Abstract
Pulmonary surfactant is a complex of phospholipids and proteins lining the alveolar walls of the lung. It reduces surface tension in the alveoli, and is critical for normal respiration. Pulmonary surfactant phospholipids consist mainly of phosphatidylcholine (PC) and phosphatidylglycerol (PG). Although the phospholipid composition of pulmonary surfactant is well known, the enzyme(s) involved in its biosynthesis have remained obscure. We previously reported the cloning of murine lysophosphatidylcholine acyltransferase 1 (mLPCAT1) as a potential biosynthetic enzyme of pulmonary surfactant phospholipids. mLPCAT1 exhibits lysophosphatidylcholine acyltransferase (LPCAT) and lysophosphatidylglycerol acyltransferase (LPGAT) activities, generating PC and PG, respectively. However, the enzymatic activity of human LPCAT1 (hLPCAT1) remains controversial. We report here that hLPCAT1 possesses LPCAT and LPGAT activities. The activity of hLPCAT1 was inhibited by N-ethylmaleimide, indicating the importance of some cysteine residue(s) for the catalysis. We found a conserved cysteine (Cys211) in hLPCAT1 that is crucial for its activity. Evolutionary analyses of the close homologs of LPCAT1 suggest that it appeared before the evolution of teleosts and indicate that LPCAT1 may have evolved along with the lung to facilitate respiration. hLPCAT1 mRNA is highly expressed in the human lung. We propose that hLPCAT1 is the biosynthetic enzyme of pulmonary surfactant phospholipids.
Keywords: lyso-platelet-activating factor acetyltransferase, lysophosphatidylglycerol acyltransferase, pulmonary surfactant, N-ethylmaleimide, evolution, lung
Pulmonary surfactant is a complex of phospholipids (∼90%) and proteins (∼10%) that reduces the surface tension in the alveoli of the lung (1, 2). Pulmonary surfactant is crucial for normal breathing and its deficiency leads to fatal respiratory distress syndromes (3). The major phospholipids in pulmonary surfactant are phosphatidylcholine (PC) and phosphatidylglycerol (PG). Among the species of PC and PG, dipalmitoyl-PC (DPPC) and disaturated-PG (DSPG) are the most abundant (1). Although the lipid composition of pulmonary surfactant has been well characterized, the enzyme(s) involved in their biosynthesis remained unclear.
The acyl-chain composition of phospholipids is mainly determined by a deacylation-reacylation cycle named Lands' cycle (4). The last step of Lands' cycle is mediated by lysophospholipid acyltransferases, including lysophosphatidylcholine acyltransferases (LPCATs) (5, 6). We and others have previously reported the cloning of four murine LPCATs, designated LPCAT1-41 (5–10). Among them, murine LPCAT1 (mLPCAT1, AB244717) exhibits LPCAT, lysophosphatidylglycerol acyltransferase (LPGAT), and lyso-platelet-activating factor acetyltransferase (lyso-PAF AT) activities (7, 11). mLPCAT1 prefers saturated acyl-CoAs as substrates, in accordance with the acyl-chain composition of pulmonary surfactant lipids, and is highly expressed in the lung, where pulmonary surfactant is generated. These observations suggest a role of mLPCAT1 in pulmonary surfactant lipid biosynthesis. Concomitantly, Chen et al. (12) reported the cloning of rat LPCAT1 (rLPCAT1, NM_001100735). rLPCAT1 shares similar properties to mLPCAT1. rLPCAT1 is extremely highly expressed in alveolar type II cells, which secrete pulmonary surfactant (7, 12). The mRNA level of rLPCAT1 increases perinatally, which correlates with the perinatal production of surfactant by the embryo.
Following the above reports, an alternative theory regarding the activity and function of human LPCAT1 (hLPCAT1, AB244719) was proposed. Agarwal et al. (13) cloned and characterized a sequence with 88% homology to mLPCAT1, which they designated as 1-acylglycerol-3-phosphate-O-acyltransferase 9 (AGPAT9). They reported that they did not observe any LPCAT activity and proposed a role for hLPCAT1 in the immune system, rather than in pulmonary surfactant lipids biosynthesis, because its mRNA was detected not only in the lung but also in the spleen.
We report here the cloning and characterization of hLPCAT1 (AGPAT9). We first cloned hLPCAT1 from cDNA of HEK293 cells. Activity assays revealed that hLPCAT1 does in fact exhibit LPCAT, LPGAT, and lyso-PAF AT activities, similar to mLPCAT1. The activity of hLPCAT1 was inhibited by a sulfhydryl-reducing reagent, N-ethylmaleimide (NEM), suggesting the importance of at least one cysteine residue for the activity of hLPCAT1. Indeed, hLPCAT1 activity was completely abolished by mutation of Cys211 to other amino acids. This cysteine is conserved among some homologs of LPCAT1. We analyzed these cysteine-containing acyltransferases by a phylogenetic tree, and the results suggest that LPCAT1 bifurcated before the appearance of teleosts, similar to the evolution of the lung. In addition, hLPCAT1 is highly expressed in the human lung. Therefore, we propose that hLPCAT1 is involved in pulmonary surfactant lipid biosynthesis to facilitate respiration.
MATERIALS AND METHODS
Detection of hLPCAT1 expression by RT-PCR
Total RNA from A549 cells, HEK293 cells, and HeLa cells was prepared using RNeasy Mini Kit (QIAGEN; Hilden, Germany). cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen; Carlsbad, CA). A 154 bp fragment of hLPCAT1 was amplified by RT-PCR using ExTaq (TaKaRa; Shiga, Japan). The primer sequences were 5′-CCTCATGACACTGACGCTCTTC-3′ as a forward primer, and 5′-CAGGAAGTCCACAACCTTCCTC-3′ as a reverse primer.
Cloning of hLPCAT1
hLPCAT1 was cloned by RT-PCR using cDNA from HEK293 cells as a template. The primer sequences were 5′-CTAGCTAGCCACCATGGATTACAAGGATGACGATGACAAGAGGCTGCGGGGATGCGGACCCCGGGCCGC-3′ as a forward primer and 5′-CCGCTCGAGCTAATCCAGCTTCTTGCGAACAGGC-3′ as a reverse primer. A FLAG epitope was attached at the N-terminus. The PCR fragment was inserted into the NheI/XhoI restriction sites of the pCXN2.1(+) vector.
Overexpression of hLPCAT1 and detection of proteins by Western blot analysis
Transfection was performed using Lipofectamine 2000 (Invitrogen). Microsomal proteins were prepared 48 h posttransfection. Cells were scraped in a buffer containing 20 mM Tris-HCl (pH 7.4), 300 mM sucrose, and a cocktail of protease inhibitors, Complete (Roche Diagnostics; Basel, Switzerland). Cells were then sonicated twice for 30 s each time. Proteins were centrifuged for 10 min at 9,000 g, and the resulting supernatant was ultracentrifuged for 1 h at 100,000 g. The pellet (microsomes) was resuspended in a buffer containing 20 mM Tris-HCl (pH 7.4), 1 mM EDTA and 300 mM sucrose.
Western blot analyses were performed as previously described (7). Anti-LPCAT1 antiserum was generated at Immuno-Biological Laboratories (Gunma, Japan) using a C-terminal peptide (EMYPDYAEDYLYPDQTHFDS) for immunization of rabbits (11). Anti-LPCAT1 antiserum, anti-FLAG M2 antibody (IBI/Kodak; Rochester, NY), and anti-HA 3F10 antibody (Roche Diagnostics) were utilized at a dilution of 1:1000. When indicated, proteins were incubated overnight at 37°C with 1 unit of N-Glycosidase F (Roche Diagnostics) in a buffer containing 20 mM sodium phosphate, 10 mM EDTA, 0.5% Triton X-100, and 1% 2-mercaptoethanol, before electrophoresis.
Site-directed mutagenesis
The mutants of hLPCAT1 were constructed by overlap extension PCR (14). PCR was carried out using KOD -Plus- (TOYOBO; Osaka, Japan).
The mutant of platelet-activating factor receptor (PAFR) was constructed using QuikChange site-directed mutagenesis kit (Stratagene; La Jolla, CA). The primers utilized for both genes are listed in Table 1.
TABLE 1.
Primer design for constructing mutants
| hLPCAT1 mutant | Forward Primer | Reverse Primer |
|---|---|---|
| C211S | AGAAGGAACTTTTACAAACAGGACCTGC | TCCTGTTTGTAAAAGTTCCTTCTGGAAAAATC |
| C211R | AGAAGGAACTCGTACAAACAGGACCTGC | TCCTGTTTGTACGAGTTCCTTCTGGAAAAATC |
| C211F | AGAAGGAACTTCTACAAACAGGACCTGC | TCCTGTTTGTAGAAGTTCCTTCTGGAAAAATC |
| N213A |
AACTTGTACAGCCAGGACCTGCCTAATTAC |
GGCAGGTCCTGGCTGTACAAGTTCCTTCTG |
| PAFR mutant |
Forward Primer |
Reverse Primer |
| N169A | CAGTGCTGGCTCAGGCGCCGTCACTCGCTGCTTTG | CAAAGCAGCGAGTGACGGCGCCTGAGCCAGCACTG |
Measurement of lysophospholipid acyltransferase activity
Lysophospholipid acyltransferase activity was measured as previously described (7). The assay buffer contained 1 µg of microsomal protein, 110 mM Tris-HCl (pH 7.4), 1.5 mM EDTA, 150 mM sucrose, 25 µM [14C]palmitoyl-CoA or 100 µM [3H]acetyl-CoA, and 50 µM lysophospholipid in the presence or absence of 1 mg/ml PC (Sigma-Aldrich; St. Louis, MO; product no. P4279). PC was dried under dry nitrogen, Tris buffer was added, and the mixture was sonicated for 1 min in a bath type sonicator. For NEM-sensitivity assay, proteins were preincubated with various concentrations of NEM for 15 min on ice before reaction.
Evolutionary analysis
Putative lysophospholipid acyltransferases from various species were searched using Basic Local Alignment Search Tool (BLAST) (15). TBLASTN search was utilized. Multiple alignment was performed with a ClustalW analyzing system from the DNA Data Bank of Japan (16).
Tissue distribution of hLPCAT1 mRNA
Quantitative PCR was performed as previously described using a LightCycler 1.5 (Roche Diagnostics) (7). Commercially available cDNA from human tissues (human Multiple Tissue cDNA Panels I and II; TaKaRa) were used as templates. The primers were the same as those utilized for detection of hLPCAT1 by RT-PCR.
RESULTS
Cloning of hLPCAT1 from HEK293 cells
Our initial step was to isolate hLPCAT1 cDNA from human cell lines. To determine which cell lines express hLPCAT1, RT-PCR was carried out using cDNA from A549, HEK293, and HeLa cells. A 154 bp fragment of hLPCAT1 was detected in all three cell lines (Fig. 1A). The full-length hLPCAT1 cDNA (1605 bp) was cloned by RT-PCR using cDNA from HEK293 cells. The enzyme consists of 534 amino acid residues and contains four conserved acyltransferase motifs (acyltransferase motifs 1–4) (17), one putative N-glycosylation site, and a dilysine ER localization motif at the C terminus (18). hLPCAT1 has 88% homology to both mLPCAT1 and rLPCAT1. Our clone possessed 100% homology to that cloned by Agarwal et al. (13)
Fig. 1.
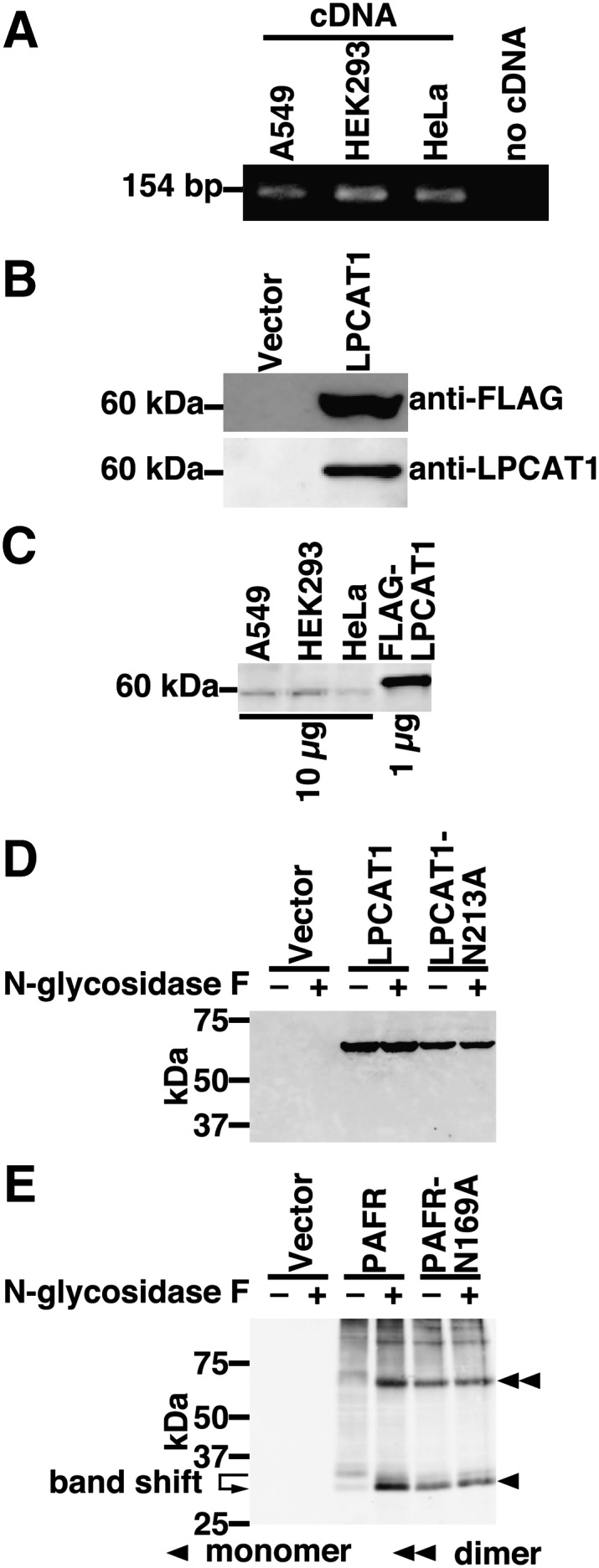
Cloning and expression of hLPCAT1. A: The expression of hLPCAT1 was examined by RT-PCR using cDNA from A549, HEK293, and HeLa cells as templates. As a negative control, RT-PCR without template (no cDNA) was performed. B: The expression of hLPCAT1 in microsomes from vector- or FLAG-hLPCAT1-transfected CHO-K1 cells was detected by Western blot using anti-FLAG antibody (upper panel, 7 μg of protein per lane) or anti-LPCAT1 antiserum (lower panel, 1 μg of protein per lane). C: The endogenous expression of hLPCAT1 in A549, HEK293, and HeLa cells was determined by Western blot (10 μg of protein per lane) using anti-LPCAT1 antiserum. 1 μg of microsomes from FLAG-hLPCAT1 overexpressing CHO-K1 cells was loaded for comparison. D: Microsomes from vector-, hLPCAT1-wild-type (WT)- or hLPCAT1-N213A-transfected HeLa cells were treated (+) or not (-) with N-glycosidase F and subjected to Western blot analysis. E: Microsomes from vector-, hPAFR-WT-, or hPAFR-N169A-transfected HeLa cells were treated (+) or not (-) with N-glycosidase F and subjected to Western blot analysis. A, C: Same experiments were performed two times independently with similar results. D, E: Same experiments were performed three times independently with similar results.
Detection of hLPCAT1 by anti-LPCAT1 antiserum
Because we previously generated an anti-mLPCAT1 antiserum (11), we investigated whether this antiserum cross-reacts with hLPCAT1. Microsomes from FLAG-hLPCAT1-overexpressing Chinese hamster ovary (CHO)-K1 cells were subjected to Western blot analysis. When using an anti-FLAG antibody, no signal was obtained from microsomes of vector-transfected cells, whereas a single band was detected from microsomes of FLAG-hLPCAT1-overexpressing cells (Fig. 1B, upper panel). The same samples were then analyzed using the anti-LPCAT1 antiserum. Similarly, a single band was observed from microsomes of hLPCAT1-transfected cells, but not from microsomes of vector-transfected cells (Fig. 1B, lower panel). This result shows that the antiserum against mLPCAT1 can also be used to detect hLPCAT1.
Because hLPCAT1 mRNA was detected in all three cell lines, we examined hLPCAT1 protein levels in these cells. Western blot was carried out using anti-LPCAT1 antiserum. Faint bands appeared in microsomal proteins (10 µg) from all three cell lines but signals were extremely low when compared with 1 µg of microsomal protein from hLPCAT1-overexpressing CHO-K1 cells (Fig. 1C).
Analysis of the putative N-glycosylation site of hLPCAT1
To investigate whether the putative N-glycosylation site of hLPCAT1 (Asn213) is glycosylated, we performed N-glycosidase treatment of microsomes from hLPCAT1-overexpressing HeLa cells, followed by Western blot analysis. N-glycosidase treatment had no effect on the molecular weight of hLPCAT1 (Fig. 1D, third lane versus fourth lane) suggesting that hLPCAT1 is expressed as a nonglycosylated protein. To further confirm that hLPCAT1 is not glycosylated, we constructed a mutant lacking the putative N-glycosylation site (mutant N213A). The mutation did not affect the molecular weight of hLPCAT1, as revealed by Western blot analysis (Fig. 1D, third lane versus fifth lane), verifying that Asn213 is not glycosylated. As expected, N-glycosidase treatment had no effect on the molecular weight of mutant N213A (Fig. 1D, fifth lane versus sixth lane). To exclude the possibility that N-glycosidase treatment was unsuccessful, microsomes from PAFR-overexpressing HeLa cells were treated similarly with N-glycosidase. N-glycosidase treatment decreased the molecular weight of PAFR, indicating that PAFR was N-glycosylated (Fig. 1E, third lane versus fourth lane), and that treatment with N-glycosidase resulted in deglycosylation. The molecular weight of N-glycosidase-treated PAFR was similar to that of a mutant lacking the N-glycosylation site (PAFR-N169A) (Fig. 1E, fourth lane versus fifth lane) (19). These results indicate that N-glycosidase treatment was successful but did not affect hLPCAT1 band size. Thus, we concluded that hLPCAT1 is expressed as a nonglycosylated protein although it possesses a putative N-glycosylation site (Asn213).
Lysophospholipid acyltransferase activity of hLPCAT1
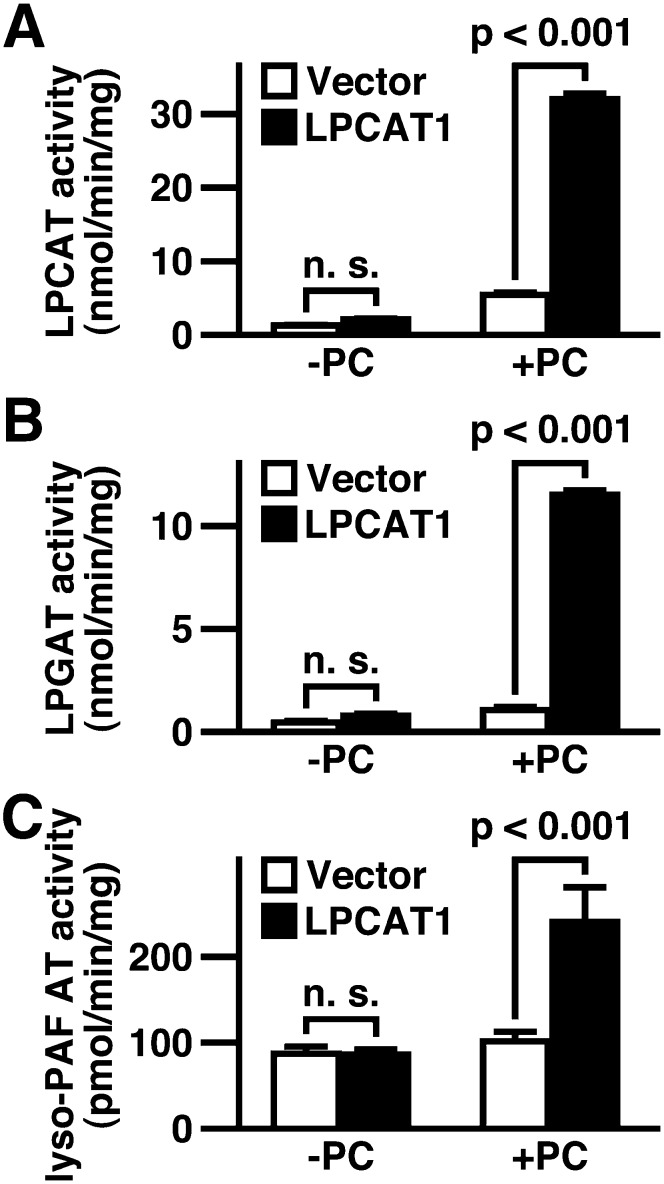
We next examined whether hLPCAT1 possesses lysophospholipid acyltransferase activity. Microsomes from hLPCAT1-transfected HeLa cells exhibited high LPCAT, LPGAT, and lyso-PAF AT activities, compared with microsomes from vector-transfected cells (Fig. 2A–C, third and fourth bars).
Fig. 2.
Enzymatic activity of hLPCAT1. A: LPCAT (using palmitoyl-LPC and palmitoyl-CoA), B: LPGAT (using palmitoyl-LPG and palmitoyl-CoA), and C: lyso-PAF AT (using lyso-PAF and acetyl-CoA) activities were measured using microsomes from vector- or hLPCAT1-transfected HeLa cells in the absence or presence of PC in the reaction mixture. n. s.: not significant. A–C: Values are means ± SD. Same experiments were performed three times independently with similar results. A–C: Two-way ANOVA, Bonferroni.
Because there is a discrepancy between our results and the report showing that hLPCAT1 has no LPCAT activity, we investigated whether the buffer composition may affect LPCAT activity. Our reaction buffer contains 1 mg/ml of PC mixture whereas those in other reports do not (13). Thus, we performed LPCAT, LPGAT, and lyso-PAF AT assays in the absence or presence of PC. In the absence of PC, no significant activity was observed, whereas in the presence of PC, hLPCAT1 exhibited LPCAT, LPGAT, and lyso-PAF AT activities (Fig. 2A–C). These results show that hLPCAT1 is indeed an LPCAT and that its activity is observed only in the presence of PC.
NEM sensitivity of hLPCAT1
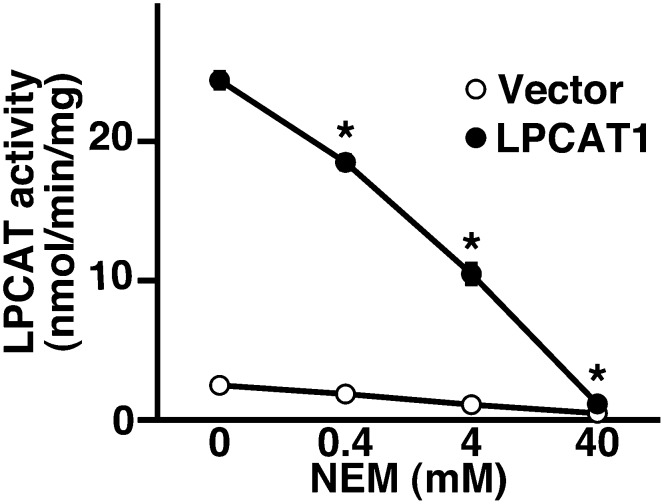
Sensitivity to NEM, a sulfhydryl-reducing reagent, is commonly used for characterizing acyltransferases. Therefore, we evaluated NEM sensitivity of hLPCAT1. Preincubation with 0.4 mM NEM [a concentration that does not inhibit the activity of glycerol-3-phosphate acyltransferase (GPAT) 1 (20)] led to a significant decrease of hLPCAT1 activity indicating that hLPCAT1 is an NEM-sensitive enzyme (Fig. 3). Higher concentrations of NEM led to a stronger inhibition, and 40 mM NEM almost completely abolished hLPCAT1 activity (Fig. 3).
Fig. 3.
NEM sensitivity of hLPCAT1 LPCAT activity of hLPCAT1 was measured after preincubation with 0-40 mM NEM. Values are means ± SD. Same experiments were performed three times independently with similar results. * P < 0.001 versus LPCAT1 with 0 M NEM, two-way ANOVA, Bonferroni.
Analysis of Cys211 of hLPCAT1
NEM sensitivity indicates the presence of at least one cysteine in hLPCAT1 critical for its activity. To investigate which cysteine is important, we compared the primary sequences of human acyltransferases (Table 2). We identified a cysteine (Cys211) conserved in the putative acyltransferase motif 3 of LPCAT1 (7, 11), LPCAT2 (8), lysophosphatidylethanolamine acyltransferase 2 (LPEAT2) (21), GPAT3 (22), and GPAT4 (23) (Table 2). Both GPAT3 and 4 are known to be NEM-sensitive enzymes (22, 23). Conversely, GPAT1 is known as a NEM-insensitive enzyme (20). In GPAT1, an arginine is located at this position. A phenylalanine and a proline are located at this position in LPGAT1 (24) and GPAT2 (25), respectively.
TABLE 2.
Comparison of motif 3 from several acyltransferases and their NEM sensitivity
| Motif 3 | NEM Sensitivity | Accession | |
|---|---|---|---|
| LPAAT1 | 175VFPEGTR181 | unknown | NP_116130 |
| LPGAT1 | 181LFPEGGF187 | unknown | NP_055688 |
| GPAT1 | 312IFLEGTR318 | resistant | NP_065969 |
| GPAT2 | 287IFLEEPP293 | sensitive | NP_997211 |
| GPAT3 | 300IFPEGT306C | sensitive | NP_116106 |
| GPAT4 | 319IFPEGT325C | sensitive | NP_848934 |
| LPCAT1 | 205IFPEGT211C | sensitive (this study) | BAE94688 |
| LPCAT2 | 217VFPEGT223C | unknown | BAF47696 |
| LPEAT2 | 200FFPEGT206C | unknown | NP_705841 |
We hypothesized that Cys211 is crucial for the activity of hLPCAT1. Therefore, we constructed three mutants, C211S (serine was selected due to its structural similarity to cysteine), C211R, and C211F. When transfected in HeLa cells, the three mutants were expressed at similar levels as revealed by Western blot analysis (Fig. 4A). Next, we measured LPCAT and LPGAT activities of these three mutants. LPCAT and LPGAT activities were completely abolished in all three mutants (Fig. 4B and C). These results show that Cys211 is essential for the activity of hLPCAT1.
Fig. 4.
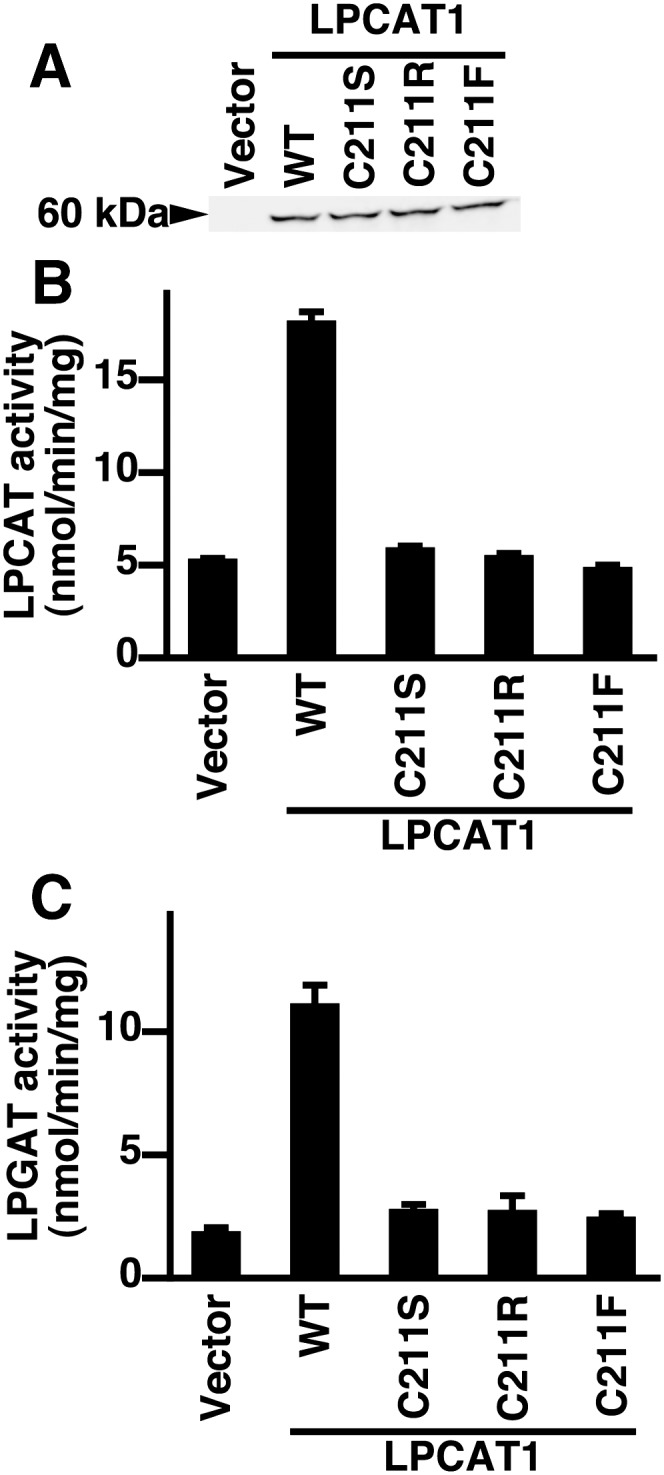
Analysis of Cys211 mutants. A: Microsomes from vector-, hLPCAT1-, or the indicated hLPCAT1 mutant-transfected HeLa cells were subjected to Western blot (5 μg of protein per lane) using anti-FLAG antibody. B: LPCAT and C: LPGAT activities of microsomes from vector-, hLPCAT1-WT-, or hLPCAT1 mutants-transfected HeLa cells were measured. B, C: Values are means ± SD. A–C: Same experiments were performed three times independently with similar results.
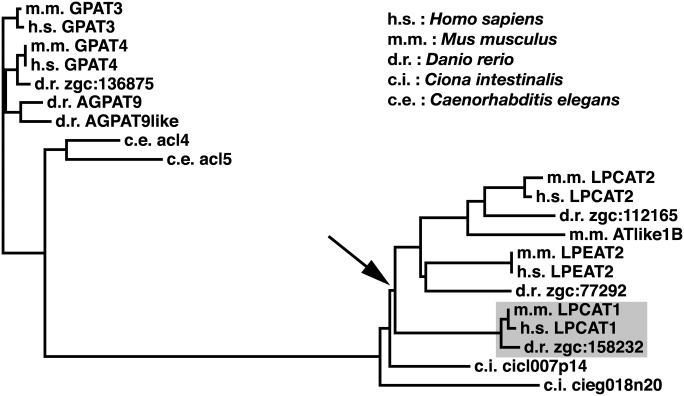
Evolutionary analysis of hLPCAT1
The importance of Cys211 and its conservation in human acyltransferases led us to the hypothesis that these enzymes that contain a cysteine at the end of motif 3 constitute a sub-family in acyltransferases, which we call “motif 3-cysteine” acyltransferases. Thus, we performed an extensive BLAST search to determine how these “motif 3-cysteine” acyltransferases have bifurcated from each other. We found 2, 2, 6, 6, and 5 “motif 3-cysteine” acyltransferases from Caenorhabditis elegans, Ciona intestinalis, Danio rerio, Mus musculus, and Homo sapiens, respectively (Table 3). Then we performed multiple alignment and drew a phylogenetic tree (Fig. 5). The result revealed that the “motif 3-cysteine” acyltransferases had first bifurcated into two groups. The first one was comprised of GPATs and the second one contained LPCATs and LPEATs. The proteins encoded by Ciona intestinalis cicl007p14 and cieg018n20 share a common ancestor with LPCATs and LPEATs of vertebrates. LPCAT1 seems to have evolved after the bifurcation of the ancestor of Ciona intestinalis and teleosts (Fig. 5, arrow). This is in agreement with the fact that the lung evolved before the appearance of teleosts (26).
TABLE 3.
Accession number of “motif 3-cysteine” acyltransferases
| Species | Enzyme | Nucleotide | Protein |
|---|---|---|---|
| Homo sapiens | LPCAT1 | AB244719 | BAE94688 |
| Homo sapiens | LPCAT2 | AB244718 | BAF47696 |
| Homo sapiens | LPEAT2 | NM_153613 | NP_705841 |
| Homo sapiens | GPAT3 | NM_032717 | NP_116106 |
| Homo sapiens | GPAT4 | NM_178819 | NP_848934 |
| Mus musculus | LPCAT1 | AB244717 | BAE94687 |
| Mus musculus | LPCAT2 | AB244716 | BAF47695 |
| Mus musculus | ATlike1B | NM_027599 | NP_081875 |
| Mus musculus | LPEAT2 | NM_207206 | NP_997089 |
| Mus musculus | GPAT3 | NM_172715 | NP_766303 |
| Mus musculus | GPAT4 | NM_018743 | NP_061213 |
| Danio rerio | zgc:158232 | NM_001044341 | NP_001037806 |
| Danio rerio | zgc:112165 | NM_001020656 | NP_001018492 |
| Danio rerio | zgc:77292 | NM_205559 | NP_991122 |
| Danio rerio | zgc:136875 | NM_001040249 | NP_001035339 |
| Danio rerio | AGPAT9 | NM_001002685 | NP_001002685 |
| Danio rerio | AGPAT9like | NM_001099450 | NP_001092920 |
| Ciona intestinalis | cicl007p14 | AK173535 | |
| Ciona intestinalis | cieg018n20 | AK114815 | |
| Caenorhabditis elegans | acl-4 | NM_075978 | NP_508379 |
| Caenorhabditis elegans | acl-5 | NM_001047817 | NP_001041282 |
Fig. 5.
Phylogenetic tree of motif 3-cysteine acyltransferases. Multiple alignment of putative lysophospholipid acyltransferases containing a cysteine at the end of motif 3 was performed. Based on the result, a phylogenetic tree was drawn. Arrow: the time of bifurcation of LPCAT1 and other proteins.
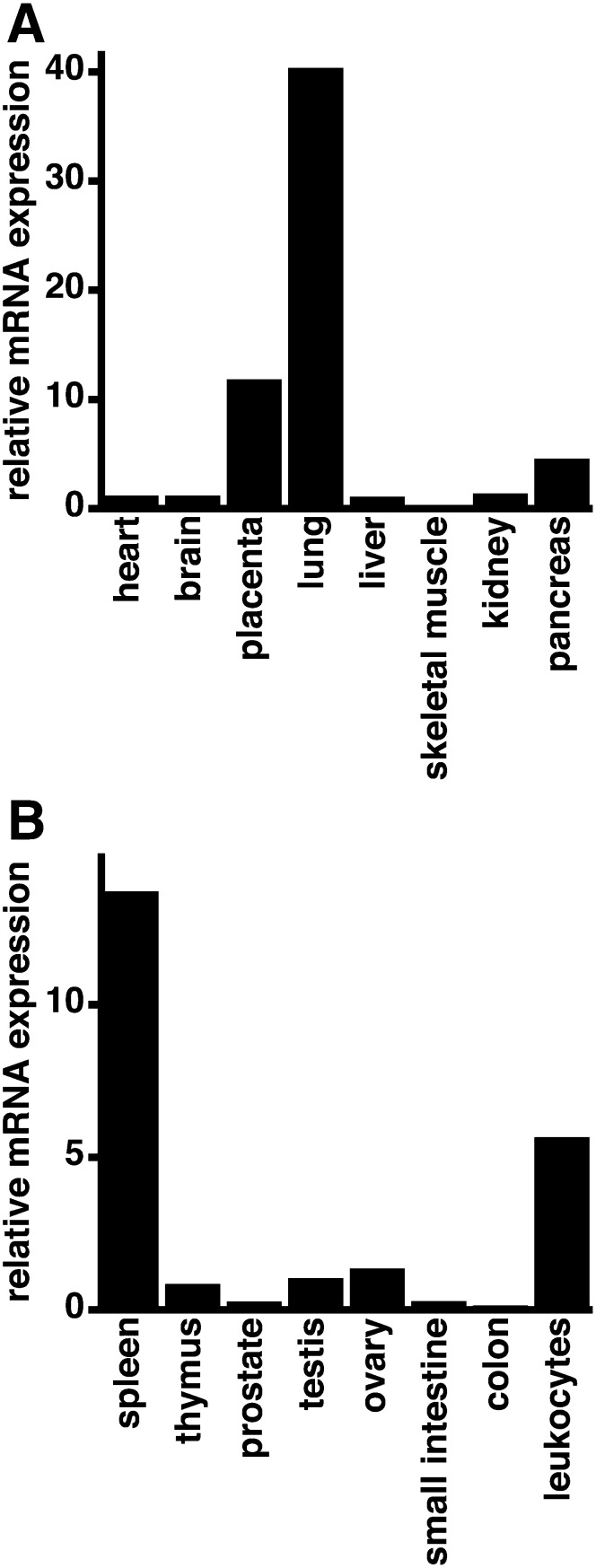
Tissue distribution of hLPCAT1 mRNA
To confirm the importance of hLPCAT1 in the human lung, we examined its mRNA expression by quantitative PCR. We used two panels of cDNA from human (panel 1: cDNA from heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas; panel 2: cDNA from spleen, thymus, prostate, testis, ovary, small intestine, colon, and leukocytes) as templates for quantitative PCR. In panel 1, the expression of hLPCAT1 was the highest in the lung, followed by the placenta (Fig. 6A). In panel 2, the expression of hLPCAT1 was the highest in the spleen, followed by leukocytes (Fig. 6B).
Fig. 6.
Tissue distribution of hLPCAT1 mRNA. A: Quantitative PCR was performed using cDNA from human heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas as templates. Values are illustrated as the relative expression level of hLPCAT1 divided by that of glyceraldehyde-3-phosphate dehydrogenase. The value for human heart was set as 1. B: Quantitative PCR was performed using cDNA from human spleen, thymus, prostate, testis, ovary, small intestine, colon, and leukocytes as templates. Values are illustrated as the relative expression level of hLPCAT1 divided by that of glyceraldehyde-3-phosphate dehydrogenase. The value for human thymus was set as 1.
DISCUSSION
LPCAT1 has been proposed to be the biosynthetic enzyme of pulmonary surfactant lipids (7, 12). This hypothesis was suggested for the following reasons. First, LPCAT1 synthesizes DPPC and DSPG in vitro, the major lipid components of pulmonary surfactant. Second, LPCAT1 is highly expressed in the lung, especially in alveolar type II cells, which secrete pulmonary surfactant. Third, LPCAT1 expression is upregulated perinatally, coincident with the production of pulmonary surfactant. However, the above observations were performed using either rats or mice and in these instances hLPCAT1 was not investigated.
Agarwal et al. (13) cloned hLPCAT1 and reported that it possesses no LPCAT activity. This observation impelled us to investigate the activity of hLPCAT1 and determine whether it has the potential to biosynthesize pulmonary surfactant lipids. In this study, we showed that hLPCAT1 possesses LPCAT, LPGAT, and lyso-PAF AT activities as previously reported in the murine homolog. The activity was dependent on the presence of PC in the reaction buffer. It is unclear how PC works in the reaction but it is likely that it acts as a detergent, because Soupene et al. (27) could observe LPCAT activity of mLPCAT1 in the presence of Tween-20 instead of PC. Consistently, we detected LPCAT activity of hLPCAT1 in the presence of 0.015% Tween-20 without PC (T. Harayama, H. Shindou, and T. Shimizu, unpublished observations). There is also possibility that lipid interaction is important for the activity of LPCAT1. The deficiency of LPCAT activity in the report of Agarwal et al. (13) could therefore be explained by a lack of detergent in the reaction mixture.
To further characterize novel properties of LPCAT1, we demonstrated that LPCAT1 was NEM sensitive, like GPAT2-4 (22, 23, 25). Interestingly, LPCAT1 shares a conserved cysteine (Cys211, at the end of motif 3) with GPAT3 and 4. Indeed, mutation of Cys211 showed that it was crucial for the activity of LPCAT1. This cysteine may be responsible for the difference between the NEM sensitivity of various acyltransferases. Further experiments are needed to directly certificate this hypothesis. At this position, GPAT1 and lysophosphatidic acyltransferase 1 (LPAAT1; also called AGPAT1) contain arginines that have been shown to be crucial for their activity (28, 29). Thus, the amino acid at the end of motif 3 seems to be essential for the activity of these acyltransferases. Although GPAT2 does not possess the described motif 3-cysteine, a cysteine at motif 2 was proposed to be responsible for its NEM sensitivity (25).
The importance of the amino acid at the end of motif 3 suggests that this residue can be utilized to classify acyltransferases. Therefore, we performed an extensive BLAST search to investigate the conservation of the LPCAT1 motif 3-cysteine. The results showed that the cysteine at the end of motif 3 is highly conserved among many enzymes of many species. We termed these enzymes “motif 3-cysteine” acyltransferases in this work. The establishment of this classification allowed us to investigate the evolution of LPCAT1. We compared the motif 3-cysteine enzymes to determine when LPCAT1 first appeared. The phylogenetic tree suggested that the evolution of LPCAT1 occurred before the appearance of teleosts. It is said that the ancestor of the lung also appeared before the evolution of teleosts as a “respiratory pharynx”, which became the air bladder in teleosts and the lung in other vertebrates (26). Thus, the evolution of LPCAT1 appears to coincide with that of the lung. Because both LPCAT1 and LPCAT2 possess lyso-PAF AT activity, the ancestor enzyme of LPCAT1 might be a PAF biosynthetic enzyme. Additionally, LPCAT1 might have evolved as the biosynthetic enzyme of pulmonary surfactant phospholipids. The high expression level of hLPCAT1 mRNA in the human lung underscores its involvement in pulmonary function.
The properties of hLPCAT1 that we have described here implicate it in the biosynthesis of pulmonary surfactant lipids. First, it synthesizes DPPC and DSPG in vitro, the major lipid components of pulmonary surfactant. Second, the evolution of LPCAT1 appears to coincide with the evolution of the lung. Third, hLPCAT1 is highly expressed in the human lung. In addition to these findings, hLPCAT1 mRNA (which was called AYTL2 in the report) was shown to be upregulated during in vitro differentiation of alveolar type II cells (30). Our preliminary data suggest that hLPCAT1 exhibits substrate specificity for palmitoyl-CoA (T. Harayama, H. Shindou, and T. Shimizu, unpublished observations). Based on the above observations by us and others, we propose that hLPCAT1 is the biosynthetic enzyme of pulmonary surfactant lipids. Further studies are needed to elucidate the roles of hLPCAT1 in surfactant production in vivo. Given the fact that mutations in numerous genes involved in pulmonary surfactant function lead to various pulmonary diseases (for example, ATP-binding cassette A3 and surfactant protein B) (3, 31), alterations in hLPCAT1 function may also be responsible for some lung dysfunctions.
Acknowledgments
The authors are grateful to Drs. R. Taguchi, M. Nakamura, S. Ishii, Y. Kita, S. M. Tokuoka, and T. Takahashi, and to D. Hishikawa, K. Kuniyeda, N. Murakami, and M. Eto for valuable suggestions. We thank F. Hamano, H. Nakanishi, N. Hirota, D. Yasuda, T. Hashidate, and S. Kobayashi for cooperation. We thank Dr. J-i. Miyazaki (Osaka University) for supplying the expression vector pCXN2.
Footnotes
Abbreviations:
- AGPAT
- 1-acylglycerol-3-phosphate-O-acyltransferase
- BLAST
- Basic Local Alignment Search Tool
- CHO
- Chinese hamster ovary
- DPPC
- dipalmitoylphosphatidylcholine
- DSPG
- disaturated phosphatidylglycerol
- GPAT
- glycerol-3-phosphate acyltransferase
- hLPCAT1
- human lysophosphatidylcholine acyltransferase 1
- LPAAT
- lysophosphatidic acid acyltransferase
- LPCAT
- lysophosphatidylcholine acyltransferase
- LPEAT2
- lysophosphatidylethanolamine acyltransferase 2
- LPGAT
- lysophosphatidylglycerol acyltransferase
- lyso-PAF AT
- lyso-platelet-activating factor acetyltransferase
- mLPCAT1
- murine lysophosphatidylcholine acyltransferase 1
- NEM
- N-ethylmaleimide
- PAF
- platelet-activating factor
- PAFR
- platelet-activating factor receptor
- PC
- phosphatidylcholine
- PG
- phosphatidylglycerol
- rLPCAT1
- rat lysophosphatidylcholine acyltransferase 1
This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (T.S.) and a Global COE Program (The University of Tokyo) from Japan Society for Promotion of Sciences (T.S.). T.S. and H.S. were supported by the Center for NanoBio Integration at The University of Tokyo. H.S. was supported by Health and Labour Sciences Research Grants (Research on Allergic Disease and Immunology) from the Ministry of Health, Labour, and Welfare of Japan, Grant-in-Aid for Young Scientists (B)(21790264) from the MEXT of Japan, Mitsubishi Pharma Research Foundation, and Ono Medical Research Foundation. T.H. was supported by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists.
REFERENCES
- 1.Veldhuizen R., Nag K., Orgeig S., Possmayer F. 1998. The role of lipids in pulmonary surfactant. Biochim. Biophys. Acta. 1408: 90–108. [DOI] [PubMed] [Google Scholar]
- 2.Daniels C. B., Orgeig S. 2003. Pulmonary surfactant: the key to the evolution of air breathing. News Physiol. Sci. 18: 151–157. [DOI] [PubMed] [Google Scholar]
- 3.Whitsett J. A., Wert S. E., Trapnell B. C. 2004. Genetic disorders influencing lung formation and function at birth. Hum. Mol. Genet. 13: R207–R215. [DOI] [PubMed] [Google Scholar]
- 4.Lands W. E. 1958. Metabolism of glycerolipides; a comparison of lecithin and triglyceride synthesis. J. Biol. Chem. 231: 883–888. [PubMed] [Google Scholar]
- 5.Shimizu T. 2009. Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu. Rev. Pharmacol. Toxicol. 49: 123–150. [DOI] [PubMed] [Google Scholar]
- 6.Shindou H., Shimizu T. 2009. Acyl-CoA: lysophospholipid acyltransferases. J. Biol. Chem. 284: 1–5. [DOI] [PubMed] [Google Scholar]
- 7.Nakanishi H., Shindou H., Hishikawa D., Harayama T., Ogasawara R., Suwabe A., Taguchi R., Shimizu T. 2006. Cloning and characterization of mouse lung-type acyl-CoA:lysophosphatidylcholine acyltransferase 1 (LPCAT1). Expression in alveolar type II cells and possible involvement in surfactant production. J. Biol. Chem. 281: 20140–20147. [DOI] [PubMed] [Google Scholar]
- 8.Shindou H., Hishikawa D., Nakanishi H., Harayama T., Ishii S., Taguchi R., Shimizu T. 2007. A single enzyme catalyzes both platelet-activating factor production and membrane biogenesis of inflammatory cells. Cloning and characterization of acetyl-CoA:LYSO-PAF acetyltransferase. J. Biol. Chem. 282: 6532–6539. [DOI] [PubMed] [Google Scholar]
- 9.Hishikawa D., Shindou H., Kobayashi S., Nakanishi H., Taguchi R., Shimizu T. 2008. Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity. Proc. Natl. Acad. Sci. USA. 105: 2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y., Chen Y. Q., Bonacci T. M., Bredt D. S., Li S., Bensch W. R., Moller D. E., Kowala M., Konrad R. J., Cao G. 2008. Identification and characterization of a major liver lysophosphatidylcholine acyltransferase. J. Biol. Chem. 283: 8258–8265. [DOI] [PubMed] [Google Scholar]
- 11.Harayama T., Shindou H., Ogasawara R., Suwabe A., Shimizu T. 2008. Identification of a novel noninflammatory biosynthetic pathway of platelet-activating factor. J. Biol. Chem. 283: 11097–11106. [DOI] [PubMed] [Google Scholar]
- 12.Chen X., Hyatt B. A., Mucenski M. L., Mason R. J., Shannon J. M. 2006. Identification and characterization of a lysophosphatidylcholine acyltransferase in alveolar type II cells. Proc. Natl. Acad. Sci. USA. 103: 11724–11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agarwal A. K., Sukumaran S., Bartz R., Barnes R. I., Garg A. 2007. Functional characterization of human 1-acylglycerol-3-phosphate-O-acyltransferase isoform 9: cloning, tissue distribution, gene structure, and enzymatic activity. J. Endocrinol. 193: 445–457. [DOI] [PubMed] [Google Scholar]
- 14.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 77: 51–59. [DOI] [PubMed] [Google Scholar]
- 15.McGinnis S., Madden T. L. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32: W20–W25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewin T. M., Wang P., Coleman R. A. 1999. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 38: 5764–5771. [DOI] [PubMed] [Google Scholar]
- 18.Jackson M. R., Nilsson T., Peterson P. A. 1993. Retrieval of transmembrane proteins to the endoplasmic reticulum. J. Cell Biol. 121: 317–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García Rodríguez C., Cundell D. R., Tuomanen E. I., Kolakowski L. F., Gerard C., Gerard N. P. 1995. The role of N-glycosylation for functional expression of the human platelet-activating factor receptor. Glycosylation is required for efficient membrane trafficking. J. Biol. Chem. 270: 25178–25184. [DOI] [PubMed] [Google Scholar]
- 20.Yet S. F., Lee S., Hahm Y. T., Sul H. S. 1993. Expression and identification of p90 as the murine mitochondrial glycerol-3-phosphate acyltransferase. Biochemistry. 32: 9486–9491. [DOI] [PubMed] [Google Scholar]
- 21.Cao J., Shan D., Revett T., Li D., Wu L., Liu W., Tobin J. F., Gimeno R. E. 2008. Molecular identification of a novel mammalian brain isoform of acyl-CoA:lysophospholipid acyltransferase with prominent ethanolamine lysophospholipid acylating activity, LPEAT2. J. Biol. Chem. 283: 19049–19057. [DOI] [PubMed] [Google Scholar]
- 22.Cao J., Li J. L., Li D., Tobin J. F., Gimeno R. E. 2006. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA. 103: 19695–19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagle C. A., Vergnes L., Dejong H., Wang S., Lewin T. M., Reue K., Coleman R. A. 2008. Identification of a novel sn-glycerol-3-phosphate acyltransferase isoform, GPAT4, as the enzyme deficient in Agpat6−/− mice. J. Lipid Res. 49: 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y., Cao J., Shi Y. 2004. Identification and characterization of a gene encoding human LPGAT1, an endoplasmic reticulum-associated lysophosphatidylglycerol acyltransferase. J. Biol. Chem. 279: 55866–55874. [DOI] [PubMed] [Google Scholar]
- 25.Wang S., Lee D. P., Gong N., Schwerbrock N. M., Mashek D. G., Gonzalez-Baró M. R., Stapleton C., Li L. O., Lewin T. M., Coleman R. A. 2007. Cloning and functional characterization of a novel mitochondrial N-ethylmaleimide-sensitive glycerol-3-phosphate acyltransferase (GPAT2). Arch. Biochem. Biophys. 465: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torday J. S., Rehan V. K., Hicks J. W., Wang T., Maina J., Weibel E. R., Hsia C. C. W., Sommer R. J., Perry S. F. 2007. Deconvoluting lung evolution: from phenotypes to gene regulatory networks. Integr. Comp. Biol. 47: 601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soupene E., Fyrst H., Kuypers F. A. 2008. Mammalian acyl-CoA:lysophosphatidylcholine acyltransferase enzymes. Proc. Natl. Acad. Sci. USA. 105: 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dircks L. K., Ke J., Sul H. S. 1999. A conserved seven amino acid stretch important for murine mitochondrial glycerol-3-phosphate acyltransferase activity. Significance of arginine 318 in catalysis. J. Biol. Chem. 274: 34728–34734. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita A., Nakanishi H., Suzuki H., Kamata R., Tanaka K., Waku K., Sugiura T. 2007. Topology of acyltransferase motifs and substrate specificity and accessibility in 1-acyl-sn-glycero-3-phosphate acyltransferase 1. Biochim. Biophys. Acta. 1771: 1202–1215. [DOI] [PubMed] [Google Scholar]
- 30.McDevitt T. M., Gonzales L. W., Savani R. C., Ballard P. L. 2007. Role of endogenous TGF-beta in glucocorticoid-induced lung type II cell differentiation. Am. J. Physiol. Lung Cell. Mol. Physiol. 292: L249–L257. [DOI] [PubMed] [Google Scholar]
- 31.Yokota T., Matsumura Y., Ban N., Matsubayashi T., Inagaki N. 2008. Heterozygous ABCA3 mutation associated with non-fatal evolution of respiratory distress. Eur. J. Pediatr. 167: 691–693. [DOI] [PubMed] [Google Scholar]