ileal-derived FGF15 might function as a signaling hormone acting on the liver via FGFR4 to repress CYP7A1 expression and bile acid synthesis (18). In addition, the FGF15/FGFR4 pathway synergizes with SHP to regulate bile acid synthesis. While the mechanism responsible for cooperation between the FGF15/FGFR4 and SHP pathways remains to be clarified, recent studies raise the possibility that they are linked by a c-Jun NH2-terminal kinase (JNK)-dependent pathway (18).
In the last few years, with the discovery of FXR, and more recently of TGR5, a bile acid membrane receptor (22, 23), the role of bile acids as signaling molecules with important paracrine and endocrine functions has become more evident (24). Apart from the regulation of their own synthesis and transport, bile acids are involved in triggering the adaptive response to several insults to the liver (25). Furthermore, their role in the control of general energy-related metabolism, and more precisely in hepatic glucose handling, has been reported (26). Finally, bile acids display central roles in modulating liver cell death pathways as well as survival transduction pathways (Fig. 1).
KEY MECHANISMS OF APOPTOSIS
Apoptotic cell death is a highly regulated mechanism that can be viewed as a program of cell suicide vital for a wide variety of biological processes. The irony of the apoptosis process is that death is the basis for establishing life. In fact, apoptosis is an integral part of normal embryonic development and tissue homeostasis during adulthood. Sculpture of fingers and toes is a classic example of apoptosis, where cells between the digits must actually die to separate them. Apoptosis has also been documented as a prominent player in immune regulation and tissue homeostasis as well as in pathological and therapeutic settings. Defects in physiological pathways of apoptosis may occur, contributing to the development of numerous medical illnesses for which adequate therapy or prevention is lacking. In fact, excessive apoptosis can lead to T-cell depletion, neurodegenerative diseases, or hepatocellular degeneration, while impaired apoptosis contributes to oncogenesis, autoimmune diseases, and persistent infections (27). Thus, it is not surprising that apoptosis has become a topic of intensive research to identify molecular targets and propose effective therapies in the management of apoptosis-associated disorders.
Cells dying by apoptosis present a number of morphological changes that include chromatin condensation, cytoplasmic shrinkage, membrane blebbing, and the generation of small apoptotic vesicles containing intact cytoplasmic organelles as well as nuclear remnants (28). These apoptotic bodies are rapidly eliminated by resident phagocytic and neighboring cells, preventing the release of cellular components into the extracellular space and resulting inflammatory and immune response. Besides the morphological changes, apoptotic cells undergo a number of distinct biochemical events involving the loss of mitochondrial membrane potential, DNA fragmentation, and protein cleavage (29).
BIOCHEMICAL PATHWAYS OF APOPTOSIS
The apoptotic process may occur by several molecular pathways. The best characterized and most prominent, however, is the intrinsic pathway, involving mitochondria (30), and the extrinsic pathway, which is activated by death receptors at the cellular membrane (31). Although apparently independent, these two apoptotic pathways may interact through a finely orchestrated crosstalk involving key proteins that are common to both pathways.
Mitochondrial pathway
The mitochondrial pathway of apoptosis is triggered by intracellular stresses that converge on mitochondria, leading to membrane permeabilization, release of apoptogenic proteins, and disruption of the mitochondrial membrane potential. These changes culminate in activation of caspases and ultimately cell death (for a review, see Ref. 32). Mitochondrial membrane permeabilization may occur by the opening of the mitochondrial permeability transition pore or through the formation of specific release channels in the outer membrane, promoted by pro-apoptotic members of the Bcl-2 family. Bcl-2 family members regulate the mitochondrial pathway in both a positive and negative fashion (33). In unstressed cells, anti-apoptotic members (e.g., Bcl-2, Bcl-xL, and Bcl-w) are integral membrane proteins localized in the mitochondrial outer membrane. In contrast, pro-apoptotic members (e.g., Bax and Bak) are usually located in the cytosol as inert forms. Following induction of apoptosis, Bax and Bak undergo conformational changes, oligomerize, and translocate to mitochondria, where they directly or indirectly form specific pores for the release of apoptogenic factors, such as cytochrome c. Once in the cytosol, cytochrome c oligomerizes with Apaf-1, recruiting procaspase-9 to form the apoptosome. This results in cleavage and activation of caspase-9, which in turn cleaves and activates downstream caspases that function as effectors of the apoptotic process.
Death receptor pathway
The classical death receptor pathway of apoptosis is triggered by ligand-induced activation of death receptors at the cell surface, followed by recruitment and oligomerization of intracellular adaptor molecules (34). Death receptors are type 1 transmembrane proteins, belonging to the tumor necrosis factor (TNF) receptor superfamily (35), and include TNF receptor-1, CD95/Fas, TRAIL (TNF-related apoptosis inducing ligand) receptors 1 and 2, and death receptors 3 and 6. As an alternative, the extrinsic pathway can be activated by dependency receptors, which are believed to be connected to rapid caspase activation. In the absence of ligand, these receptors deliver a death signal through yet unidentified mediators, thus generating a state of cellular dependence from their ligands (36).
Caspase-8 is the dominant death receptor-activated initiator caspase. In type I cells, caspase-8 directly activates effector caspases perpetuating the apoptotic process (37). Importantly, in type II signaling cells, such as hepatocytes, progression of the apoptotic cascade depends entirely on its amplification by mitochondria (38). In this case, caspase-8 cleaves inactive cytoplasmic Bid. Once activated, cytoplasmic Bid induces conformational changes in pro-apoptotic Bax and Bak, which in turn transduce the death stimuli to mitochondria (39). This coordination between both pathways reinforces the critical role of mitochondria during programmed cell death.
Cell cycle and apoptosis
Apoptosis and proliferation are intimately coupled. In fact, tissue homeostasis is dependent on the precise balance between cell proliferation and cell death; thus, it is not surprising that some cell cycle factors modulate both processes. In eukaryotic cells, the key trigger for the progression from one phase of the cell cycle to the next and, ultimately, cell cycle progression results from a complex series of interactions between cyclins, cyclin-dependent kinases (CDKs), and cyclin-dependent kinase inhibitors (40). Different cyclins act at distinct phases of the cell cycle by binding to and stimulating the activities of CDKs. Cyclin D1, for instance, specifically regulates the G1-to-S progression of the cell cycle. In addition to their CDK-binding activities, D-type cyclins possess CDK-independent properties and form physical associations with several transcription factors (41). Although the role of cyclins and CDKs in stimulating cell cycle progression and proliferation is indisputable, there is increasing evidence to suggest that these, as well as other cell-cycle-related proteins, may also play critical roles in cell growth inhibition, senescence, and apoptosis (42). For example, it was initially shown that cyclin D1 mRNA levels were strongly increased during the death of neurons (43). However, the mechanisms by which cyclin D1 induces apoptosis are far from completely understood.
Additional regulatory controls that can initiate apoptosis include the E2F-1 transcription factor. As a member of the E2F family, it not only regulates the expression of many genes necessary for normal proliferation, in a cell-cycle-specific manner, but has also been implicated in inducing apoptosis (44). E2F-1 is typically bound to the unphosphorylated retinoblastoma protein (pRb). However, under certain stress conditions and during cell cycle, inactivation of pRb by phosphorylation releases E2F-1 to transactivate genes necessary for apoptosis. Curiously, some studies suggest that ectopic expression of cyclin D1 may cause apoptosis by phosphorylating pRb, resulting in deregulation of the pRb/E2F complex (45). In addition, activation of p53 is one of the mechanisms involved in E2F-1-induced apoptosis. In the absence of stress signals, p53 is kept in check to allow normal cell proliferation and/or maintenance of cell viability. The p53 protein is mainly regulated at the posttranslational level by Mdm-2, or in humans, Hdm-2, which has been shown to inhibit p53 activity by regulating its stability, cellular localization, and ability to activate transcription (46). Interestingly, E2F-1 stabilizes p53 by induction of alternate reading frame (ARF), which directly binds to Mdm-2 and prevents p53 proteasomal degradation.
The link between cell cycle and apoptosis has also been recognized in proteins such as c-Myc, Ras, and other cell cycle regulators, including p21 and p27. In fact, some of these proteins are targets for cleavage by caspases, underscoring the absence of cell cycle progression during apoptosis (47). A relationship between bile acids and cell proliferation also exists (48). Taken together, induction of apoptosis depends on the cellular environment where conflicting signals for cell proliferation and cell cycle arrest may result in cell death.
BILE ACID INDUCTION OF APOPTOSIS
The mechanisms by which bile acids induce apoptosis in hepatocytes are still not entirely known. It was thought that hydrophobic bile acids, such as glycochenodeoxycholic (GCDCA) and taurochenodeoxycholic acids, could induce cytotoxicity by acting as detergents on cell membranes. Nevertheless, serum bile acid levels in cholestatic patients are insufficient to produce a significant detergent effect; therefore, bile acid cytotoxicity appears to result from other processes. In fact, basic cellular mechanisms of hepatocyte injury might be primarily involved, ultimately causing cell death by either necrosis or apoptosis. There are several studies showing association between caspase activation, mitochondrial dysfunction, and cellular distribution of Bcl-2-related proteins in models of cholestasis and hepatocyte injury (49). Indeed, pathophysiological concentrations of bile acids induce apoptosis both by directly activating death receptors (50) and inducing oxidative damage and mitochondrial dysfunction (51, 52), a combination that strongly sensitizes cells to apoptosis (Fig. 1). In addition, the activation of death receptors invariably signals the mitochondrial pathway of apoptosis in hepatocytes. Thus, hydrophobic bile acids are unique among naturally occurring apoptotic agents due to their potential to induce apoptosis through both nonspecific detergent effects and receptor-mediated interactions.
Toxic bile acids have been shown to induce apoptosis in a Fas- and TRAIL-dependent manner (50, 53), involving recruitment of the Fas-associated death domain (FADD), activation of caspase-8 and Bid, as well as downstream effector caspases. Activation of death receptors by bile acids involves induction of their transport from the Golgi (54). Spontaneous receptor oligomerization then occurs at the cell membrane. As an example, lithocholic-acid-induced apoptosis in colon cancer cell lines proceeds through a mitochondrial/caspase-9-dependent pathway initiated by caspase-8 (55). Bile acids are also known to cause ligand-independent activation of Fas in hepatocytes, resulting in subsequent caspase-8 activation. The mechanism of Fas activation has been studied in hepatocytes and is thought to occur through generation of oxidative stress by dihydroxy bile acids followed by epidermal growth factor receptor-dependent Fas translocation and oligomerization (56, 57). Bile acid oxidative stress, which ultimately leads to Fas activation, can be generated through a multifactorial mechanism involving NADPH oxidase activation and interaction of bile acids with mitochondria to directly generate reactive oxygen species (ROS) (58). More recently, G-protein-coupled receptor TGR5 activation has also been implicated in Fas translocation and oligomerization (59), which is in agreement with previous results showing TGR5 activation by lithocholic and chenodeoxycholic acids (60). Alternatively, Fas activation by Fas ligand or other agonists may also be increased; interestingly, this process is stimulated by p53. The transcription factor was shown to transiently increase Fas expression at the cell surface via transport from the Golgi complex and to induce Fas-FADD binding (61). In addition, p53 was implicated in cyclin kinase inhibitor-enhanced, bile-acid-induced apoptosis via Fas (62).
Novel findings in the field point to a critical role of caspase-6 in bile-acid-mediated apoptosis of human liver cell lines and primary rat hepatocytes (63). Caspase-6, generally considered an executioner caspase, has recently been identified as a potential activator of caspase-8 in models of bile-acid-induced apoptosis. This protease appears to be part of a feedback loop in bile-acid-induced apoptosis of hepatocytes, thereby enhancing the toxicity of pro-apoptotic bile acids. Curiously, caspase-6 activation is less important in apoptosis induced by other stimuli in the same cells. Finally, although mitochondrial cytochrome c release is FADD/caspase-8 dependent during hepatocyte apoptosis, pro-apoptotic bile acids, at concentrations similar to those found in cholestatic liver injury, are still capable of inducing apoptosis by the intrinsic pathway when death receptor activation is inhibited (63). Recently, apoptosis via the endoplasmic reticulum (ER) stress-mediated pathway was also found in GCDCA-induced apoptosis of liver cells, although to a lesser extent (64, 65). Treatment with the bile acid resulted in ER-related Ca2+ release, increased calpain and caspase-12 activities, and induction of the ER stress biomarkers Bip and Chop mRNA expression levels. Therefore, inhibition of death receptor signaling alone is probably not sufficient to reduce hepatocyte apoptosis and consecutive bile-acid-induced liver damage in cholestatic diseases.
BILE ACID INHIBITION OF APOPTOSIS
Cell survival signaling
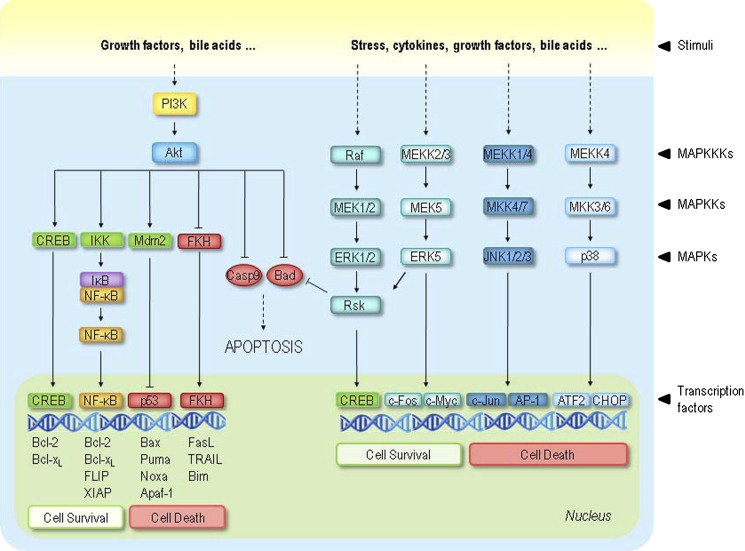
Although bile acids are inherently toxic compounds, they are also potent signaling agents that modulate intracellular molecular pathways (Fig. 1). Interestingly, these molecules are capable of inhibiting their own cytotoxicity by triggering survival signals (66). The so-called survival signaling pathways have evolved to protect cells from pathologic apoptosis and include nuclear factor κB (NF-κB), phosphatidylinositol 3-kinase (PI3K), and mitogen-activated protein kinase (MAPK) pathways (Fig. 2). Survival stimuli generally trigger intracellular signaling through activation of transmembrane receptors. The liver has the ability to limit apoptosis during cholestasis by triggering specific mechanisms. Although it has been shown that Bcl-2, Bcl-XL, and Bax are expressed in the liver, only cholangiocytes and not hepatocytes normally express anti-apoptotic Bcl-2. However, induction of cholestasis by bile duct ligation leads to Bcl-2 expression in hepatocytes, which may represent an adaptative phenomenon to protect hepatocytes. In addition, the activation of NF-κB and subsequent regulation of anti-apoptotic genes (67), as well as the cytoplasmic sequestration of p53 (68), are mechanisms triggered by the liver to inhibit or modulate apoptosis induced by toxic bile acids.
Fig. 2.
Schematic representation of the interplay between cellular survival signaling and apoptosis regulators. Cell survival requires the active inhibition of apoptosis, which is accomplished by reducing the expression of pro-apoptotic factors as well as promoting expression of anti-apoptotic factors. Survival pathways may be triggered by a wide variety of extracellular signals, such as growth factors, stress, proinflammatory cyto kines, and bile acids. Activation of Akt results in inhibition of pro-apoptotic factors, such as Bad and caspase-9, and activation of FKH, CREB, and NF-κB transcription factors, all involved in cell survival regulation. The MAPK pathways include ERK, JNK, and p38 cascades, all of which contain the same series of kinases. Although ERK signaling is considered mainly cytoprotective, JNK and p38 are referred to primarily as stress-activated proteins. The effect of MAPK pathways on survival is mediated, at least partly, by activation of ribosomal S6 kinase (RSK) family members. Much like Akt, RSKs inactivate Bad and activate CREB, ultimately leading to cell survival.
Several studies have linked bile-acid-induced extracellular-regulated kinase (ERK) signaling to a cytoprotective response against bile-acid-induced Fas receptor/caspase activations (56, 69, 70). In addition, JNK signaling may have both pro- and anti-apoptotic effects, including those that are downstream of the TNF-α receptor in hepatocytes (71). In this regard, the bile acid deoxycholic acid (DCA) activates both ERK and JNK pathways. While DCA-induced ERK1/2 and JNK2 signaling appears to be mainly cytoprotective, DCA-induced JNK1 is cytotoxic. Triggering of the ERK cascade by DCA results in increased DNA binding of CCAAT enhancer binding protein β (C/EBPβ) and cyclic AMP-response element binding protein (CREB), two putative cytoprotective transcription factors. Paradoxically, it has been determined that DCA-induced JNK1/2 activation is dependent on Fas signaling, which implies that bile-acid-induced activation of Fas can generate opposing signals. On the one hand, it activates caspase and JNK1 activation, resulting in cell death; on the other hand, it induces JNK2 leading to protection (72). Other studies have demonstrated that some bile acids activate PI3K-dependent survival pathways to prevent their otherwise inherent toxicity (73–75) or protect against more hydrophobic bile acids species (66, 76). In this respect, bile acids appear to dance with both death receptors and cell survival cascades; the balance likely lies within the strength and fidelity of the dancing partner (77).
UDCA modulation of apoptosis
In contrast to the toxic effects of hydrophobic bile acids, UDCA is a hydrophilic bile acid widely used as a therapeutic drug for patients with cholestatic liver diseases (78, 79). At present, it is the only drug approved by the United States Food and Drug Administration for the treatment of primary biliary cirrhosis. Despite its clinical efficacy, the precise mechanism by which UDCA improves liver function is still not entirely understood. It is now well established that inhibition of liver cell apoptosis is one of the main routes for the protective effect of UDCA, along with protection of cholangiocytes against cytotoxicity of hydrophobic bile acids and stimulation of impaired biliary secretion (80). Most likely, UDCA protective function results from a coordinated process involving several effects, depending on the type and stage of the disease.
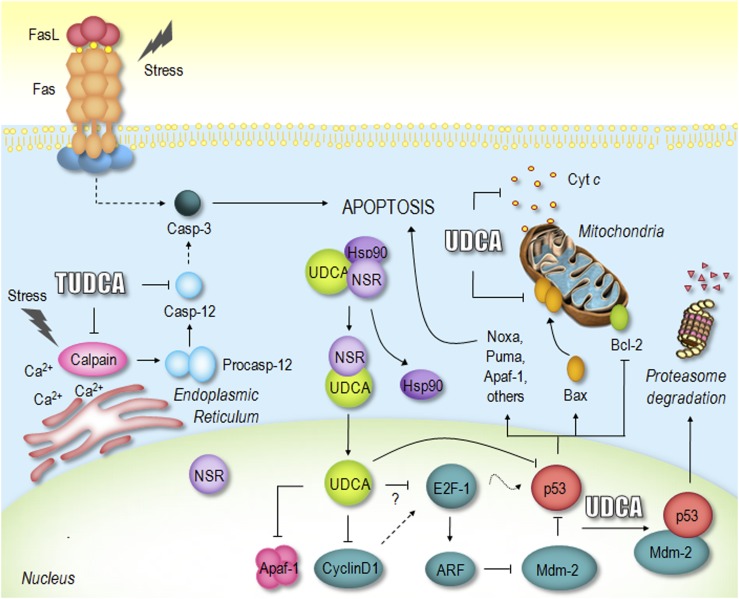
The anti-apoptotic effects of UDCA have been demonstrated both in rat liver (81, 82) and isolated human hepatocytes (83). While toxic bile acids fed to rats induce apoptosis in the liver, UDCA inhibits this effect by preventing formation of ROS and translocation of the pro-apoptotic protein Bax from the cytosol to the mitochondria. More importantly, these effects were also observed in nonhepatic cells, indicating that UDCA acts as a pleiotropic agent playing a unique role in modulating the classic mitochondrial pathway of apoptosis in different cell types (84). UDCA and its amidated conjugates, tauroursodeoxycholic acid (TUDCA) and glycoursodeoxycholic acid, were shown to modulate mitochondrial membrane perturbation, pore formation, Bax translocation, cytochrome c release, caspase activation, and subsequent substrate cleavage (85). In addition, TUDCA can directly stabilize mitochondrial membranes, having a profound effect on Bax channel formation in isolated mitochondria (86) (Fig. 3).
Fig. 3.
Proposed mechanisms of UDCA and TUDCA inhibition of apoptosis. UDCA negatively modulates the mitochondrial pathway by inhibiting Bax translocation, ROS formation, cytochrome c release, and caspase-3 activation. UDCA can also interfere with the death receptor pathway, inhibiting caspase-3 activation. Moreover, TUDCA inhibits apoptosis associated with ER stress by modulating intracellular calcium levels and inhibiting calpain and caspase-12 activation. Importantly, UDCA interacts with NSR, leading to NSR/hsp90 dissociation and nuclear translocation of the UDCA/NSR complex. Once in the nucleus, UDCA modulates the E2F-1/p53/Bax pathway, thus preventing apoptosis. Finally, UDCA downregulates cyclin D1 and Apaf-1, further inhibiting the mitochondrial apoptotic cascade. See text for more complete description. Cyt c, cytochrome c; Hsp90, heat shock protein 90.
UDCA was also shown to partially prevent apoptosis via the death receptor pathway in primary mouse hepatocytes cocultured with fibroblasts expressing Fas ligand. The protective effect was not associated with reductions in Fas trimerization but rather appeared to involve a direct effect on the mitochondrial membrane (87). Although not fully understood, TUDCA may also regulate the ER stress-mediated pathway, reducing calcium efflux and activation of caspase-12, which plays an important role in several liver diseases (88) (Fig. 3). Curiously, a recent study described TUDCA as a chemical chaperone of the ER in mice, alleviating ER stress, and thereby acting as a potential agent in the treatment of type 2 diabetes and the metabolic syndrome (89).
Currently, the major interest in UDCA cytoprotection is on identifying the primary molecular targets that ultimately mediate UDCA signaling to the mitochondria. It has been reported that bile acids are detectable within the nuclei of rat hepatocytes (90, 91) where they may play an important role in controlling gene expression. More recently, nuclear translocation of UDCA mediated by nuclear steroid receptors (NSRs) was shown to be essential for its anti-apoptotic properties (92) (Fig. 3). Once in the nucleus, UDCA might regulate gene expression by interacting directly with chromatin or alternatively through interaction with nuclear proteins, such as transcription factors. These findings have opened new avenues for UDCA protection from apoptosis. Microarray studies revealed that UDCA per se modulates the expression of at least 96 genes in primary rat hepatocytes, most of them involved in apoptosis and cell cycle regulation (93). UDCA was shown to reduce transcriptional activation and expression of cyclin D1 in hepatocytes incubated with DCA, which appears to contribute to the anti-apoptotic effects of the hydrophilic bile acid (94). On the other hand, TUDCA also appears to modulate apoptosis induced by toxic bile acids through inhibition of the human transcription factor activating protein-1 (AP-1) (95). Importantly, AP-1 is a dimer consisting of proteins belonging to Jun, Fos, ATF, or MAF families, which is involved in the regulation of cell proliferation, transformation, and death (96).
UDCA inhibition of apoptosis also involves modulation of other transcription factors, such as E2F-1 and p53, independently of its effect on mitochondria and/or caspases (97). Interestingly, p53 is a key molecular target in UDCA prevention of cell death, where the finely tuned, complex control of p53 by Mdm-2 represents a prime target for UDCA modulation of p53-mediated cell death (98). Of note, UDCA has been the first-line treatment of several liver diseases for decades, without increasing patients' predisposition to cancer. On the contrary, a recent study examined the effects of prolonged UDCA administration on the prevalence and recurrence of colorectal adenoma in 114 patients with primary biliary cirrhosis (99). The results revealed that long-term administration of UDCA did not increase the risk of colon adenoma and, in fact, acted as a chemopreventive agent by significantly decreasing the rates of colorectal adenoma recurrence following removal. Other authors have presented data linking UDCA to a lower rate of colorectal dysplasia in patients with ulcerative colitis and primary sclerosing cholangitis (100). In addition, UDCA prevented tumor development in a rat azoxymethane model of colon cancer, in part by inhibiting the growth-enhancing alterations in cyclin D1 and E-cadherin induced by this carcinogen (101–103). Finally, it has recently been reported that UDCA abrogated cisplatin-induced apoptosis of mouse sensory neurons via downregulation of the p53 signaling pathway (104). Together, these findings support a beneficial role of UDCA in the fine-tuning modulation of deregulated apoptosis, representing perhaps a therapeutic option to overcome p53-mediated alterations in cancer as well as other p53-associated disorders.
Activation of survival pathways may represent important additional mechanisms by which UDCA inhibits cell death. For example, UDCA can stimulate the MAPK and the PI3K signaling pathways. In fact, it has been demonstrated that the p38/ERK/MAPK and PI3K pathways are involved in protection of TUDCA against GCDCA-induced apoptosis in rat hepatocytes (66). TUDCA also significantly inhibited each of the amyloid β (Aβ)-induced apoptotic events in rat neuronal cells, in part, through activation of the PI3K pathway (105). In addition, it protected cardiomyocytes in culture against reperfusion injury in a PI3K/Akt-dependent pathway (106).
It is interesting to note that UDCA may also be cytoprotective through mechanisms other than inhibition of apoptosis. In fact, UDCA has the ability to suppress the NF-κB pathway and subsequent cytokine expression via activation of the glucocorticoid receptor (GR), thereby playing an important anti-inflammatory role in the liver (107).
Crosstalk between bile acids and NSRs
During the past several years, it has become evident that the NSR family of transcription factors regulates apoptosis in different cell systems. Curiously, NSRs play a heterogeneous function in apoptosis, which is affected by tissue-specific parameters, such as alternative initiation sites within nuclear receptor genes as well as the varying effects of comodulators. However, the mechanism by which NSR can differentially affect the expression of the same subset of genes for different cells is still a matter of debate. Bile acids, as cholesterol-derived molecules, share chemical and structural similarities to steroid hormones and can potentially bind and modulate NSR activation. In fact, they are now considered cognate ligands of certain nuclear receptors originally classified as orphan nuclear receptors.
The link between UDCA and the GR was primarily correlated with anti-inflammatory properties of the bile acid. Interestingly, UDCA has also been described as a regulator of the immune response, and its immunomodulatory effects in cholestatic liver diseases show similarities with glucocorticoid (GC)-mediated immunomodulation.
Several attempts have been made to identify GC-induced genes by using advent microarray technologies. Nevertheless, many of the target genes whose transactivation or transrepression by GR somehow modulates apoptosis remain uncertain. Although GR acts primarily as a transcription factor, it also exhibits distinct nongenomic effects. As an example, GR has been shown to be located within mitochondria in some cell types, which may be a crucial step in triggering apoptosis (108). The upregulation of GR-mediated gene transcription appears to enhance apoptosis of myeloma cells, osteoblasts, lymphoid cells, and neuronal cells. Importantly, GR activation has been correlated with excessive cell death pathological conditions, such as Alzheimer's and Parkinson's diseases (109, 110). At odds with its pro-apoptotic effects in certain cell types, GR also mediates prosurvival signals in others. Strikingly, profound protective effects against apoptosis have been continuously attributed to GR in several liver-derived cells (111, 112). Rat liver regeneration is also thought to involve elevated expression of GR (113). Interestingly, GR was also found to physically interact with the tumor suppressor p53 and its dual role in inducing apoptosis or cell cycle arrest. The functional interactions between GR and p53 in different physiological conditions may significantly alter the effects of each transcription factor (114).
Studies from our lab have shown that GR is required for UDCA anti-apoptotic function. For example, the endogenous silencing of GR abolished the protective effect of UDCA against transforming growth factor β1 (TGF-β1)-induced apoptosis in primary rat hepatocytes (115). During TGF-β1-induced apoptosis, significant degradation of GR occurs in liver cells. However, pretreatment of cells with UDCA not only markedly upregulated GR but also resulted in increased receptor nuclear translocation, when compared with TGF-β1 alone. In addition, the transcription factor E2F-1 was found to be a potent target for UDCA-induced GR activation. In fact, changes in E2F-1 protein levels induced by TGF-β1 were no longer prevented by UDCA after transfection of hepatocytes with small interfering RNA to GR. Furthermore, the subsequent increase in levels of the p53 repressor Mdm-2 and the decrease of p53 induced by UDCA were abrogated with GR silencing, thus supporting the notion of E2F-1 modulation (Fig. 3). However, the precise link between NSR and UDCA-regulated E2F-1, Mdm-2, and p53 remains to be determined. Other authors have also suggested that GR interferes with the expression of mitogenic factors, such as cyclins, CDKs, and E2F-1 (116).
More recently, there have been additional insights on how NSRs are involved in UDCA as an anti-apoptotic agent. Indeed, we demonstrated that UDCA promotes the dissociation between GR and its molecular chaperone, hsp90, thus inducing subsequent GR translocation into the nucleus of hepatocytes (92). However, when the C-terminal region of GR was deleted, UDCA no longer induced GR/hsp90 dissociation or GR nuclear translocation nor protected against apoptosis, indicating that GR ligand binding domain (LBD) is at least one target domain required for the anti-apoptotic function of UDCA. By using a fluorescently labeled UDCA molecule, we clearly demonstrated that UDCA reaches the nucleus of primary rat hepatocytes in a GR-dependent mechanism. Nuclear translocation of NSR by bile acids may not always result in transactivation or transrepression of NSR. In fact, bile acids might only require NSR to reach the nucleus, where they themselves regulate genes through modulation of other transcription factors. Indeed, although GCs are well-known inhibitors of apoptotic cell death, we showed that the protective role of UDCA does not require GR transactivation in liver cells. UDCA increases the activation of GC response elements in hepatocytes that overexpress GR but does not protect against TGF-β1-induced apoptosis by further increasing GR transactivation. Therefore, it is conceivable that UDCA requires NSR for translocation to the cell nucleus as part of a ligand-receptor complex, using a mechanism similar to that of steroid hormones (Fig. 3).
Interestingly, other studies have demonstrated that UDCA interacts with GR in the absence of specific ligands, as a novel and selective GR modifier (117). In addition, it was reported that UDCA promotes DNA binding of GR through interaction with its LBD, but without eliciting GR transactivation (107). Only under certain conditions may UDCA enhance GR-responsive gene expression. For example, in the presence of dexamethasone, UDCA increases GR-induced tyrosine aminotransferase, a hepatocyte-specific marker of GR (118). Although there is no structural evidence for the direct binding of UDCA to GR, it is thought that UDCA acts at a region of the LBD distinct from that of dexamethasone. UDCA then possibly modulates GR structure into a unique conformation that can translocate into the nucleus and bind DNA, but no longer interacts with coactivators to elicit transcriptional activation. These data may, in part, explain the anti-inflammatory properties of UDCA. In fact, UDCA pretreatment was shown to suppress DNA binding activity of AP-1 and repressed NF-κB-dependent transcription via interaction of GR with NF-κB. It is generally felt that transrepression of such transcription factors is the primary mechanism by which GCs mediate their anti-inflammatory activity. Moreover, different ligands induce various conformations of NSRs leading to unique regulatory properties of the receptors. It is therefore possible that UDCA may regulate several apoptosis-related genes by functionally modulating GR. Other mechanisms of GR activation by UDCA might be membrane related and involve an unidentified secondary signal that activates other signal transduction pathways. Although GRs share similar ligands and target the same genes as the mineralcorticoid receptor (MR), they may also form heteromeric complexes in which MR represses GR function (119). Thus, although there is no evidence that MR and UDCA interact directly, it is conceivable that UDCA-mediated modulation of GR may also affect MR.
The function of MR during apoptosis is thought to be primarily protective in nature. In the brain, stress-induced GR activation leads to an arrest of neurogenesis and increased apoptosis, which in turn is inhibited by MR activation. In addition, MR increases neuronal survival by decreasing p53 levels as well as the ratio of pro-apoptotic to anti-apoptotic members of the Bcl-2 family (120). This is thought to result from repression of GR by MR. The anti-apoptotic action of MR has been extensively described in neuronal cells, but less is known about its role during hepatocyte apoptosis. Interestingly, our results have suggested a relevant mechanistic function for MR during hepatocyte apoptosis. We demonstrated that much like GR, MR contributes to the protective effect of UDCA against the E2F-1/Mdm-2/p53 apoptotic pathway induced by TGF-β1 in primary rat hepatocytes (115). In addition, E2F-1, Mdm-2, and p53 were also found to be potent targets for UDCA-induced MR activation in liver cells. However, in the presence of Aβ peptide, TUDCA appears to differentially modulate MR and GR nuclear translocation in rat cortical neurons (121). In fact, TUDCA preferentially interacted with MR LBD, thus promoting MR dissociation from its cytosolic chaperones and translocation to the nucleus. It is possible that specific comodulators in neuronal cells may somehow induce TUDCA-mediated MR nuclear translocation, while reducing GR nuclear traffic. Surprisingly, and in contrast to hepatocytes, pretreatment of rat pheochromocytoma (PC12) cells with TUDCA significantly modulated Aβ-induced changes in MR and GR transactivation. In fact, the transcriptional activity of MR was increased while GR transactivation was reduced by TUDCA. This differential modulation of GR and MR in hepatocytes and PC12 cells suggests that additional pathways exist for the anti-apoptotic function of UDCA and TUDCA involving NSR as key factors. Whether UDCA and TUDCA may also regulate apoptosis through differential mechanisms of action is unclear.
UDCA has also been shown to regulate activity of other nuclear receptors. In fact, UDCA is a relatively strong pregnane X receptor (PXR) agonist and a weaker FXR agonist. Although UDCA does not itself bind FXR, it does inhibit receptor activation by more hydrophobic bile acid species (122). Moreover, during liver regeneration, increased levels of UDCA were shown to inhibit bile acid-FXR mediated repression of several genes (123). Other studies have shown that UDCA activates PXR to induce CYP3A4 (124). PXR is transcriptionally active in stellate cells by inhibiting transformation and proliferation. It is possible then that the antifibrotic effects of UDCA are also mediated via PXR.
BILE ACIDS AS TREATMENT OF EXPERIMENTAL NEURODEGENERATIVE DISORDERS
Inhibition of neuronal apoptosis by bile acids
Neurodegeneration corresponds to any pathological condition primarily affecting neurons (125). Neurodegenerative diseases are associated with a number of insults that may trigger the apoptotic process, including misfolded proteins, reactive oxygen and nitrogen species, mitochondrial complex inhibition, calcium imbalance, excitotoxicity, trophic factor withdrawal, and death receptor activation (126).
The therapeutic role of UDCA has been established in the treatment of certain liver diseases. Importantly, UDCA has the ability to modulate apoptosis at several levels, suggesting a common mechanism for cell survival that is independent of cell type (82). Thus, the use of UDCA for nonliver diseases associated with increased levels of apoptosis, specifically neurological disorders, is now an active area of study. Interestingly, after conjugation with taurine, UDCA administrated in high doses can be delivered to other tissues, including the brain (127). TUDCA inhibits apoptosis induced by several stimuli in isolated neuronal cells (105, 128).
The protective role of TUDCA has been extended to several mouse models of neurological disorders, including Huntington's disease. In fact, TUDCA markedly reduced the mitochondrial perturbations associated with apoptosis induction in cultured neuronal cells incubated with 3-nitropropionic acid (128). Consistently, the systemic administration of TUDCA in the 3-nitropropionic acid rat model of Huntington's disease was shown to reduce the associated morphologic striatal lesions (127). Moreover, behavioral studies correlated with histopathological findings, since the neuroprotection resulted in almost prevention of hyperactive behavior. Initial studies were extended to the R6/2 transgenic mouse model of Huntington's disease, where it was demonstrated that administration of TUDCA significantly reduced striatal neurodegeneration and ameliorated locomotor and sensorimotor deficits (129).
Interestingly, it was also shown that TUDCA can improve the survival and function of nigral transplants in a rat model of Parkinson's disease (130). In fact, TUDCA significantly reduced apoptosis in ventral mesencephalic tissue cultures and within the transplants, suggesting that the bile acid exerts beneficial effects on dopamine neuron survival mainly through neuronal death inhibition. In addition, TUDCA was shown to partially rescue a Parkinson's disease model of Caenorhabditis elegans from mitochondrial dysfunction (131).
The anti-apoptotic role of TUDCA was extended to acute conditions, particularly to a rat model of transient focal cerebral ischemia (132). Importantly, it was demonstrated that intravenous administration of TUDCA reduced infarct volume by ∼50%, modulated the levels of apoptosis, and inhibited the neurobehavioral impairment. In addition, TUDCA was capable of reducing the degree of brain injury and improving neurologic performance in a collagenase-induced hemorrhagic model of stroke by preserving mitochondrial membrane stability and inhibiting caspase activation (133).
Impressively, it has been recently shown that TUDCA preserves photoreceptor structure and function in the rd10 retinitis pigmentosa mouse model through postnatal day 30 (134). TUDCA was able to preserve rods and cones through postnatal day 30, the stage at which photoreceptor cell loss peaks. Thus, these experiments tested the efficacy of TUDCA at a critical stage of degeneration in retinitis pigmentosa. In addition, intraperitoneal injections of TUDCA were neuroprotective in a murine model of spinal cord injury (135). In fact, TUDCA reduced the number of necrotic and apoptotic cells by mitochondria stabilization, inducing a better functional outcome after spinal cord injury. Interestingly, pretreatment with TUDCA also significantly reduced glutamate-induced apoptosis of rat cortical neurons. TUDCA was capable of modulating glutamate-induced caspase activation and cytochrome c release, reducing the apoptotic threshold (136).
Prompted by these results, the anti-apoptotic role of TUDCA has recently been investigated in experimental models of Alzheimer's disease. Alzheimer's disease is the most common age-related neurodegenerative disorder (137). Amyloid plaques, extracellular deposits of mainly 40–42 Aβ peptides, and neurofibrillary tangles, intraneuronal aggregates comprised primarily of hyperphosphorylated insoluble tau, are common brain lesions in Alzheimer's disease. These aggregates are now strongly implicated in the onset and/or progression of Alzheimer's disease, inducing several neurodegenerative changes, such as neuronal apoptotic death. Initial studies using primary rat neurons and astrocytes incubated with Aβ showed increased levels of apoptosis, which was prevented by TUDCA and UDCA (138). The anti-apoptotic effects of TUDCA were further confirmed in PC12 neuronal cells incubated with Aβ (139). Similar results were seen in in vitro models of familial Alzheimer's disease consisting of mouse neuroblastoma cells expressing APP with the Swedish mutation, or double-mutated human APP and PS1 (140), and in primary cortical neurons incubated with fibrillar Aβ1-42 (141).
Due to their potential role in Aβ-induced toxicity, mitochondria were isolated and coincubated with TUDCA to assess its protective effect against apoptosis. In fact, UDCA, and more efficiently TUDCA, inhibited Aβ-induced mitochondrial membrane permeabilization and consequent cytochrome c release in isolated neuronal mitochondria (138). Moreover, using electron paramagnetic resonance spectroscopy analysis, TUDCA prevented Aβ-driven modifications in mitochondrial membrane redox status, lipid polarity, and protein order (142), in addition to decreasing Bax translocation (105). Similar results were obtained in primary human cerebral endothelial cells incubated with the vasculotropic E22Q Aβ mutant that is associated with hereditary cerebral hemorrhage in amyloidosis Dutch type (143). Interestingly, TUDCA was not capable of modulating the secondary structure and the fibrillogenic propensities of Aβ, suggesting dissociation between the capacity to induce apoptosis and the mechanisms of aggregation and fibrillization.
It was also shown that, in PC12 neuronal cells, TUDCA interferes with upstream targets of the mitochondria, including the E2F-1/p53/Bax apoptotic pathway (139). The role of p53 in mediating the effects of TUDCA was further confirmed using in vitro models of familial Alzheimer's disease (140). It is still not clear, however, how UDCA and TUDCA interfere with p53 expression and activation. It was demonstrated that UDCA reduced p53 transcriptional and DNA binding activities in hepatocytes undergoing p53-induced apoptosis by favoring its interaction with Mdm-2 (98). Thus, it is tempting to speculate that a similar effect would occur in neurons exposed to Aβ.
TUDCA was also shown to mitigate the toxic downstream effects of Aβ. In primary rat cortical neurons incubated with fibrillar Aβ1-42, TUDCA inhibited the levels of apoptosis and caspase-3 activation and abrogated the caspase-3-cleavage of tau into a toxic species (141). Cleavage of tau by caspase-3 at Asp421 in the C-terminal region is detected in primary cortical neurons, transgenic mice, and brains affected by Alzheimer's disease, promoting tau assembly (141, 144). Thus, by interfering with apoptotic pathways, both at the mitochondrial and transcriptional levels, TUDCA not only increased the survival of neurons but also prevented downstream abnormal conformations of tau. Finally, it is tempting to speculate that TUDCA may also modulate neuronal apoptosis by interfering with ER stress. Given the growing evidence supporting the implication of this mechanism in Alzheimer's disease pathology, it would be of much interest to evaluate the chaperone capacity of TUDCA in Alzheimer's disease models. Importantly, it was recently demonstrated that TUDCA administration to a transgenic mouse model of familial amyloidotic polyneuropathy significantly reduced transthyretin (TTR) toxic aggregates and decreased apoptotic and oxidative biomarkers usually associated with TTR deposition (145). These studies suggested that TUDCA modulated TTR aggregation by interfering with cellular responses, including ER stress.
CONCLUSIONS
In health, bile acids are essential for emulsifying lipids in the intestinal lumen, and their synthesis and transport drive bile formation and provide the major degradation pathway for cholesterol. Despite the beneficial effects, bile acids are always amphipatic molecules with inherent toxicity. With the realization that bile acids induce apoptosis, it was subsequently demonstrated that UDCA, the current and primary treatment of cholestasis, was a potent inhibitor of apoptosis by interrupting classic pathways of apoptosis. These studies provided a new framework to further understand and exploit the potential of this therapeutic agent. During the last decade, great progress has been made in characterizing the biochemical and molecular mechanisms underlying UDCA and its effects on cell survival. Most notably, it was protective from a long-list of apoptosis-inducing agents, and the effect was independent of cell type. In addition, given its clinical safety, UDCA and/or its conjugate derivatives may be potentially powerful therapeutic tools in several injuries associated with greater susceptibility to apoptotic cell death. For instance, TUDCA is readily water soluble, can be administrated orally or intravenously, and is associated with minimal toxicity. Nevertheless, its clinical application to human diseases outside of liver has yet to be vigorously tested.
Importantly, apoptosis plays a role in many pathological conditions, affecting a wide range of tissues, including liver, kidney, central nervous system, and heart, among others. As such, the field continues to be enormously active at both the basic science and clinical levels. The future will certainly focus on documenting new associations between apoptosis and disease and developing more effective therapeutic strategies based on modulation of the apoptotic process.
Acknowledgments
The authors thank all members of the laboratory for critical reading of the manuscript.
Footnotes
Abbreviations:
- Aβ
- amyloid β
- AP-1
- activating protein-1
- CDK
- cyclin-dependent kinase
- C/EBPβ
- CCAAT enhancer binding protein β
- CREB
- cyclic AMP-response element binding protein
- CYP7A1
- cholesterol 7α-hydroxylase
- CYP8B1
- cholesterol 12α-hydrolase
- DCA
- deoxycholic acid
- ER
- endoplasmic reticulum
- ERK
- extracellular-regulated kinase
- FADD
- Fas-associated death domain
- FGF19
- fibroblast growth factor intestine 19
- FGFR4
- fibroblast growth factor receptor 4
- FXR
- farnesoid X receptor
- GC
- glucocorticoid
- GCDCA
- glycochenodeoxycholic acid
- GR
- glucocorticoid receptor
- JNK
- c-Jun NH2-terminal kinase
- LBD
- ligand binding domain
- LRH-1
- liver receptor homolog-1
- MAPK
- mitogen-activated protein kinase
- MR
- mineralcorticoid receptor
- NF-κB
- nuclear factor κB
- NSR
- nuclear steroid receptor
- PC12
- pheochromocytoma
- PI3K
- phosphatidylinositol 3-kinase
- pRb
- retinoblastoma protein
- PXR
- pregnane X receptor
- ROS
- reactive oxygen species
- RSK
- ribosomal S6 kinase
- SHP
- small heterodimer partner
- TGF-β1
- transforming growth factor β1
- TNF
- tumor necrosis factor
- TRAIL
- TNF-related apoptosis inducing ligand
- TTR
- transthyretin
- TUDCA
- tauroursodeoxycholic acid
- UDCA
- ursodeoxycholic acid
REFERENCES
- 1.Bayerdorffer E., Mannes G. A., Richter W. O., Ochsenkuhn T., Wiebecke B., Kopcke W., Paumgartner G. 1993. Increased serum deoxycholic acid levels in men with colorectal adenomas. Gastroenterology. 104: 145–151. [DOI] [PubMed] [Google Scholar]
- 2.Patel T., Gores G. J. 1995. Apoptosis and hepatobiliary disease. Hepatology. 21: 1725–1741. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann A. F., Roda A. 1984. Physicochemical properties of bile acids and their relationship to biological properties: an overview of the problem. J. Lipid Res. 25: 1477–1489. [PubMed] [Google Scholar]
- 4.Babu P., Sangeetha N., Maitra U. 2006. Supramolecular chemistry of bile acids derivatives: formation of gels. Macromol. Symp. 241: 60–67. [Google Scholar]
- 5.Russell D. W., Setchell K. D. 1992. Bile acid biosynthesis. Biochemistry. 31: 4737–4749. [DOI] [PubMed] [Google Scholar]
- 6.Rodrigues C. M., Steer C. J. 2000. The neuroprotective characteristic of ursodeoxycholic acid and its conjugates. In Biology of Bile Acids in Health and Disease van Berge Henegouwen G., Keppler D., Leuschner U., Paumgartner G., Stiehl A., Kluwer Academic Publishers, Dordrecht, The Netherlands: 255–270. [Google Scholar]
- 7.Chiang J. Y. 2004. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J. Hepatol. 40: 539–551. [DOI] [PubMed] [Google Scholar]
- 8.Li T., Chen W., Chiang J. Y. 2007. PXR induces CYP27A1 and regulates cholesterol metabolism in the intestine. J. Lipid Res. 48: 373–384. [DOI] [PubMed] [Google Scholar]
- 9.Han S., Chiang J. Y. 2009. Mechanism of vitamin D receptor inhibition of cholesterol 7α-hydroxylase gene transcription in human hepatocytes. Drug Metab. Dispos. 37: 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lew J. L., Zhao A., Yu J., Huang L., De Pedro N., Pelaez F., Wright S. D., Cui J. 2004. The farnesoid X receptor controls gene expression in a ligand- and promoter-selective fashion. J. Biol. Chem. 279: 8856–8861. [DOI] [PubMed] [Google Scholar]
- 11.Russell D. W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72: 137–174. [DOI] [PubMed] [Google Scholar]
- 12.Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., et al. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 6: 517–526. [DOI] [PubMed] [Google Scholar]
- 13.Lu T. T., Makishima M., Repa J. J., Schoonjans K., Kerr T. A., Auwerx J., Mangelsdorf D. J. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 6: 507–515. [DOI] [PubMed] [Google Scholar]
- 14.Mataki C., Magnier B. C., Houten S. M., Annicotte J. S., Argmann C., Thomas C., Overmars H., Kulik W., Metzger D., Auwerx J., et al. 2007. Compromised intestinal lipid absorption in mice with a liver-specific deficiency of liver receptor homolog 1. Mol. Cell. Biol. 27: 8330–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee Y. K., Schmidt D. R., Cummins C. L., Choi M., Peng L., Zhang Y., Goodwin B., Hammer R. E., Mangelsdorf D. J., Kliewer S. A. 2008. Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis. Mol. Endocrinol. 22: 1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerr T. A., Saeki S., Schneider M., Schaefer K., Berdy S., Redder T., Shan B., Russell D. W., Schwarz M. 2002. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell. 2: 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L., Lee Y. K., Bundman D., Han Y., Thevananther S., Kim C. S., Chua S. S., Wei P., Heyman R. A., Karin M., et al. 2002. Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell. 2: 721–731. [DOI] [PubMed] [Google Scholar]
- 18.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., et al. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2: 217–225. [DOI] [PubMed] [Google Scholar]
- 19.Holt J. A., Luo G., Billin A. N., Bisi J., McNeill Y. Y., Kozarsky K. F., Donahee M., Wang D. Y., Mansfield T. A., Kliewer S. A., et al. 2003. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 17: 1581–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim I., Ahn S. H., Inagaki T., Choi M., Ito S., Guo G. L., Kliewer S. A., Gonzalez F. J. 2007. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 48: 2664–2672. [DOI] [PubMed] [Google Scholar]
- 21.Rao A., Haywood J., Craddock A. L., Belinsky M. G., Kruh G. D., Dawson P. A. 2008. The organic solute transporter α-β, Ostα-Ostß, is essential for intestinal bile acid transport and homeostasis. Proc. Natl. Acad. Sci. USA. 105: 3891–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maruyama T., Miyamoto Y., Nakamura T., Tamai Y., Okada H., Sugiyama E., Nakamura T., Itadani H., Tanaka K. 2002. Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 298: 714–719. [DOI] [PubMed] [Google Scholar]
- 23.Kawamata Y., Fujii R., Hosoya M., Harada M., Yoshida H., Miwa M., Fukusumi S., Habata Y., Itoh T., Shintani Y., et al. 2003. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 278: 9435–9440. [DOI] [PubMed] [Google Scholar]
- 24.Keitel V., Kubitz R., Haussinger D. 2008. Endocrine and paracrine role of bile acids. World J. Gastroenterol. 14: 5620–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geier A., Wagner M., Dietrich C. G., Trauner M. 2007. Principles of hepatic organic anion transporter regulation during cholestasis, inflammation and liver regeneration. Biochim. Biophys. Acta. 1773: 283–308. [DOI] [PubMed] [Google Scholar]
- 26.Ma K., Saha P. K., Chan L., Moore D. D. 2006. Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Invest. 116: 1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daniel P. T. 2000. Dissecting the pathways to death. Leukemia. 14: 2035–2044. [DOI] [PubMed] [Google Scholar]
- 28.Kerr J. F., Wyllie A. H., Currie A. R. 1972. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 26: 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reed J. C. 2002. Apoptosis-based therapies. Nat. Rev. Drug Discov. 1: 111–121. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y. 2002. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell. 9: 459–470. [DOI] [PubMed] [Google Scholar]
- 31.Ashkenazi A. 2002. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat. Rev. Cancer. 2: 420–430. [DOI] [PubMed] [Google Scholar]
- 32.Kroemer G., Galluzzi L., Brenner C. 2007. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 87: 99–163. [DOI] [PubMed] [Google Scholar]
- 33.Youle R. J., Strasser A. 2008. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9: 47–59. [DOI] [PubMed] [Google Scholar]
- 34.Krammer P. H. 1999. CD95(APO-1/Fas)-mediated apoptosis: live and let die. Adv. Immunol. 71: 163–210. [DOI] [PubMed] [Google Scholar]
- 35.Locksley R. M., Killeen N., Lenardo M. J. 2001. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 104: 487–501. [DOI] [PubMed] [Google Scholar]
- 36.Mehlen P., Bredesen D. E. 2004. The dependence receptor hypothesis. Apoptosis. 9: 37–49. [DOI] [PubMed] [Google Scholar]
- 37.Peter M. E., Krammer P. H. 2003. The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ. 10: 26–35. [DOI] [PubMed] [Google Scholar]
- 38.Li S., Zhao Y., He X., Kim T. H., Kuharsky D. K., Rabinowich H., Chen J., Du C., Yin X. M. 2002. Relief of extrinsic pathway inhibition by the Bid-dependent mitochondrial release of Smac in Fas-mediated hepatocyte apoptosis. J. Biol. Chem. 277: 26912–26920. [DOI] [PubMed] [Google Scholar]
- 39.Eskes R., Desagher S., Antonsson B., Martinou J. C. 2000. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol. Cell. Biol. 20: 929–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reed S. I. 1997. Control of the G1/S transition. Cancer Surv. 29: 7–23. [PubMed] [Google Scholar]
- 41.Wang C., Li Z., Fu M., Bouras T., Pestell R. G. 2004. Signal transduction mediated by cyclin D1: from mitogens to cell proliferation: a molecular target with therapeutic potential. Cancer Treat. Res. 119: 217–237. [DOI] [PubMed] [Google Scholar]
- 42.Han E. K., Ng S. C., Arber N., Begemann M., Weinstein I. B. 1999. Roles of cyclin D1 and related genes in growth inhibition, senescence and apoptosis. Apoptosis. 4: 213–219. [DOI] [PubMed] [Google Scholar]
- 43.Freeman R. S., Estus S., Johnson E. M., Jr 1994. Analysis of cell cycle-related gene expression in postmitotic neurons: selective induction of cyclin D1 during programmed cell death. Neuron. 12: 343–355. [DOI] [PubMed] [Google Scholar]
- 44.Nevins J. R. 1998. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 9: 585–593. [PubMed] [Google Scholar]
- 45.Sofer-Levi Y., Resnitzky D. 1996. Apoptosis induced by ectopic expression of cyclin D1 but not cyclin E. Oncogene. 13: 2431–2437. [PubMed] [Google Scholar]
- 46.Michael D., Oren M. 2003. The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 13: 49–58. [DOI] [PubMed] [Google Scholar]
- 47.Jacotot E., Ferri K. F., Kroemer G. 2000. Apoptosis and cell cycle: distinct checkpoints with overlapping upstream control. Pathol. Biol. 48: 271–279. [PubMed] [Google Scholar]
- 48.Barone M., Francavilla A., Polimeno L., Ierardi E., Romanelli D., Berloco P., Di Leo A., Panella C. 1996. Modulation of rat hepatocyte proliferation by bile salts: in vitro and in vivo studies. Hepatology. 23: 1159–1166. [DOI] [PubMed] [Google Scholar]
- 49.Maher J. J. 2004. What doesn't kill you makes you stronger: how hepatocytes survive prolonged cholestasis. Hepatology. 39: 1141–1143. [DOI] [PubMed] [Google Scholar]
- 50.Faubion W. A., Guicciardi M. E., Miyoshi H., Bronk S. F., Roberts P. J., Svingen P. A., Kaufmann S. H., Gores G. J. 1999. Toxic bile salts induce rodent hepatocyte apoptosis via direct activation of Fas. J. Clin. Invest. 103: 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodrigues C. M., Fan G., Wong P. Y., Kren B. T., Steer C. J. 1998. Ursodeoxycholic acid may inhibit deoxycholic acid-induced apoptosis by modulating mitochondrial transmembrane potential and reactive oxygen species production. Mol. Med. 4: 165–178. [PMC free article] [PubMed] [Google Scholar]
- 52.Yerushalmi B., Dahl R., Devereaux M. W., Gumpricht E., Sokol R. J. 2001. Bile acid-induced rat hepatocyte apoptosis is inhibited by antioxidants and blockers of the mitochondrial permeability transition. Hepatology. 33: 616–626. [DOI] [PubMed] [Google Scholar]
- 53.Higuchi H., Yoon J. H., Grambihler A., Werneburg N., Bronk S. F., Gores G. J. 2003. Bile acids stimulate cFLIP phosphorylation enhancing TRAIL-mediated apoptosis. J. Biol. Chem. 278: 454–461. [DOI] [PubMed] [Google Scholar]
- 54.Sodeman T., Bronk S. F., Roberts P. J., Miyoshi H., Gores G. J. 2000. Bile salts mediate hepatocyte apoptosis by increasing cell surface trafficking of Fas. Am. J. Physiol. Gastrointest. Liver Physiol. 278: G992–G999. [DOI] [PubMed] [Google Scholar]
- 55.Katona B. W., Anant S., Covey D. F., Stenson W. F. 2009. Characterization of enantiomeric bile acid-induced apoptosis in colon cancer cell lines. J. Biol. Chem. 284: 3354–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiao L., Studer E., Leach K., McKinstry R., Gupta S., Decker R., Kukreja R., Valerie K., Nagarkatti P., El Deiry W., et al. 2001. Deoxycholic acid (DCA) causes ligand-independent activation of epidermal growth factor receptor (EGFR) and FAS receptor in primary hepatocytes: inhibition of EGFR/mitogen-activated protein kinase-signaling module enhances DCA-induced apoptosis. Mol. Biol. Cell. 12: 2629–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sokol R. J., Devereaux M., Dahl R., Gumpricht E. 2006. “Let there be bile”–understanding hepatic injury in cholestasis. J. Pediatr. Gastroenterol. Nutr. 43 (Suppl. 1): S4–S9. [DOI] [PubMed] [Google Scholar]
- 58.Reinehr R., Becker S., Eberle A., Grether-Beck S., Haussinger D. 2005. Involvement of NADPH oxidase isoforms and Src family kinases in CD95-dependent hepatocyte apoptosis. J. Biol. Chem. 280: 27179–27194. [DOI] [PubMed] [Google Scholar]
- 59.Yang J. I., Yoon J. H., Myung S. J., Gwak G. Y., Kim W., Chung G. E., Lee S. H., Lee S. M., Kim C. Y., Lee H. S. 2007. Bile acid-induced TGR5-dependent c-Jun-N terminal kinase activation leads to enhanced caspase 8 activation in hepatocytes. Biochem. Biophys. Res. Commun. 361: 156–161. [DOI] [PubMed] [Google Scholar]
- 60.Katona B. W., Cummins C. L., Ferguson A. D., Li T., Schmidt D. R., Mangelsdorf D. J., Covey D. F. 2007. Synthesis, characterization, and receptor interaction profiles of enantiomeric bile acids. J. Med. Chem. 50: 6048–6058. [DOI] [PubMed] [Google Scholar]
- 61.Bennett M., Macdonald K., Chan S. W., Luzio J. P., Simari R., Weissberg P. 1998. Cell surface trafficking of Fas: a rapid mechanism of p53-mediated apoptosis. Science. 282: 290–293. [DOI] [PubMed] [Google Scholar]
- 62.Zhang G., Park M. A., Mitchell C., Walker T., Hamed H., Studer E., Graf M., Rahmani M., Gupta S., Hylemon P. B., et al. 2008. Multiple cyclin kinase inhibitors promote bile acid-induced apoptosis and autophagy in primary hepatocytes via p53–CD95-dependent signaling. J. Biol. Chem. 283: 24343–24358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rust C., Wild N., Bernt C., Vennegeerts T., Wimmer R., Beuers U. 2009. Bile acid-induced apoptosis in hepatocytes is caspase-6-dependent. J. Biol. Chem. 284: 2908–2916. [DOI] [PubMed] [Google Scholar]
- 64.Tsuchiya S., Tsuji M., Morio Y., Oguchi K. 2006. Involvement of endoplasmic reticulum in glycochenodeoxycholic acid-induced apoptosis in rat hepatocytes. Toxicol. Lett. 166: 140–149. [DOI] [PubMed] [Google Scholar]
- 65.Iizaka T., Tsuji M., Oyamada H., Morio Y., Oguchi K. 2007. Interaction between caspase-8 activation and endoplasmic reticulum stress in glycochenodeoxycholic acid-induced apoptotic HepG2 cells. Toxicology. 241: 146–156. [DOI] [PubMed] [Google Scholar]
- 66.Schoemaker M. H., Conde de la Rosa L., Buist-Homan M., Vrenken T. E., Havinga R., Poelstra K., Haisma H. J., Jansen P. L., Moshage H. 2004. Tauroursodeoxycholic acid protects rat hepatocytes from bile acid-induced apoptosis via activation of survival pathways. Hepatology. 39: 1563–1573. [DOI] [PubMed] [Google Scholar]
- 67.Schoemaker M. H., Gommans W. M., de la Rosa L. C., Homan M., Klok P., Trautwein C., van Goor H., Poelstra K., Haisma H. J., Jansen P. L., et al. 2003. Resistance of rat hepatocytes against bile acid-induced apoptosis in cholestatic liver injury is due to nuclear factor-κB activation. J. Hepatol. 39: 153–161. [DOI] [PubMed] [Google Scholar]
- 68.Oh S. H., Nan J. X., Sohn D. W., Kim Y. C., Lee B. H. 2002. Salvia miltiorrhiza inhibits biliary obstruction-induced hepatocyte apoptosis by cytoplasmic sequestration of p53. Toxicol. Appl. Pharmacol. 182: 27–33. [DOI] [PubMed] [Google Scholar]
- 69.Rao Y. P., Studer E. J., Stravitz R. T., Gupta S., Qiao L., Dent P., Hylemon P. B. 2002. Activation of the Raf-1/MEK/ERK cascade by bile acids occurs via the epidermal growth factor receptor in primary rat hepatocytes. Hepatology. 35: 307–314. [DOI] [PubMed] [Google Scholar]
- 70.Qiao L., Han S. I., Fang Y., Park J. S., Gupta S., Gilfor D., Amorino G., Valerie K., Sealy L., Engelhardt J. F., et al. 2003. Bile acid regulation of C/EBPβ, CREB, and c-Jun function, via the extracellular signal-regulated kinase and c-Jun NH2-terminal kinase pathways, modulates the apoptotic response of hepatocytes. Mol. Cell. Biol. 23: 3052–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liedtke C., Plumpe J., Kubicka S., Bradham C. A., Manns M. P., Brenner D. A., Trautwein C. 2002. Jun kinase modulates tumor necrosis factor-dependent apoptosis in liver cells. Hepatology. 36: 315–325. [DOI] [PubMed] [Google Scholar]
- 72.Gupta S., Natarajan R., Payne S. G., Studer E. J., Spiegel S., Dent P., Hylemon P. B. 2004. Deoxycholic acid activates the c-Jun N-terminal kinase pathway via FAS receptor activation in primary hepatocytes. Role of acidic sphingomyelinase-mediated ceramide generation in FAS receptor activation. J. Biol. Chem. 279: 5821–5828. [DOI] [PubMed] [Google Scholar]
- 73.Rust C., Karnitz L. M., Paya C. V., Moscat J., Simari R. D., Gores G. J. 2000. The bile acid taurochenodeoxycholate activates a phosphatidylinositol 3-kinase-dependent survival signaling cascade. J. Biol. Chem. 275: 20210–20216. [DOI] [PubMed] [Google Scholar]
- 74.Torchia E. C., Stolz A., Agellon L. B. 2001. Differential modulation of cellular death and survival pathways by conjugated bile acids. BMC Biochem. 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qiao L., Yacoub A., Studer E., Gupta S., Pei X. Y., Grant S., Hylemon P. B., Dent P. 2002. Inhibition of the MAPK and PI3K pathways enhances UDCA-induced apoptosis in primary rodent hepatocytes. Hepatology. 35: 779–789. [DOI] [PubMed] [Google Scholar]
- 76.Im E., Martinez J. D. 2004. Ursodeoxycholic acid (UDCA) can inhibit deoxycholic acid (DCA)-induced apoptosis via modulation of EGFR/Raf-1/ERK signaling in human colon cancer cells. J. Nutr. 134: 483–486. [DOI] [PubMed] [Google Scholar]
- 77.Guicciardi M. E., Gores G. J. 2002. Ursodeoxycholic acid cytoprotection: dancing with death receptors and survival pathways. Hepatology. 35: 971–973. [DOI] [PubMed] [Google Scholar]
- 78.Beuers U., Boyer J. L., Paumgartner G. 1998. Ursodeoxycholic acid in cholestasis: potential mechanisms of action and therapeutic applications. Hepatology. 28: 1449–1453. [DOI] [PubMed] [Google Scholar]
- 79.Lazaridis K. N., Gores G. J., Lindor K. D. 2001. Ursodeoxycholic acid ‘mechanisms of action and clinical use in hepatobiliary disorders’. J. Hepatol. 35: 134–146. [DOI] [PubMed] [Google Scholar]
- 80.Paumgartner G., Beuers U. 2002. Ursodeoxycholic acid in cholestatic liver disease: mechanisms of action and therapeutic use revisited. Hepatology. 36: 525–531. [DOI] [PubMed] [Google Scholar]
- 81.Benz C., Angermuller S., Tox U., Kloters-Plachky P., Riedel H. D., Sauer P., Stremmel W., Stiehl A. 1998. Effect of tauroursodeoxycholic acid on bile-acid-induced apoptosis and cytolysis in rat hepatocytes. J. Hepatol. 28: 99–106. [DOI] [PubMed] [Google Scholar]
- 82.Rodrigues C. M., Fan G., Ma X., Kren B. T., Steer C. J. 1998. A novel role for ursodeoxycholic acid in inhibiting apoptosis by modulating mitochondrial membrane perturbation. J. Clin. Invest. 101: 2790–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Benz C., Angermuller S., Otto G., Sauer P., Stremmel W., Stiehl A. 2000. Effect of tauroursodeoxycholic acid on bile acid-induced apoptosis in primary human hepatocytes. Eur. J. Clin. Invest. 30: 203–209. [DOI] [PubMed] [Google Scholar]
- 84.Castro R., Solá S., Steer C., Rodrigues C. 2007. Bile acids as modulators of apoptosis. In Hepatotoxicity: From Genomics to in-Vitro and in-Vivo Models Sahu S., editor John Wiley & Sons, West Sussex, UK: 391–419. [Google Scholar]
- 85.Rodrigues C. M., Ma X., Linehan-Stieers C., Fan G., Kren B. T., Steer C. J. 1999. Ursodeoxycholic acid prevents cytochrome c release in apoptosis by inhibiting mitochondrial membrane depolarization and channel formation. Cell Death Differ. 6: 842–854. [DOI] [PubMed] [Google Scholar]
- 86.Rodrigues C. M., Sola S., Sharpe J. C., Moura J. J., Steer C. J. 2003. Tauroursodeoxycholic acid prevents Bax-induced membrane perturbation and cytochrome C release in isolated mitochondria. Biochemistry. 42: 3070–3080. [DOI] [PubMed] [Google Scholar]
- 87.Azzaroli F., Mehal W., Soroka C. J., Wang L., Lee J., Crispe N., Boyer J. L. 2002. Ursodeoxycholic acid diminishes Fas-ligand-induced apoptosis in mouse hepatocytes. Hepatology. 36: 49–54. [DOI] [PubMed] [Google Scholar]
- 88.Xie Q., Khaoustov V. I., Chung C. C., Sohn J., Krishnan B., Lewis D. E., Yoffe B. 2002. Effect of tauroursodeoxycholic acid on endoplasmic reticulum stress-induced caspase-12 activation. Hepatology. 36: 592–601. [DOI] [PubMed] [Google Scholar]
- 89.Ozcan U., Yilmaz E., Ozcan L., Furuhashi M., Vaillancourt E., Smith R. O., Gorgun C. Z., Hotamisligil G. S. 2006. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 313: 1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Setchell K. D., Rodrigues C. M., Clerici C., Solinas A., Morelli A., Gartung C., Boyer J. 1997. Bile acid concentrations in human and rat liver tissue and in hepatocyte nuclei. Gastroenterology. 112: 226–235. [DOI] [PubMed] [Google Scholar]
- 91.Monte M. J., Martinez-Diez M. C., El-Mir M. Y., Mendoza M. E., Bravo P., Bachs O., Marin J. J. 2002. Changes in the pool of bile acids in hepatocyte nuclei during rat liver regeneration. J. Hepatol. 36: 534–542. [DOI] [PubMed] [Google Scholar]
- 92.Solá S., Amaral J. D., Castro R. E., Ramalho R. M., Borralho P. M., Kren B. T., Tanaka H., Steer C. J., Rodrigues C. M. 2005. Nuclear translocation of UDCA by the glucocorticoid receptor is required to reduce TGF-β1-induced apoptosis in rat hepatocytes. Hepatology. 42: 925–934. [DOI] [PubMed] [Google Scholar]
- 93.Castro R. E., Solá S., Ma X., Ramalho R. M., Kren B. T., Steer C. J., Rodrigues C. M. 2005. A distinct microarray gene expression profile in primary rat hepatocytes incubated with ursodeoxycholic acid. J. Hepatol. 42: 897–906. [DOI] [PubMed] [Google Scholar]
- 94.Castro R. E., Amaral J. D., Sola S., Kren B. T., Steer C. J., Rodrigues C. M. 2007. Differential regulation of cyclin D1 and cell death by bile acids in primary rat hepatocytes. Am. J. Physiol. Gastrointest Liver Physiol. 293: G327–G334. [DOI] [PubMed] [Google Scholar]
- 95.Pusl T., Vennegeerts T., Wimmer R., Denk G. U., Beuers U., Rust C. 2008. Tauroursodeoxycholic acid reduces bile acid-induced apoptosis by modulation of AP-1. Biochem. Biophys. Res. Commun. 367: 208–212. [DOI] [PubMed] [Google Scholar]
- 96.Shaulian E., Karin M. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4: E131–E136. [DOI] [PubMed] [Google Scholar]
- 97.Solá S., Ma X., Castro R. E., Kren B. T., Steer C. J., Rodrigues C. M. 2003. Ursodeoxycholic acid modulates E2F-1 and p53 expression through a caspase-independent mechanism in transforming growth factor β1-induced apoptosis of rat hepatocytes. J. Biol. Chem. 278: 48831–48838. [DOI] [PubMed] [Google Scholar]
- 98.Amaral J. D., Castro R. E., Sola S., Steer C. J., Rodrigues C. M. 2007. p53 is a key molecular target of ursodeoxycholic acid in regulating apoptosis. J. Biol. Chem. 282: 34250–34259. [DOI] [PubMed] [Google Scholar]
- 99.Serfaty L., De Leusse A., Rosmorduc O., Desaint B., Flejou J. F., Chazouilleres O., Poupon R. E., Poupon R. 2003. Ursodeoxycholic acid therapy and the risk of colorectal adenoma in patients with primary biliary cirrhosis: an observational study. Hepatology. 38: 203–209. [DOI] [PubMed] [Google Scholar]
- 100.Tung B. Y., Emond M. J., Haggitt R. C., Bronner M. P., Kimmey M. B., Kowdley K. V., Brentnall T. A. 2001. Ursodiol use is associated with lower prevalence of colonic neoplasia in patients with ulcerative colitis and primary sclerosing cholangitis. Ann. Intern. Med. 134: 89–95. [DOI] [PubMed] [Google Scholar]
- 101.Earnest D. L., Holubec H., Wali R. K., Jolley C. S., Bissonette M., Bhattacharyya A. K., Roy H., Khare S., Brasitus T. A. 1994. Chemoprevention of azoxymethane-induced colonic carcinogenesis by supplemental dietary ursodeoxycholic acid. Cancer Res. 54: 5071–5074. [PubMed] [Google Scholar]
- 102.Wali R. K., Khare S., Tretiakova M., Cohen G., Nguyen L., Hart J., Wang J., Wen M., Ramaswamy A., Joseph L., et al. 2002. Ursodeoxycholic acid and F(6)-D(3) inhibit aberrant crypt proliferation in the rat azoxymethane model of colon cancer: roles of cyclin D1 and E-cadherin. Cancer Epidemiol. Biomarkers Prev. 11: 1653–1662. [PubMed] [Google Scholar]
- 103.Wali R. K., Stoiber D., Nguyen L., Hart J., Sitrin M. D., Brasitus T., Bissonnette M. 2002. Ursodeoxycholic acid inhibits the initiation and postinitiation phases of azoxymethane-induced colonic tumor development. Cancer Epidemiol. Biomarkers Prev. 11: 1316–1321. [PubMed] [Google Scholar]
- 104.Park H., Kim M. K., Kim S. U. 2008. Ursodeoxycholic acid prevents apoptosis of mouse sensory neurons induced by cisplatin by reducing P53 accumulation. Biochem Biophys Res Commun. 377: 1025–1030. [DOI] [PubMed] [Google Scholar]
- 105.Solá S., Castro R. E., Laires P. A., Steer C. J., Rodrigues C. M. 2003. Tauroursodeoxycholic acid prevents amyloid-β peptide-induced neuronal death via a phosphatidylinositol 3-kinase-dependent signaling pathway. Mol. Med. 9: 226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rajesh K. G., Suzuki R., Maeda H., Yamamoto M., Yutong X., Sasaguri S. 2005. Hydrophilic bile salt ursodeoxycholic acid protects myocardium against reperfusion injury in a PI3K/Akt dependent pathway. J. Mol. Cell. Cardiol. 39: 766–776. [DOI] [PubMed] [Google Scholar]
- 107.Miura T., Ouchida R., Yoshikawa N., Okamoto K., Makino Y., Nakamura T., Morimoto C., Makino I., Tanaka H. 2001. Functional modulation of the glucocorticoid receptor and suppression of NF-κB-dependent transcription by ursodeoxycholic acid. J. Biol. Chem. 276: 47371–47378. [DOI] [PubMed] [Google Scholar]
- 108.Sionov R. V., Cohen O., Kfir S., Zilberman Y., Yefenof E. 2006. Role of mitochondrial glucocorticoid receptor in glucocorticoid-induced apoptosis. J. Exp. Med. 203: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rasmuson S., Andrew R., Nasman B., Seckl J. R., Walker B. R., Olsson T. 2001. Increased glucocorticoid production and altered cortisol metabolism in women with mild to moderate Alzheimer's disease. Biol. Psychiatry. 49: 547–552. [DOI] [PubMed] [Google Scholar]
- 110.Kim S., Jeon B. S., Heo C., Im P. S., Ahn T. B., Seo J. H., Kim H. S., Park C. H., Choi S. H., Cho S. H., et al. 2004. A-synuclein induces apoptosis by altered expression in human peripheral lymphocyte in Parkinson's disease. FASEB J. 18: 1615–1617. [DOI] [PubMed] [Google Scholar]
- 111.Yamamoto M., Fukuda K., Miura N., Suzuki R., Kido T., Komatsu Y. 1998. Inhibition by dexamethasone of transforming growth factor β1-induced apoptosis in rat hepatoma cells: a possible association with Bcl-xL induction. Hepatology. 27: 959–966. [DOI] [PubMed] [Google Scholar]
- 112.Bailly-Maitre B., de Sousa G., Boulukos K., Gugenheim J., Rahmani R. 2001. Dexamethasone inhibits spontaneous apoptosis in primary cultures of human and rat hepatocytes via Bcl-2 and Bcl-xL induction. Cell Death Differ. 8: 279–288. [DOI] [PubMed] [Google Scholar]
- 113.Karabelyos C., Dobozy O., Szalai C., Klenjanszki K., Varju K., Hadhazi A., Kiss A., Fulop A. K., Madarasz B., Falus A. 1999. Elevated hepatic glucocorticoid receptor expression during liver regeneration in rats. Pathol. Oncol. Res. 5: 107–109. [DOI] [PubMed] [Google Scholar]
- 114.Sengupta S., Wasylyk B. 2004. Physiological and pathological consequences of the interactions of the p53 tumor suppressor with the glucocorticoid, androgen, and estrogen receptors. Ann. N. Y. Acad. Sci. 1024: 54–71. [DOI] [PubMed] [Google Scholar]
- 115.Solá S., Castro R. E., Kren B. T., Steer C. J., Rodrigues C. M. 2004. Modulation of nuclear steroid receptors by ursodeoxycholic acid inhibits TGF-β1-induced E2F-1/p53-mediated apoptosis of rat hepatocytes. Biochemistry. 43: 8429–8438. [DOI] [PubMed] [Google Scholar]
- 116.Rogatsky I., Trowbridge J. M., Garabedian M. J. 1997. Glucocorticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol. Cell. Biol. 17: 3181–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tanaka H., Makino Y., Miura T., Hirano F., Okamoto K., Komura K., Sato Y., Makino I. 1996. Ligand-independent activation of the glucocorticoid receptor by ursodeoxycholic acid. Repression of IFN-gamma-induced MHC class II gene expression via a glucocorticoid receptor-dependent pathway. J. Immunol. 156: 1601–1608. [PubMed] [Google Scholar]
- 118.Mitsuyoshi H., Nakashima T., Inaba K., Ishikawa H., Nakajima Y., Sakamoto Y., Matsumoto M., Okanoue T., Kashima K. 1997. Ursodeoxycholic acid enhances glucocorticoid-induced tyrosine aminotransferase-gene expression in cultured rat hepatocytes. Biochem. Biophys. Res. Commun. 240: 732–736. [DOI] [PubMed] [Google Scholar]
- 119.Planey S. L., Derfoul A., Steplewski A., Robertson N. M., Litwack G. 2002. Inhibition of glucocorticoid-induced apoptosis in 697 pre-B lymphocytes by the mineralocorticoid receptor N-terminal domain. J. Biol. Chem. 277: 42188–42196. [DOI] [PubMed] [Google Scholar]
- 120.Almeida O. F., Conde G. L., Crochemore C., Demeneix B. A., Fischer D., Hassan A. H., Meyer M., Holsboer F., Michaelidis T. M. 2000. Subtle shifts in the ratio between pro- and antiapoptotic molecules after activation of corticosteroid receptors decide neuronal fate. FASEB J. 14: 779–790. [DOI] [PubMed] [Google Scholar]
- 121.Solá S., Amaral J. D., Borralho P. M., Ramalho R. M., Castro R. E., Aranha M. M., Steer C. J., Rodrigues C. M. 2006. Functional modulation of nuclear steroid receptors by tauroursodeoxycholic acid reduces amyloid β-peptide-induced apoptosis. Mol. Endocrinol. 20: 2292–2303. [DOI] [PubMed] [Google Scholar]
- 122.Howard W. R., Pospisil J. A., Njolito E., Noonan D. J. 2000. Catabolites of cholesterol synthesis pathways and forskolin as activators of the farnesoid X-activated nuclear receptor. Toxicol. Appl. Pharmacol. 163: 195–202. [DOI] [PubMed] [Google Scholar]
- 123.Chiang J. Y., Kimmel R., Weinberger C., Stroup D. 2000. Farnesoid X receptor responds to bile acids and represses cholesterol 7α-hydroxylase gene (CYP7A1) transcription. J. Biol. Chem. 275: 10918–10924. [DOI] [PubMed] [Google Scholar]
- 124.Stedman C., Robertson G., Coulter S., Liddle C. 2004. Feed-forward regulation of bile acid detoxification by CYP3A4: studies in humanized transgenic mice. J. Biol. Chem. 279: 11336–11343. [DOI] [PubMed] [Google Scholar]
- 125.Przedborski S., Vila M., Jackson-Lewis V. 2003. Neurodegeneration: what is it and where are we? J. Clin. Invest. 111: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bredesen D. E., Rao R. V., Mehlen P. 2006. Cell death in the nervous system. Nature. 443: 796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Keene C. D., Rodrigues C. M., Eich T., Linehan-Stieers C., Abt A., Kren B. T., Steer C. J., Low W. C. 2001. A bile acid protects against motor and cognitive deficits and reduces striatal degeneration in the 3-nitropropionic acid model of Huntington's disease. Exp. Neurol. 171: 351–360. [DOI] [PubMed] [Google Scholar]
- 128.Rodrigues C. M., Stieers C. L., Keene C. D., Ma X., Kren B. T., Low W. C., Steer C. J. 2000. Tauroursodeoxycholic acid partially prevents apoptosis induced by 3-nitropropionic acid: evidence for a mitochondrial pathway independent of the permeability transition. J. Neurochem. 75: 2368–2379. [DOI] [PubMed] [Google Scholar]
- 129.Keene C. D., Rodrigues C. M., Eich T., Chhabra M. S., Steer C. J., Low W. C. 2002. Tauroursodeoxycholic acid, a bile acid, is neuroprotective in a transgenic animal model of Huntington's disease. Proc. Natl. Acad. Sci. USA. 99: 10671–10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Duan W. M., Rodrigues C. M., Zhao L. R., Steer C. J., Low W. C. 2002. Tauroursodeoxycholic acid improves the survival and function of nigral transplants in a rat model of Parkinson's disease. Cell Transplant. 11: 195–205. [PubMed] [Google Scholar]
- 131.Ved R., Saha S., Westlund B., Perier C., Burnam L. G., Sluder A., Hoener M., Rodrigues C. M., Alfonso A., Steer C., et al. 2005. Similar patterns of mitochondrial vulnerability and rescue induced by genetic modification of α -synuclein, parkin and DJ-1 in C. elegans. J. Biol. Chem. 280: 42655–42668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rodrigues C. M., Spellman S. R., Sola S., Grande A. W., Linehan-Stieers C., Low W. C., Steer C. J. 2002. Neuroprotection by a bile acid in an acute stroke model in the rat. J. Cereb. Blood Flow Metab. 22: 463–471. [DOI] [PubMed] [Google Scholar]
- 133.Rodrigues C. M., Sola S., Nan Z., Castro R. E., Ribeiro P. S., Low W. C., Steer C. J. 2003. Tauroursodeoxycholic acid reduces apoptosis and protects against neurological injury after acute hemorrhagic stroke in rats. Proc. Natl. Acad. Sci. USA. 100: 6087–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Phillips M. J., Walker T. A., Choi H. Y., Faulkner A. E., Kim M. K., Sidney S. S., Boyd A. P., Nickerson J. M., Boatright J. H., Pardue M. T. 2008. Tauroursodeoxycholic acid preservation of photoreceptor structure and function in the rd10 mouse through postnatal day 30. Invest. Ophthalmol. Vis. Sci. 49: 2148–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Colak A., Kelten B., Sagmanligil A., Akdemir O., Karaoglan A., Sahan E., Celik O., Barut S. 2008. Tauroursodeoxycholic acid and secondary damage after spinal cord injury in rats. J. Clin. Neurosci. 15: 665–671. [DOI] [PubMed] [Google Scholar]
- 136.Castro R. E., Sola S., Ramalho R. M., Steer C. J., Rodrigues C. M. 2004. The bile acid tauroursodeoxycholic acid modulates phosphorylation and translocation of bad via phosphatidylinositol 3-kinase in glutamate-induced apoptosis of rat cortical neurons. J. Pharmacol. Exp. Ther. 311: 845–852. [DOI] [PubMed] [Google Scholar]
- 137.LaFerla F. M., Green K. N., Oddo S. 2007. Intracellular amyloid-β in Alzheimer's disease. Nat. Rev. Neurosci. 8: 499–509. [DOI] [PubMed] [Google Scholar]
- 138.Rodrigues C. M., Sola S., Silva R., Brites D. 2000. Bilirubin and amyloid-β peptide induce cytochrome c release through mitochondrial membrane permeabilization. Mol. Med. 6: 936–946. [PMC free article] [PubMed] [Google Scholar]
- 139.Ramalho R. M., Ribeiro P. S., Solá S., Castro R. E., Steer C. J., Rodrigues C. M. 2004. Inhibition of the E2F-1/p53/Bax pathway by tauroursodeoxycholic acid in amyloid β-peptide-induced apoptosis of PC12 cells. J. Neurochem. 90: 567–575. [DOI] [PubMed] [Google Scholar]
- 140.Ramalho R. M., Borralho P. M., Castro R. E., Solá S., Steer C. J., Rodrigues C. M. 2006. Tauroursodeoxycholic acid modulates p53-mediated apoptosis in Alzheimer's disease mutant neuroblastoma cells. J. Neurochem. 98: 1610–1618. [DOI] [PubMed] [Google Scholar]
- 141.Ramalho R. M., Viana R. J., Castro R. E., Steer C. J., Low W. C., Rodrigues C. M. 2008. Apoptosis in transgenic mice expressing the P301L mutated form of human tau. Mol. Med. 14: 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rodrigues C. M., Solá S., Brito M. A., Brondino C. D., Brites D., Moura J. J. 2001. Amyloid β-peptide disrupts mitochondrial membrane lipid and protein structure: protective role of tauroursodeoxycholate. Biochem. Biophys. Res. Commun. 281: 468–474. [DOI] [PubMed] [Google Scholar]
- 143.Viana R. J., Nunes A. F., Castro R. E., Ramalho R. M., Meyerson J., Fossati S., Ghiso J., Rostagno A., Rodrigues C. M. 2009. Tauroursodeoxycholic acid prevents E22Q Alzheimer's Aβ toxicity in human cerebral endothelial cells. Cell. Mol. Life Sci. 66: 1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gamblin T. C., Chen F., Zambrano A., Abraha A., Lagalwar S., Guillozet A. L., Lu M., Fu Y., Garcia-Sierra F., LaPointe N., et al. 2003. Caspase cleavage of tau: linking amyloid and neurofibrillary tangles in Alzheimer's disease. Proc. Natl. Acad. Sci. USA. 100: 10032–10037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Macedo B., Batista A. R., Ferreira N., Almeida M. R., Saraiva M. J. 2008. Anti-apoptotic treatment reduces transthyretin deposition in a transgenic mouse model of familial amyloidotic polyneuropathy. Biochim. Biophys. Acta. 1782: 517–522. [DOI] [PubMed] [Google Scholar]