Abstract
Previous studies have shown that MALDI-imaging mass spectrometry (IMS) can be used to visualize the distribution of various biomolecules, especially lipids, in the cells and tissues. In this study, we report the cell-selective distribution of PUFA-containing glycerophospholipids (GPLs) in the mouse brain. We established a practical experimental procedure for the IMS of GPLs. We demonstrated that optimization of the composition of the matrix solution and spectrum normalization to the total ion current (TIC) is critical. Using our procedure, we simultaneously differentiated and visualized the localizations of specific molecular species of GPLs in mouse brain sections. The results showed that PUFA-containing phosphatidylcholines (PCs) were distributed in a cell-selective manner: arachidonic acid- and docosahexaenoic acid-containing PCs were seen in the hippocampal neurons and cerebellar Purkinje cells, respectively. Furthermore, these characteristic localizations of PUFA-PCs were formed during neuronal maturation. The phenomenon of brain cell-selective production of specific PUFA-GPLs will help elucidate the potential physiological functions of PUFAs in specific brain regions.
Keywords: MALDI, imaging MS, LC ESI-MS/MS, phospholipids, development
Much progress has recently been made in studies of brain lipids. Neural lipids play a role in synaptic vesicle fusion (1, 2), regulation of receptors and ion channels (3, 4), and formation of raft microdomains for neuronal cellular communications (5–7). Among brain lipids, glycerophospholipids (GPLs) are the most abundant, comprising a large molecular family in which phosphoric acid in the ester form is bound to a glycerolipid. GPLs are subdivided into distinct classes [e.g., phosphatidylcholines (PCs) and phosphatidylinositols] based on the structure of the head group linked to the phosphate, attached at the sn-3 position of the glycerol backbone. In brain membrane GPLs, many PUFAs, such as arachidonic acid (AA) and docosahexaenoic acid (DHA), are stored at the sn-2 position in an asymmetrical manner (8). Such PUFAs are mainly released from GPLs by the activity of phospholipase A2 (PLA2) (9), and are metabolized into various important bioactive lipids (7) involved in PUFA-mediated signaling pathways (10, 11). The systemic analysis of diverse lipids, “lipidomics,” plays a crucial role in understanding lipid signaling (12–14). It is also crucial to determine the neuronal distributions of GPLs, especially PUFAs, to understand their possible roles in brain function. Until recently, however, lack of lipid visualization systems has made lipid distributions difficult to determine.
Imaging mass spectrometry (IMS) is a unique detection technique that allows visualization of the distribution of various biomolecules in tissue sections (15, 16). Several studies have shown that MALDI-IMS can be used to visualize the distribution of endogenous metabolites in cells and tissues (17–19), especially GPLs (20–25) and drug metabolites (26–28), as well as imaging with SIMS based IMS (29–32). One of the advantages of IMS is the ability to discriminate among structurally heterogeneous molecules by their masses (33, 34) and image them simultaneously. A recent breakthrough in statistical IMS (35–38) has enabled comprehensive imaging analysis of GPL molecular families.
This article presents and demonstrates an improved experimental procedure that further enables highly qualitative IMS. This technical improvement in IMS contributes toward in situ lipidomics, allowing profiling of large-scale changes in lipid composition or determining the topographical distribution of individual lipid species in neural cells. Using our procedure, we simultaneously differentiated and visualized the localizations of specific molecular species of GPLs in mouse brain sections. In particular, our novel observations on PUFA-PCs suggest the brain cell-selective production of specific PUFA-GPLs in specific brain regions.
MATERIALS AND METHODS
Chemicals
Methanol, trifluoroacetic acid, potassium acetate, and lithium acetate were purchased from Wako Chemical (Tokyo, Japan). Calibration standard peptide and 2,5-dihydroxybenzoic acid (DHB) were purchased from Bruker Daltonics (Leipzig, Germany). Synthesized 17:0 lyso-PC was purchased from Avanti Polar Lipid (Alabaster, AL).
Euthanization of animals and tissue extraction
All experiments with mice were conducted according to the protocols approved by the Animal Care and Use Committee of the Mitsubishi Kagaku Institute of Life Sciences. The brains of male C57BL/6J Cr mice at the indicated ages (1 and 12 postnatal days, 7 and 96 postnatal weeks) were used. Mouse brains were extracted within a minute (typically in 40 s) after euthanization. Postmortem degradation of GPLs was observed within 15 min by IMS in a series of mouse brains extracted at different times (15, 30, 60, and 120 min, see also supplementary Fig. I), presumably because of stimulation by phospholipase A (PLA) under ischemic conditions (39, 40). The trimmed tissue blocks were immediately frozen in powdered dry ice, allowing tissues to be frozen without cracks, and stored at −80°C until use.
Tissue section preparation
Tissues blocks were sectioned at −16°C using a cryostat (CM 3050; Leica, Germany) to a thickness of 5 µm, as described in previous reports (41, 42). Although brain blocks were held by an optimum cutting temperature polymer, they were not embedded into it, because it was thought that any residual polymer on the tissue slices might degrade the mass spectra (42). The frozen sections were thaw-mounted on indium-tin-oxide (ITO)-coated glass slides (Bruker Daltonics) and ITO-coated sheets (Tobi Co., Ltd., Kyoto, Japan). The slides were used for tandem time-of-flight (TOF/TOF) measurements and the sheets were used for quadrupole ion trap (QIT)-TOF measurements. Prepared sections were subjected to matrix application within 5 min.
Spray-coating of the matrix solution
A DHB solution (40 mg/ml DHB, 20 mM potassium acetate, 70% MetOH, 0.1% trifluoroacetic acid) was used as the matrix solution for imaging of PCs. The matrix solution was sprayed over the tissue surface using a 0.2-mm nozzle caliber airbrush (Procon Boy FWA Platinum; Mr. Hobby, Tokyo, Japan). Matrices were applied simultaneously to the tissue sections that were to be compared with equalized analyte extraction and cocrystallization conditions. The distance between the nozzle tip and the tissue surface was held at 10 cm and the spraying period was fixed at 5 min. Approximately 100 μl of matrix solution was sprayed onto each brain section.
A properly handled spray-coating step created a uniform matrix crystal layer on the surface (supplementary Fig. IA), and awareness of some technical points contributed to success. First, too small a distance between the airbrush and the tissues (<10 cm) often resulted in large droplets of matrix solution and inhomogeneous crystals (supplementary Fig. IIB). Second, humidity is also important factor. Room humidity was held under 25% at room temperature (25°C). High humidity tends to cause formation of needle-like crystals, as shown in supplementary Figure IIC (>80%, at room temperature).
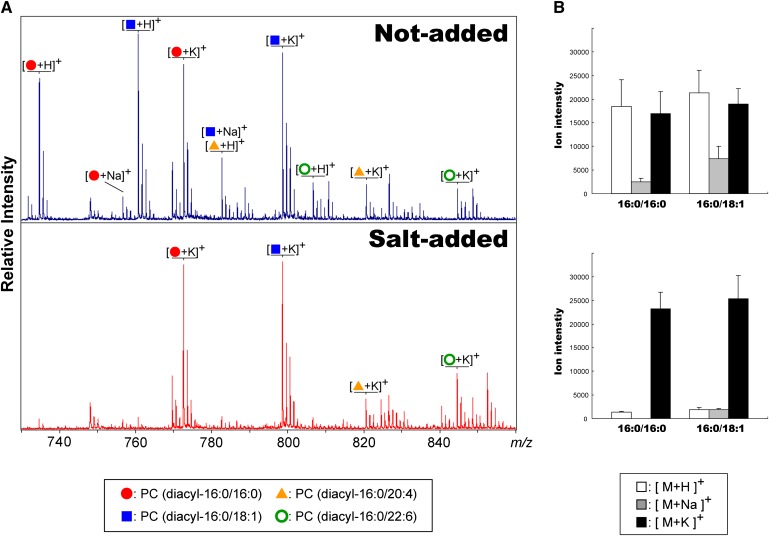
Reduction of the generation of multiple molecular ions from a single PC molecular species by addition of an alkali-metal salt to the matrix solution
PCs undergo preferential cationization with alkalis to form metal-adduct molecules (22, 43, 44). Because tissue sections are rich in sodium and potassium salts, such alkali-metal adduct phospholipids, rather than protonated molecules, are preferentially generated (Fig. 1A). Molecular ionization with such multiple ion forms from a single species often hampers IMS experiments for several reasons. First, the distribution image of GPLs might not reflect the actual distribution of GPLs but rather the heterogeneous distribution of salts. Second, because GPLs have many molecular species, a single peak might contain multiple types of ions. Indeed, for example, a protonated PC (diacyl-16:0/20:4) molecule is detected as having the same mass as a sodiated PC (diacyl-16:0/18:1) ion at m/z 782 (Fig. 1A). To overcome this problem, it is important to reduce the potential ion forms to a single alkali-metal adduct form. In this study, potassium salt was added to the matrix solution (20 mM potassium acetate). As a result, the molecular ion forms were limited to potassium adducts and the spectra were simplified (Fig. 1A, B). Consequently, the two types of PCs detected at m/z 782 described above could be separated at m/z 820 and m/z 798.
Fig. 1.
Generation of multiple molecular ions from a single PC molecular species was reduced by adding an alkali-metal salt to the matrix solution. A: Spectra obtained from sections of mouse brain-homogenate using matrix solution with/without potassium acetate. The use of the salt-added matrix solution allowed multiple molecular ion-forms of PCs to be reduced to a single potassiated molecular ion form. B: Ion intensities of two PC species, PC (diacyl-16:0/16:0) and PC (diacyl-16:0/18:1), in three molecular ion forms, protonated, sodiated, and potassiated molecules. Error bars indicate SD.
Tandem mass spectrometry
Molecular identification was performed with tandem mass spectrometry (MSn) analysis using a QIT-TOF mass spectrometer (AXIMA-QIT; Shimadzu, Kyoto, Japan) to ensure molecular assignment only by mass. The MSn analysis was performed directly on the mouse brain sections. Acquisition was performed in the mid-mass range mode (m/z 750–2000) at a stage voltage of 18 V in the positive-ion detection mode. Calibration was performed using an external calibration method. To perform accurate molecular identification of PC molecular species by using the QIT-TOF instrument, we performed MS3 analysis following MS2 analysis by using ions at [M-59 u]+, which correspond to the ions with neutral loss of trimethylamine groups. We assigned ions to PC molecules that showed neutral loss of phosphate acid groups (–124 u) during MS3 analysis, because this loss indicates that the analyzed molecules contain trimethylamine and phosphate groups. Owing to the instrumental specificity of QIT devices, they cannot trap small ions (m/z < 200) in the “mid-range mode” and therefore cannot detect signals in the small m/z region during MSn analysis.
IMS conditions
IMS was performed using a MALDI TOF/TOF-type instrument (Ultraflex 2 TOF/TOF; Bruker Daltonics) with modified laser focusing system. This instrument was equipped with a 355 nm Nd:YAG laser. The data were acquired in the positive reflectron mode under an accelerating potential of 20 kV using an external calibration method. Signals between m/z 400 and 1000 were collected. Raster scans on tissue surfaces were performed automatically using FlexControl and FlexImaging 2.0 software (Bruker Daltonics). The number of laser irradiations was 100 shots in each spot. Image reconstruction was performed using FlexImaging 2.0 software.
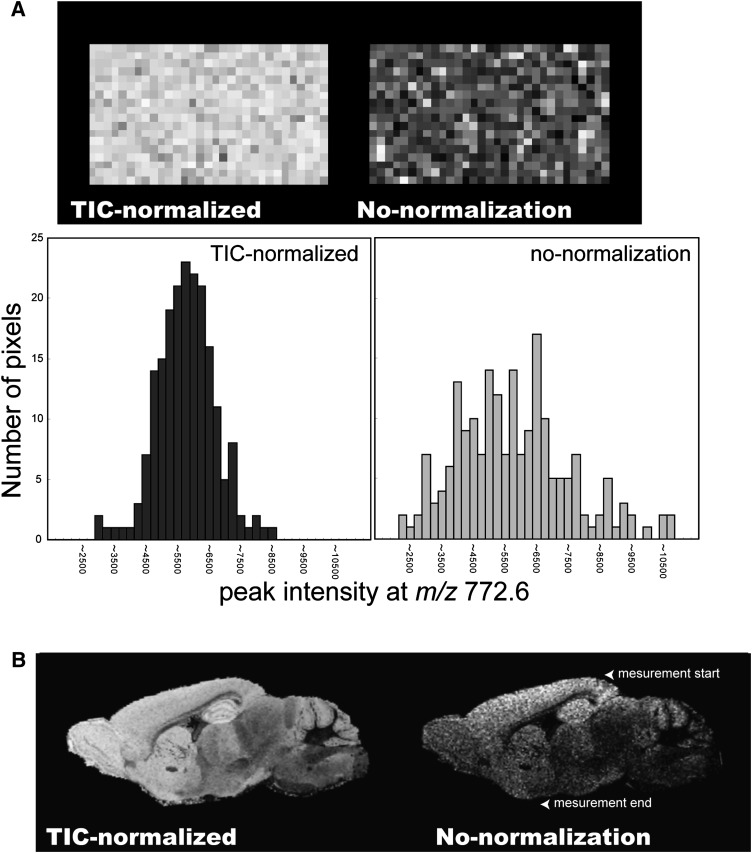
Spectrum normalization with TIC improves quantitation and visualization quality in IMS
The addition of salt to the matrix solution effectively reduces the molecular ion forms of GPLs. However, salts interfere with the matrix-analyte crystallization process and thus, lead to the development of heterogeneous crystals, which in turn results in spot-to-spot variance of signal intensities (45, 46). This problem was solved using a spectrum-normalization procedure with TIC (Fig. 2).
Fig. 2.
Spectrum normalization using TIC improves the visualization quality of IMS. A: IMS results for PC (diacyl-16:0/16:0) on a section of mouse brain homogenate, processed with/without TIC-normalization (upper panel), and plot of ion-intensity distribution for PC (diacyl-16:0/16:0) obtained from a brain-homogenate section, with/without TIC-normalization (lower panel). B: Ion images of PC (diacyl-16:0/16:0) on an adult mouse brain section, in which spectra were processed with/without TIC-normalization.
We performed spectrum normalization with TIC for each mass spectrum by using software that we developed. The obtained spectra were multiplied with arbitrary variables such that all spectra had equal TIC values [i.e., equal integral values of the measured m/z region (400 < m/z < 900)]. Such TIC normalization is also available with the “Normalize Spectra” function of FlexImaging 2.0 software with filter function to exclude a number of noise spectra from the normalization process (see details in the software manual).
To evaluate the effect of the normalization procedure, we prepared a section of mouse brain homogenate that had a uniform distribution of biomolecules. Fig. 2A shows the ion images for m/z 772.6 corresponding to PC (diacyl-16:0/16:0), with and without spectrum normalization. After the normalization procedure, the image was corrected such that the ion distribution was uniform throughout the section. The signal intensity was then plotted and found to have a Gaussian distribution. Spectrum normalization with TIC improved the results of the IMS of mouse brain sections. Fig. 2B shows the ion images of a mouse brain section for PC (diacyl-16:0/16:0), with and without spectrum normalization. In the ion image without normalization, the ion distribution was heterogeneous, even between adjacent pixels. Furthermore, the signal intensity was found to decrease with time (arrowhead). In contrast, when the normalization procedure was used, a clear ion-distribution pattern that correlated well with the anatomical features of the brain section was obtained.
LC/ESI-MS/MS
To identify the PC species, precursor ion scanning was performed using the 4000Q-TRAP instrument; these methods are effective for detecting PCs because characteristic fragment ions are generated by collision-induced dissociation (47) (for details regarding the experimental conditions, see the supplementary information).
Multivariate statistical analysis
An N-by-P data matrix was constructed from the intensity of N pixels and P peaks and then principle component analysis (PCA) was performed using MATLAB™ software (Mathworks, Inc., Sherborn, MA) (for details, see the supplementary information).
RESULTS
Determination of the composition of PC molecular species in the mouse brain
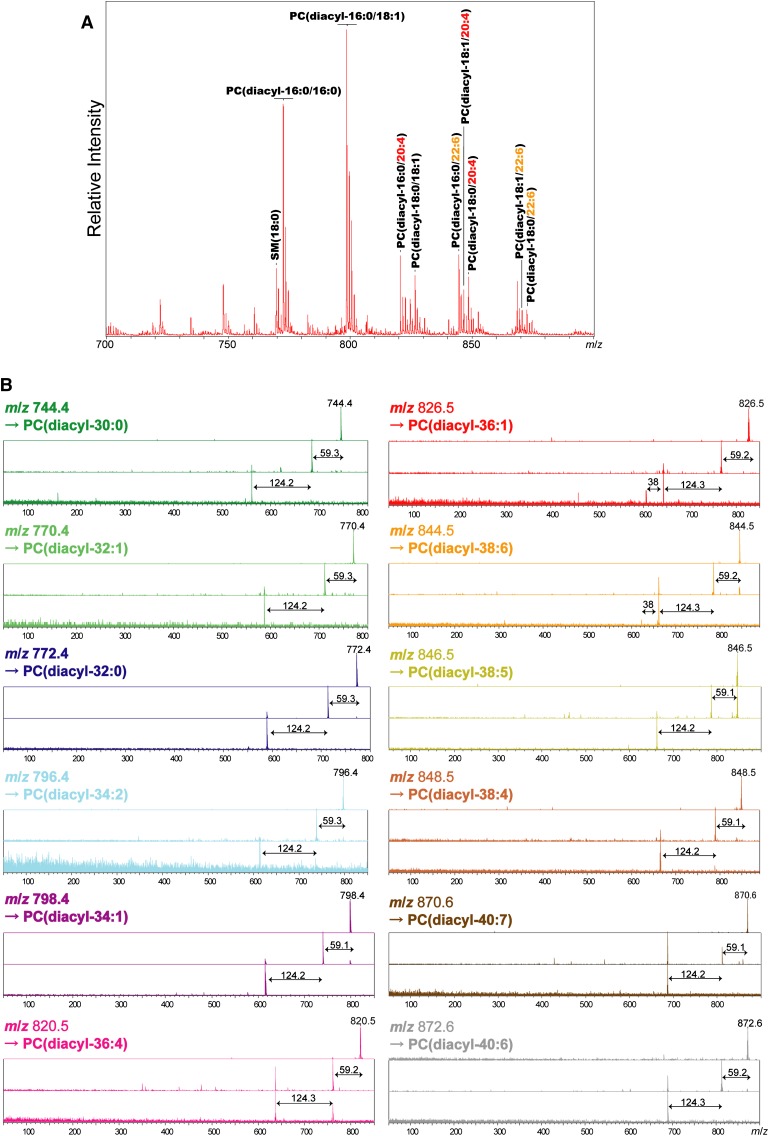
First, IMS of sagittal mouse brain sections was performed using the positive ion detection mode, and approximately 150 mass peaks in the mass range of 700 < m/z < 900 (Fig. 3A) were detected. Among these, nine intense mass peaks were assigned using their masses to abundant PC molecular species and one sphingomyelin, all of which contain a trimethylamine head group. In addition, these assignments were verified with structural analysis of each peak using MSn (Fig. 3B). For this purpose, a QIT-TOF mass spectrometer was used. This instrument can identify molecules using a highly sensitive MSn from mixture ions generated on the tissue surface (48). Table 1 summarizes the results of MS and MSn. In each mass peak, the presence of a trimethylamine head group and phosphate was confirmed (neutral losses of 59 u and 124 u from precursor ions, corresponding to trimethylamine and cyclophosphate, respectively), and these were used as diagnostic ions in product ion mass spectra (48–50) (Fig. 3).
Fig. 3.
Primary PC molecular species simultaneously detected in mouse brain sections by IMS. A: An averaged mass spectrum obtained from an entire mouse brain section. In the spectrum, intense mass peaks corresponding to nine abundant PCs were assigned according to mass. B: Results of MSn structural analysis of ions corresponding to the PCs. Both MS2 and MS3 product ion spectra show that the mass peaks are derived from the PCs. Neutral losses (NL) of 59 u and 124 u from precursor ions, corresponding to trimethylamine and cyclophosphate, respectively, were used as diagnostic ions. Also refer to Table 2.
TABLE 1.
Results of MS and MSn with the intense mass peaks obtained directly from the mouse brain sections
| Observed Mass | Observed Ions in MS2 | Observed Ions in MS3 | Assigned Molecular Species |
|---|---|---|---|
| 772.4 | 713.1 | 588.9 | [PC(diacyl-16:0/16:0)+K]+ |
| 798.4 | 739.3 | 615.1 | [PC(diacyl-16:0/18:1)+K]+ |
| 820.5 | 761.3, 637.0 | 637.0 | [PC(diacyl-16:0/20:4)+K]+ |
| 826.5 | 767.3 | 643.0, 605.0 | [PC(diacyl-18:0/18:1)+K]+ |
| 844.5 | 785.3 | 661.0, 623.0 | [PC(diacyl-16:0/22:6)+K]+ |
| 846.5 | 787.4 | 663.2 | [PC(diacyl-18:1/20:4)+K]+ |
| 848.5 | 789.4 | 665.2 | [PC(diacyl-18:0/20:4)+K]+ |
| 870.6 | 811.5, 687.3 | 687.3 | [PC(diacyl-18:1/22:6)+K]+ |
| 872.6 | 813.4 | 689.1 | [PC(diacyl-18:0/22:6)+K]+ |
In addition, we determined the accurate abundance ratios for PC-molecular species in the mouse brain. For this purpose, LC/ESI-MS/MS was employed, which enabled quantitative analysis of GPLs and identification of the fatty acids linked at the sn-1 and sn-2 positions (51). Table 2 summarizes the quantitative composition of PC molecular species in the total brain lysate.
TABLE 2.
Molecular species of PCs from mouse whole brain lysate identified by UPLC/ESI-MS/MS (n = 3)
| Molecular Species | Relative Abundance | SD |
|---|---|---|
| 16:0/18:1 | 100 | |
| 16:0/16:0 | 58.5 | 4.4 |
| 18:0/18:1 | 48.2 | 2.5 |
| 16:0/22:6 | 35.1 | 3.2 |
| 16:0/20:4 | 29.3 | 3.8 |
| 18:0/22:6 | 19.8 | 1.8 |
| 18:0/20:4 | 14.7 | 0.8 |
| 18:1/20:4 | 10.2 | 1.5 |
| 18:1/22:6 | 8.5 | 0.7 |
Distinct distributions of PC molecular species in the mouse brain
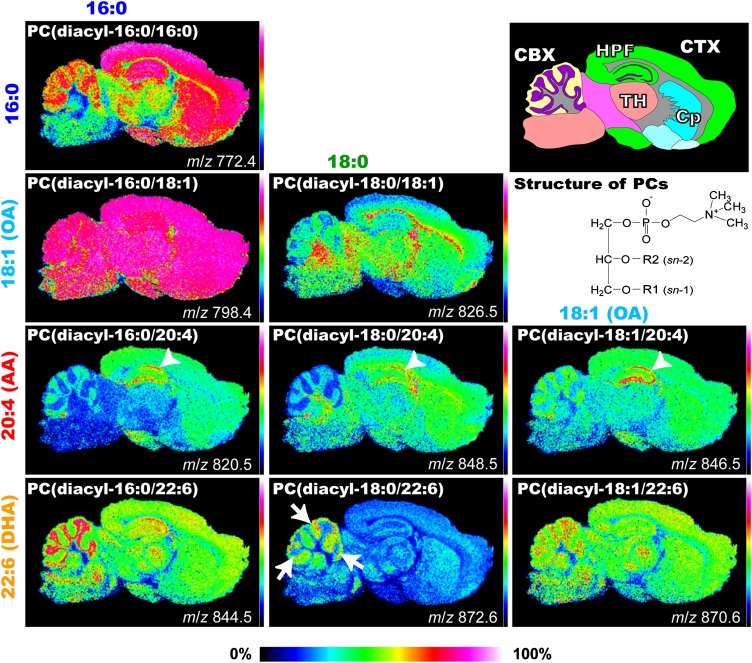
Next, the tissue distributions of the nine major PC molecular species in the entire and various regions of the brain sections were visualized (see also supplementary Figs. III–V). For this purpose, a TOF/TOF mass spectrometer was used that had high sensitivity in single-stage MS. Figure 4 shows the IMS results for the primary brain PCs. Whereas the most abundant molecular PC species, PC (diacyl-16:0/18:1), was uniformly distributed across the entire gray matter region of each section, other PC molecular species showed rather heterogeneous distribution patterns. In particular, the PUFA-PCs, which are both AA-PCs and DHA-PCs, showed characteristic patterns. Three AA-containing species, PC (diacyl-16:0/20:4), PC (diacyl-18:0/20:4), and PC (diacyl-18:1/20:4), were abundant and commonly concentrated in the pyramidal and granular cell layer regions in the hippocampal formation (arrowheads). Two DHA-containing species, PC (diacyl-16:0/22:6) and PC (diacyl-18:1/22:6), were abundant and commonly enriched in the granule cell layer of the cerebellum, whereas PC (diacyl-18:0/22:6) showed a characteristic dotted distribution pattern near the same layer. Among PCs with monounsaturated fatty acids (MUFA), PC (diacyl-16:0/18:1) was enriched in the white matter or in the region containing the myelin sheath.
Fig. 4.
Differential distribution of PC molecular species in sagittal mouse brain sections. MALDI-IMS spectra of a brain section simultaneously identified the heterogeneous distributions of several PCs. Schema of the mouse brain sagittal section and ion images of PCs obtained by IMS are shown. Ion images of PCs are arranged according to their fatty acid (FA) composition. PCs with identical FA compositions at the sn-1 position are arranged lengthwise, whereas those with identical FA compositions at the sn-2 position are arranged sideways. The structures of PCs are also presented. Among the PCs, AA-PCs showed characteristic localization in the hippocampal cell layers (arrowheads). Among DHA-containing species, two abundant species, PC (diacyl-16:0/22:6) and PC (diacyl-18:1/22:6), were commonly enriched in the granule layer of the cerebellum, whereas PC (diacyl-18:0/22:6) showed a characteristic dotted distribution pattern near the cell layer (arrows). CBX, cerebellar cortex; CP, corpus striatum; CTX, cerebral cortex; HPF, hippocampal formation; TH, thalamus.
PUFA-containing PCs were highly detected in specific neural cell types
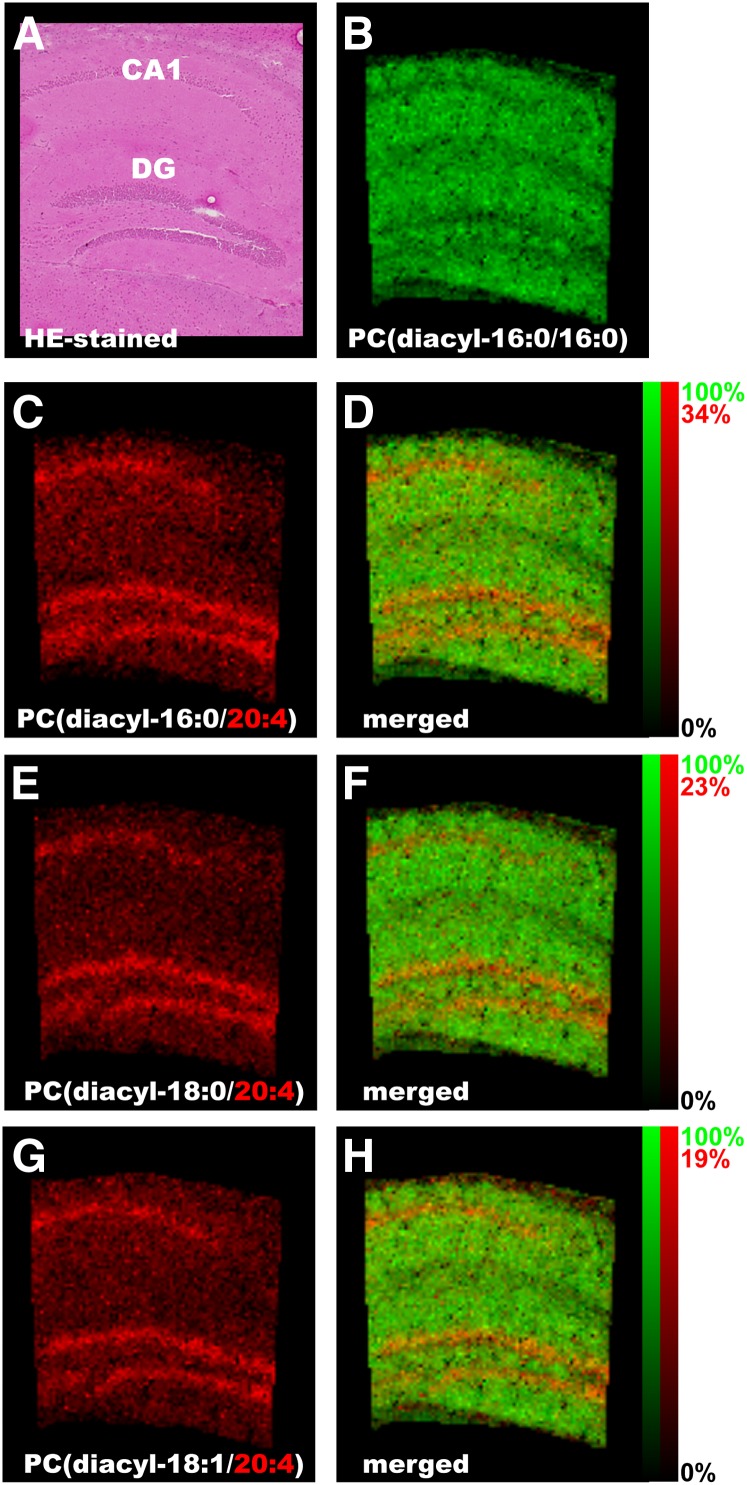
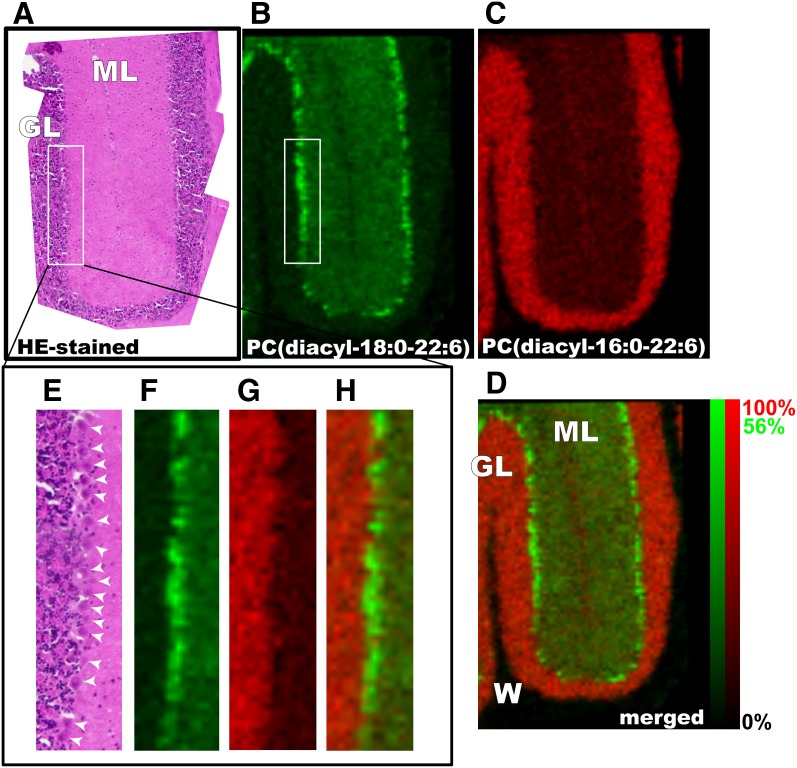
The characteristic localization of PUFA-containing PCs was then determined. To study their distribution in more detail, high-magnification IMS was performed at an instrumental raster step size (raster size) of 15 μm. In the magnified images of the hippocampal formation, all three abundant AA-PCs were found in the regions corresponding to the hippocampal cell layers, i.e., the pyramidal and granule cell layers of the dentate gyrus (Fig. 5). On the other hand, PC (diacyl-16:0/16:0) was enriched outside of the pyramidal and granule cell layers (Fig. 5B). Also, an intensive dotted distribution pattern of PC (diacyl-18:0/22:6) was observed in the cerebellum (Fig. 4). Using high-magnification IMS of the cerebellar cortex, PC (diacyl-18:0/22:6) was found to be enriched in the Purkinje cell layer (Fig. 6). Optical observation of successive hematoxylin-eosin (HE)-stained brain sections also suggested that PC (diacyl-18:0/22:6) was selectively detected in Purkinje cells (Fig. 6, arrowheads) and in molecular layers in which dendrites of Purkinje cells exist. In contrast, granule cells were impoverished in PC (diacyl-18:0/22:6). Interestingly, a complementary distribution of two other abundant DHA-PCs, PC (diacyl-16:0/22:6) and PC (diacyl-18:1/22:6), was observed in the granule cells of the cerebellum (Fig. 6C, D).
Fig. 5.
AA-containing PCs are localized in the cell layers in the hippocampal formation. High-magnification IMS at a raster size of 15 μm (C, E, and G), and successive section stained with HE (A) revealed that AA-containing PCs were enriched in the cell layer regions of the hippocampal formation, which are the pyramidal cell layer of CA1 and the granule cell layer of DG, whereas PC (16:0/16:0) was widely distributed in the dendritic region of the hippocampal formation (B). D, F, and H: merged images. The relative abundance of the 2 ions is indicated in the color scale bar.
Fig. 6.
Purkinje cells selectively contained a DHA-containing PC. High-magnification IMS at a raster size of 15 μm revealed the Purkinje cell-selective distribution of PC (diacyl-18:0/22:6) in the cerebellum. Both optical observation of HE-stained successive brain sections (A and E) and ion images of DHA-PCs (B and F) clearly suggest that the PC was enriched in the Purkinje cell layer (arrowheads). Interestingly, a complementary distribution of another abundant DHA-PC, PC (diacyl-16:0/22:6), was enriched in the granule layer of the cerebellum (C and G). D: merged image. ML, molecular layer; GL, granule layer; W, white matter. The relative abundance of the two ions is indicated in the color scale bar.
Statistical analysis revealed temporal and spatial changes in PCs with age
Having demonstrated the cell-selective distribution of PUFA-PCs in the adult mouse brain, changes in the distribution and amount of PC molecular species at various ages were evaluated. Studies of humans and several other mammalian species have shown that some PUFAs, including AA and DHA, are selectively incorporated and accumulated in GPL bilayers during brain development (52–55), but information on the locations of these PUFAs in the brain has not been available. Knowledge of when and where the changes occur would provide insight into the possible functions of PUFAs at specific brain locations. PC molecular species were comprehensively analyzed and their PUFA-PC distribution patterns and amounts in a developmental series of brain sections were obtained postnatally at 1 day, 12 days, 7 weeks, and 96 weeks.
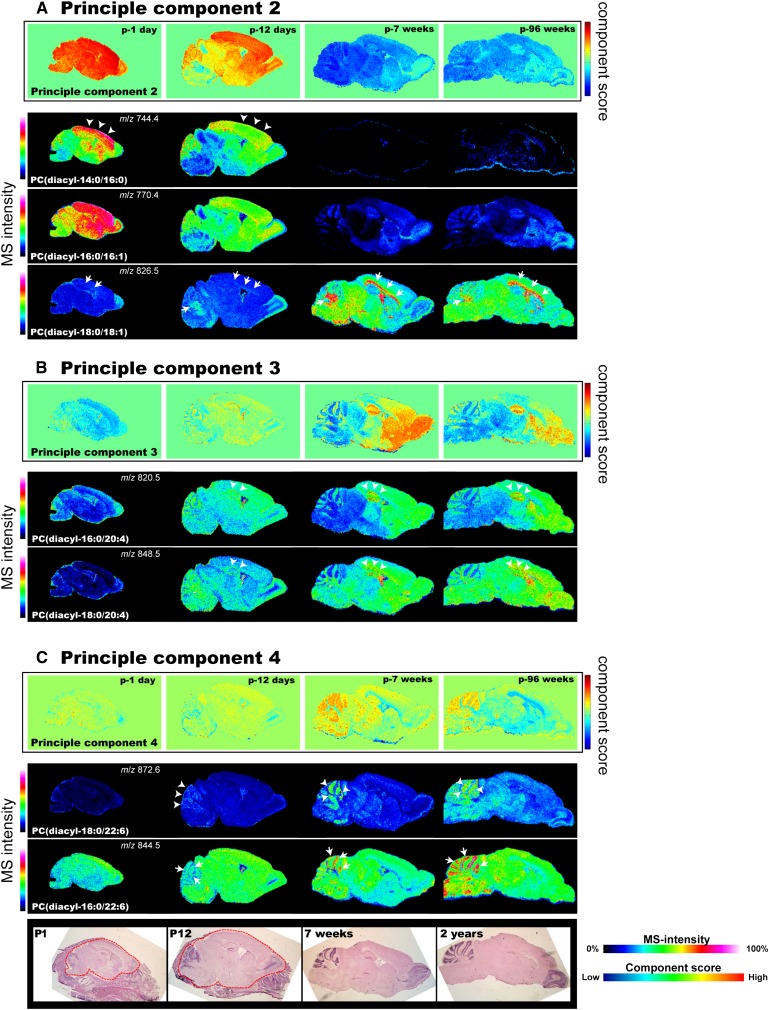
To extract important information from such large IMS data sets (40,000 data points for the four developing brain sections), PCA, an unsupervised multivariate data analysis technique (37, 38, 56), was used. PCA is useful for obtaining an overview of entire data sets (i.e., general trends in fluctuations in GPLs' expression with development) before proceeding to a more detailed data analysis (i.e., visualization of individual GPLs). For a detailed description of the procedure and results of PCA, see the supplementary information. Table 3 summarizes the results and interpretation of the PCA. To locate the brain regions or developmental time points with spectral fluctuations, the principal components were scored by color at each data point (Fig. 7, top line of each panel). It was assumed that components 2, 3, and 4 contained important information because of their heterogeneous distributions of component scores among sections. Eventually, it was determined that they represented three significant trends in PC expression changes.
TABLE 3.
Results and interpretation of PCA of the developing mouse brain
| Primary Contributing PCs |
Interpretation of the Component | |||
|---|---|---|---|---|
| Component 2 | PC(diacyl-14:0/16:0) | PC(diacyl-16:0/16:1) | PC(diacyl-18:0/18:1) | Reduction in PCs with short-chain fatty acids and accumulation of myelin-constituted PCs |
| Component 3 | PC(diacyl-16:0/20:4) | PC(diacyl-18:0/20:4) | PC(diacyl-16:0/18:1) | Accumulation of two abundant AA-PCs in the hippocampus, cerebral cortex, corpus striatum, and granule cell layer of the cerebellum |
| Component 4 | PC(diacyl-18:0/22:6) | PC(diacyl-16:0/22:6) | PC(diacyl-18:1/22:6) | Accumulation of DHA-PCs in the cerebellum |
Fig. 7.
IMS-PCA analysis of the developmental mouse brains revealed dynamic changes in PC expression. IMS was performed on a developmental series of four mouse brain sections, obtained postnatally on days 1 and 12 and in weeks 7 and 96, and then PCA was performed to extract general trends in PC expression fluctuation during development. Three trends emerged. A: PC2 represents a reduction in PCs with short-chain fatty acids and an accumulation of myelin-constituted PCs with development. B: PC3 represents an accumulation of AA-PCs in the hippocampal formation, cerebral cortex, corpus striatum, and granule cell layer of the cerebellum. C: PC4 represents an accumulation of DHA-PCs in the cell layers of the cerebellum. In each panel, the upper row illustrates the principal component score for each pixel, and the lower rows show the results of individual visualizations of PCs that have large loading factors for each principal component. The brain sections were HE-stained and optical images of the sections are shown.
Characteristic saturated fatty acid/MUFA-PC expression patterns during brain development
As the first trend, component 2 represents the reduction of PCs with C14-16 fatty acids (myristic and palmitoyl acid) and the accumulation of myelin-constituting PCs with development. The inset of Fig. 7A shows that the component 2 scores for spectra in the infant brains (P1 and P12; P, postnatal day) were higher than those in the adult brains (7 and 96 weeks old). As Table 3 shows, PCs with C14-16 fatty acids, namely PC (diacyl-14:0/16:0) and PC (diacyl-16:0/16:1), along with a myelin-constituting PC, namely PC (diacyl-18:0/18:1), were suggested to be associated with the statistical difference in the component score. As expected, the visualization of each molecular species showed drastic reductions in PC (diacyl-14:0/16:0) and PC (diacyl-16:0/16:1), especially in the cerebral cortex, and their levels in the adult brains were quite low (arrowheads). On the other hand, PC (diacyl-18:0/18:1) accumulated in the white matter with myelin formation, especially in the callous corpus and in the medulla and white matter of the cerebellum (arrows).
Characteristic PUFA-PC distribution patterns during brain development
Second, component 3 represents the accumulation of AA-PCs during the formation of the hippocampus, cerebral cortex, corpus striatum, and granule cell layer of the cerebellum. As with the component 2 analysis, the focus was on the increase in component 3 score with development in the above-mentioned regions (Fig. 7B inset). In this principal component, AA-PCs, namely PC (diacyl-16:0/20:4) and PC (diacyl-18:0/20:4), largely contributed to the statistical changes (Table 3). Visualization confirmed that the expression levels of the AA-PCs were certainly increased, especially in the hippocampal cell layers and corpus striatum. As the arrowheads indicate, a faint localization observed at P1 had progressed in P12 and was clearly identified in the adult brains.
Finally, component 4 represents the accumulation of DHA-PCs in the cell layers of the cerebellum. Component 4 scores clearly trended higher in the cerebellar cortex, especially in the molecular and granule cell layers. In this case, DHA-PCs, namely, PC (diacyl-16:0/22:6) and PC (diacyl-18:0/22:6), were expected to be associated with the statistical difference in the component score (Table 3). In particular, PC (diacyl-18:0/22:6), which was contained in the soma and dendrites of Purkinje cells, progressively became enriched and formed a dotted distribution pattern in the cerebellum, whereas at P1 it was distributed at the surface of the cerebellar cortex (arrows) where immature Purkinje cells can be found. Also, PC (diacyl-18:0/22:6) progressively accumulated in the granule cell layer of the cerebellum (arrowheads). In addition to PCA, other multivariate analyses were performed, namely ICA (supplementary Fig. VI and Table 4). Although PCA extracted the information most efficiently, ICA also identified other trends in expression changes summarized in Table 4. As a consequence, both multivariate analyses characterized the accumulation of PUFA-PCs in specific regions from total expression changes.
TABLE 4.
Results and interpretation of ICA of the developing mouse brain
| Primary Contributing PCs |
Interpretation of the Component | |||
|---|---|---|---|---|
| Component 1 | PC(diacyl-16:0/18:1) | PC(diacyl-16:0/16:0) | PC(diacyl-16:0/18:0) | Accumulation of PCs with SFA and MUFA |
| Component 2 | PC(diacyl-18:0/18:1) | PC(diacyl-14:0/16:0) | PC(diacyl-16:0/16:1) | Reduction in PCs with short-chain fatty acids and accumulation of myelin-constituted PCs |
| Component 3 | PC(diacyl-16:0/20:4) | PC(diacyl-18:0/20:4) | PC(diacyl-16:0/16:1) | Accumulation of two abundant AA-PCs in the hippocampus, cerebral cortex, corpus striatum, and granule cell layer of the cerebellum |
DISCUSSION
IMS is an effective tool for lipid imaging
In this report, a practical method using IMS was introduced for visualizing GPL distribution in mouse brains, and the capability of IMS was demonstrated as a potential standard technology for lipid imaging. Applying the technique, the distribution patterns of GPL molecular species in various regions of the mouse brain were comprehensively determined and an atlas of distribution maps was generated.
IMS is effective for detecting (21, 22, 57) and visualizing GPLs (20, 58–60). A large amount of GPLs in the brain (more than 50% dry weight) and an easily charged structure (e.g., phosphate ester) yield high-ionization efficiency. In particular, in the positive-ion detection mode, molecular species of choline-containing lipids (e.g., PC, lyso-PC, and sphingomyelin) are highly ionized due to their trimethylamine head group (61). Despite the promising capability of IMS for GPL imaging, improvement in its quantitative ability and reproducibility are necessary due to the nature of MALDI-direct MS. For example, excess endogenous salts and their tissue localizations perturb the ion images of specific molecules. The present study demonstrated that adding an alkali-metal salt to the matrix solution is an effective way to minimize molecular ion adducts (Fig. 1). Also, complex analyte-extraction and crystallization processes on the tissue surface generate spot-to-spot variances in analyte ionization efficiency. In addition, an IMS experiment with a large number of data points takes a long time (<8 h), and therefore, the sublimation of matrix crystals occurs (especially in a high vacuum chamber). This results in a decrease in signal intensity, as shown in Fig. 2C. The study results demonstrated that spectrum normalization using TIC is effective in correcting heterogeneous ionization efficiency (Fig. 2). In a protein study, Norris et al. (62) previously reported that spectrum processing, including a normalization process, is effective in improving IMS-data. The present results demonstrated that, in lipid study, the normalization process eliminated spot-to-spot variances in signal intensity on the brain homogenate section (Fig. 2A), and improved both qualitative and the semi-quantitative ability (Fig. 2B) even among different sections. For the absolute quantification of micro-region GPLs, further studies, such as those using microdissection technology and LC/ES-MS/MS, are required.
Visualization of GPL distributions revealed characteristic localizations of PUFA-PCs
A properly performed IMS experiment can generate ion distribution maps that are informative for exploring GPLs in a tissue section. By employing this technique, a number of PC molecular species were visualized simultaneously in a mouse brain section and the distribution patterns of the species were found to be quite heterogeneous (Fig. 4). The most impressive finding is that some PUFA-PCs were selectively obtained by specific cells (Figs. 3 and 4).
Among the classes of GPLs, PCs are the most abundant structural component of neural and glial cell membranes, and the fatty acid constituents of PCs (i.e., molecular species) influence the membranes' physical properties, including fluidity and curvature (7, 30, 63, 64).
Thus, the cell-type heterogeneity of the fatty-acid constituent in part reflects the cells' heterogeneous membrane properties. Figure 6 shows that PC (diacyl-18:0/22:6) was contained in Purkinje cells in the cerebellum. Both ion and optical images indicate that this DHA-containing PC molecular species was highly contained not only in the soma but also in the dendrites of Purkinje cells. Because of their high degree of unsaturation, DHA-GPLs increase membrane fluidity and even regulate the functions of membrane-associated proteins (65–67). Purkinje cells are the largest neurons in the brain, with intricately elaborate dendritic arbors. Thus, higher membrane fluidity may be required for effective transport of membrane-associated proteins via the plasma membrane. Therefore, the high-level expression of DHA-PC may contribute to the transportation of membrane proteins in the cells.
Also, it is well known that GPLs, including PCs, serve as reservoirs of PUFAs, which are released and converted into bioactive lipids in response to extracellular stimuli, and which mediate various important biological processes (9). Thus, the cell-type specificity of PUFA storage constituents is worth describing. Figure 5 shows that AA-containing PCs are enriched in the cell layers of pyramidal neurons of CA1 and of granule neurons of the dentate gyrus. The prominent detection of AA-PCs in such cells suggests the storage of large amounts of AA. Judging from their localization pattern, they are presumably enriched in the nuclear membranes of the cells. In fact, AA is known to be selectively incorporated and enriched in the PCs of the nuclear membrane (68). From nuclear PCs, AA is released into the cytoplasm by PLA2 activity in response to extracellular stimuli, facilitating the initiation of the AA cascade (9). Thus, a high concentration of AA-PCs in the hippocampal cell layer indicates the necessity of producing large amounts of eicosanoids for neural activity in the hippocampal formation.
Visualization of spatio-temporal changes of GPL composition by IMS
Neural cell membranes are dynamic entities whose lipid constituents are continuously adjusted (55). The fatty acid composition of tissues is known to change throughout development and the changes differ among the classes of GPLs (52–55). The novelty of the present study is that, by visualizing these changes, functional insight was obtained into these biochemical processes. For example, a previously reported developmental increase in oleic acid (18:1) was revealed by IMS to result from the accumulation of PC (diacyl-18:0/18:1) in the myelin-containing region during maturation (Fig. 7A). Furthermore, even alterations in microdomains could be identified, which are difficult to separate using conventional biochemical approaches. Although previous studies have shown that increases in the levels of AA and DHA are relatively small in the total brain lysate, the present study found that enrichment occurred in a quite region-specific manner. AA and DHA were observed to accumulate primarily in the cell layers of the hippocampal formation and cerebellum, respectively (Fig. 7B), presumably in the nuclear membrane, during functional maturation. Such dynamic changes in GPL composition may be attributed to the temporal and heterogeneous expression of the lysophospholipid acyltransferase family, which have been recently characterized (69). Further study, focused on fluctuations in expression of these enzymes with time and location, coupled with the presented distribution map, is needed to understand how GPL diversity is established with development.
This article focused on PCs because of the space limitations; however, analyses of other GPLs and glycosphingolipids, including phosphatidylethanolamine, phosphatidylinositol, phosphatidylserine, and sulfatides, are also available, as shown in supplementary Figs. VII and VIII. As a result of the technical improvement described in this article, an atlas could be provided of these GPLs over time and space. For PCs, distribution maps were generated that cover multiple molecular species at various ages and brain regions. This data set indicates the necessity of region-specific studies in brain lipidomics and provides important information for performing such analyses.
Supplementary Material
Acknowledgments
We thank members of the Mitsubishi Kagaku Institute of Life Sciences (MITILS), particularly Dr. Maki K. Yamada, for valuable discussions.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- DHA
- docosahexaenoic acid
- DHB
- dihydroxybenzoic acid
- GPL
- glycerol phospholipid
- IMS
- imaging mass spectrometry
- ITO
- indium-tin-oxide
- PC
- phosphatidylcholine
- PCA
- principle component analysis
- PLA2
- phospholipase A2
- QIT
- quadrupole ion trap
- SIMS
- secondary IMS
- TIC
- total ion current
- TOF
- time-of-flight
This work was supported by the SENTAN program of the Japan Science and Technology Agency and a WAKATE-S grant from the Japan Society for the Promotion of Science to M.S.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of eight tables.
REFERENCES
- 1.Takamori S., Holt M., Stenius K., Lemke E. A., Gronborg M., Riedel D., Urlaub H., Schenck S., Brugger B., Ringler P., et al. 2006. Molecular anatomy of a trafficking organelle. Cell. 127: 831–846. [DOI] [PubMed] [Google Scholar]
- 2.Rohrbough J., Broadie K. 2005. Lipid regulation of the synaptic vesicle cycle. Nat. Rev. Neurosci. 6: 139–150. [DOI] [PubMed] [Google Scholar]
- 3.Kim D. 2003. Fatty acid-sensitive two-pore domain K+ channels. Trends Pharmacol. Sci. 24: 648–654. [DOI] [PubMed] [Google Scholar]
- 4.Miller B., Sarantis M., Traynelis S. F., Attwell D. 1992. Potentiation of NMDA receptor currents by arachidonic acid. Nature. 355: 722–725. [DOI] [PubMed] [Google Scholar]
- 5.Jacobson K., Mouritsen O. G., Anderson R. G. 2007. Lipid rafts: at a crossroad between cell biology and physics. Nat. Cell Biol. 9: 7–14. [DOI] [PubMed] [Google Scholar]
- 6.Allen J. A., Halverson-Tamboli R. A., Rasenick M. M. 2007. Lipid raft microdomains and neurotransmitter signalling. Nat. Rev. Neurosci. 8: 128–140. [DOI] [PubMed] [Google Scholar]
- 7.Piomelli D., Astarita G., Rapaka R. 2007. A neuroscientist's guide to lipidomics. Nat. Rev. Neurosci. 8: 743–754. [DOI] [PubMed] [Google Scholar]
- 8.Yamashita A., Sugiura T., Waku K. 1997. Acyltransferases and transacylases involved in fatty acid remodeling of phospholipids and metabolism of bioactive lipids in mammalian cells. J. Biochem. 122: 1–16. [DOI] [PubMed] [Google Scholar]
- 9.Murakami M., Nakatani Y., Atsumi G., Inoue K., Kudo I. 1997. Regulatory functions of phospholipase A2. Crit. Rev. Immunol. 17: 225–283. [DOI] [PubMed] [Google Scholar]
- 10.Williams J. H., Errington M. L., Lynch M. A., Bliss T. V. 1989. Arachidonic acid induces a long-term activity-dependent enhancement of synaptic transmission in the hippocampus. Nature. 341: 739–742. [DOI] [PubMed] [Google Scholar]
- 11.Dinh T. P., Carpenter D., Leslie F. M., Freund T. F., Katona I., Sensi S. L., Kathuria S., Piomelli D. 2002. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc. Natl. Acad. Sci. USA. 99: 10819–10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han X., Gross R. W. 2003. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 44: 1071–1079. [DOI] [PubMed] [Google Scholar]
- 13.Yoshikawa K., Kita Y., Kishimoto K., Shimizu T. 2006. Profiling of eicosanoid production in the rat hippocampus during kainic acid-induced seizure: dual phase regulation and differential involvement of COX-1 and COX-2. J. Biol. Chem. 281: 14663–14669. [DOI] [PubMed] [Google Scholar]
- 14.Guan X. L., He X., Ong W. Y., Yeo W. K., Shui G., Wenk M. R. 2006. Non-targeted profiling of lipids during kainate-induced neuronal injury. FASEB J. 20: 1152–1161. [DOI] [PubMed] [Google Scholar]
- 15.Stoeckli M., Chaurand P., Hallahan D. E., Caprioli R. M. 2001. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat. Med. 7: 493–496. [DOI] [PubMed] [Google Scholar]
- 16.Todd P. J., Schaaff T. G., Chaurand P., Caprioli R. M. 2001. Organic ion imaging of biological tissue with secondary ion mass spectrometry and matrix-assisted laser desorption/ionization. J. Mass Spectrom. 36: 355–369. [DOI] [PubMed] [Google Scholar]
- 17.Shimma S., Setou M. 2007. Mass microscopy to reveal distinct localization of heme B (m/z 616) in colon cancer liver metastasis. J. Mass Spectrom. Soc. Jpn. 55: 145–148. [Google Scholar]
- 18.Burrell M., Earnshaw C., Clench M. 2007. Imaging matrix assisted laser desorption ionization mass spectrometry: a technique to map plant metabolites within tissues at high spatial resolution. J. Exp. Bot. 58: 757–763. [DOI] [PubMed] [Google Scholar]
- 19.Sugiura Y., Shimma S., Konishi Y., Yamada M. K., Setou M. 2008. Imaging mass spectrometry technology and application on ganglioside study; visualization of age-dependent accumulation of C20-ganglioside molecular species in the mouse hippocampus. PLoS One. 3: e3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrett T. J., Prieto-Conaway M. C., Kovtoun V., Bui H., Izgarian N., Stafford G., Yost R. A. 2006. Imaging of small molecules in tissue sections with a new intermediate-pressure MALDI linear ion trap mass spectrometer. Int. J. Mass Spectrom. 260: 166–176. [Google Scholar]
- 21.Rujoi M., Estrada R., Yappert M. C. 2004. In situ MALDI-TOF MS regional analysis of neutral phospholipids in lens tissue. Anal. Chem. 76: 1657–1663. [DOI] [PubMed] [Google Scholar]
- 22.Jackson S. N., Wang H. Y., Woods A. S. 2005. In situ structural characterization of phosphatidylcholines in brain tissue using MALDI-MS/MS. J. Am. Soc. Mass Spectrom. 16: 2052–2056. [DOI] [PubMed] [Google Scholar]
- 23.Jackson S. N., Wang H. Y., Woods A. S. 2005. Direct profiling of lipid distribution in brain tissue using MALDI-TOFMS. Anal. Chem. 77: 4523–4527. [DOI] [PubMed] [Google Scholar]
- 24.Puolitaival S. M., Burnum K. E., Cornett D. S., Caprioli R. M. 2008. Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J. Am. Soc. Mass Spectrom. 19: 882–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimma S., Sugiura Y., Hayasaka T., Hoshikawa Y., Noda T., Setou M. 2007. MALDI-based imaging mass spectrometry revealed abnormal distribution of phospholipids in colon cancer liver metastasis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 855: 98–103. [DOI] [PubMed] [Google Scholar]
- 26.Cornett D. S., Frappier S. L., Caprioli R. M. 2008. MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Anal. Chem. 80: 5648–5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khatib-Shahidi S., Andersson M., Herman J. L., Gillespie T. A., Caprioli R. M. 2006. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal. Chem. 78: 6448–6456. [DOI] [PubMed] [Google Scholar]
- 28.Stoeckli M., Staab D., Schweitzer A. 2006. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. Int. J. Mass Spectrom. 260: 195–202. [Google Scholar]
- 29.Colliver T. L., Brummel C. L., Pacholski M. L., Swanek F. D., Ewing A. G., Winograd N. 1997. Atomic and molecular imaging at the single-cell level with TOF-SIMS. Anal. Chem. 69: 2225–2231. [DOI] [PubMed] [Google Scholar]
- 30.Ostrowski S. G., Van Bell C. T., Winograd N., Ewing A. G. 2004. Mass spectrometric imaging of highly curved membranes during Tetrahymena mating. Science. 305: 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monroe E. B., Jurchen J. C., Lee J., Rubakhin S. S., Sweedler J. V. 2005. Vitamin E imaging and localization in the neuronal membrane. J. Am. Chem. Soc. 127: 12152–12153. [DOI] [PubMed] [Google Scholar]
- 32.Touboul D., Brunelle A., Halgand F., De La Porte S., Laprevote O. 2005. Lipid imaging by gold cluster time-of-flight secondary ion mass spectrometry: application to Duchenne muscular dystrophy. J. Lipid Res. 46: 1388–1395. [DOI] [PubMed] [Google Scholar]
- 33.Ikegami K., Heier R. L., Taruishi M., Takagi H., Mukai M., Shimma S., Taira S., Hatanaka K., Morone N., Yao I., et al. 2007. Loss of alpha-tubulin polyglutamylation in ROSA22 mice is associated with abnormal targeting of KIF1A and modulated synaptic function. Proc. Natl. Acad. Sci. USA. 104: 3213–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sweedler J. V., Li L., Rubakhin S. S., Alexeeva V., Dembrow N. C., Dowling O., Jing J., Weiss K. R., Vilim F. S. 2002. Identification and characterization of the feeding circuit-activating peptides, a novel neuropeptide family of aplysia. J. Neurosci. 22: 7797–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hosokawa N., Sugiura Y., Setou M. 2008. Spectrum normalization method using an external standard in mass spectrometric imaging. J. Mass Spectrom. Soc. Jpn. 56: 77–81. [Google Scholar]
- 36.Yanagisawa K., Shyr Y., Xu B. J., Massion P. P., Larsen P. H., White B. C., Roberts J. R., Edgerton M., Gonzalez A., Nadaf S., et al. 2003. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet. 362: 433–439. [DOI] [PubMed] [Google Scholar]
- 37.Yao I., Sugiura Y., Matsumoto M., Setou M. 2008. In situ proteomics with imaging mass spectrometry and principal component analyses in the Scrapper-knockout mouse brain. Proteomics. 8: 3692–3701. [DOI] [PubMed] [Google Scholar]
- 38.Altelaar A. F., Luxembourg S. L., McDonnell L. A., Piersma S. R., Heeren R. M. 2007. Imaging mass spectrometry at cellular length scales. Nat. Protoc. 2: 1185–1196. [DOI] [PubMed] [Google Scholar]
- 39.Umemura A., Mabe H., Nagai H., Sugino F. 1992. Action of phospholipases A2 and C on free fatty acid release during complete ischemia in rat neocortex. Effect of phospholipase C inhibitor and N-methyl-D-aspartate antagonist. J. Neurosurg. 76: 648–651. [DOI] [PubMed] [Google Scholar]
- 40.Rehncrona S., Westerberg E., Akesson B., Siesjo B. K. 1982. Brain cortical fatty acids and phospholipids during and following complete and severe incomplete ischemia. J. Neurochem. 38: 84–93. [DOI] [PubMed] [Google Scholar]
- 41.Sugiura Y., Shimma S., Setou M. 2006. Thin sectioning improves the peak intensity and signal-to-noise ratio in direct tissue mass spectrometry. J. Mass Spectrom. Soc. Jpn. 54: 45–48. [Google Scholar]
- 42.Schwartz S. A., Reyzer M. L., Caprioli R. M. 2003. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass Spectrom. 38: 699–708. [DOI] [PubMed] [Google Scholar]
- 43.Han X., Gross R. W. 2001. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 295: 88–100. [DOI] [PubMed] [Google Scholar]
- 44.Hsu F. F., Turk J. 2001. Structural determination of glycosphingolipids as lithiated adducts by electrospray ionization mass spectrometry using low-energy collisional-activated dissociation on a triple stage quadrupole instrument. J. Am. Soc. Mass Spectrom. 12: 61–79. [DOI] [PubMed] [Google Scholar]
- 45.Sugiura Y., Shimma S., Setou M. 2006. Two-step matrix application technique to improve ionization efficiency for matrix-assisted laser desorption/ionization in imaging mass spectrometry. Anal. Chem. 78: 8227–8235. [DOI] [PubMed] [Google Scholar]
- 46.Mock K. K., Sutton C. W., Cottrell J. S. 1992. Sample immobilization protocols for matrix-assisted laser-desorption mass spectrometry. Rapid Commun. Mass Spectrom. 6: 233–238. [DOI] [PubMed] [Google Scholar]
- 47.Taguchi R., Houjou T., Nakanishi H., Yamazaki T., Ishida M., Imagawa M., Shimizu T. 2005. Focused lipidomics by tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 823: 26–36. [DOI] [PubMed] [Google Scholar]
- 48.Shimma S., Sugiura Y., Hayasaka T., Zaima N., Matsumoto M., Setou M. 2008. Mass imaging and identification of biomolecules with MALDI-QIT-TOF-based system. Anal. Chem. 80: 878–885. [DOI] [PubMed] [Google Scholar]
- 49.Stubiger G., Pittenauer E., Allmaier G. 2008. MALDI seamless postsource decay fragment ion analysis of sodiated and lithiated phospholipids. Anal. Chem. 80: 1664–1678. [DOI] [PubMed] [Google Scholar]
- 50.Touboul D., Piednoel H., Voisin V., De La Porte S., Brunelle A., Halgand F., Laprevote O. 2004. Changes of phospholipid composition within the dystrophic muscle by matrix-assisted laser desorption/ionization mass spectrometry and mass spectrometry imaging. Eur. J. Mass Spectrom. (Chichester, Eng.). 10: 657–664. [DOI] [PubMed] [Google Scholar]
- 51.Houjou T., Yamatani K., Imagawa M., Shimizu T., Taguchi R. 2005. A shotgun tandem mass spectrometric analysis of phospholipids with normal-phase and/or reverse-phase liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 19: 654–666. [DOI] [PubMed] [Google Scholar]
- 52.Sinclair A. J., Crawford M. A. 1972. The accumulation of arachidonate and docosahexaenoate in the developing rat brain. J. Neurochem. 19: 1753–1758. [DOI] [PubMed] [Google Scholar]
- 53.Svennerholm L. 1968. Distribution and fatty acid composition of phosphoglycerides in normal human brain. J. Lipid Res. 9: 570–579. [PubMed] [Google Scholar]
- 54.Svennerholm L., Vanier M. T. 1973. The distribution of lipids in the human nervous system. 3. Fatty acid composition of phosphoglycerides of human foetal and infant brain. Brain Res. 50: 341–351. [DOI] [PubMed] [Google Scholar]
- 55.Martinez M., Mougan I. 1998. Fatty acid composition of human brain phospholipids during normal development. J. Neurochem. 71: 2528–2533. [DOI] [PubMed] [Google Scholar]
- 56.McCombie G., Staab D., Stoeckli M., Knochenmuss R. 2005. Spatial and spectral correlations in MALDI mass spectrometry images by clustering and multivariate analysis. Anal. Chem. 77: 6118–6124. [DOI] [PubMed] [Google Scholar]
- 57.Jones J. J., Borgmann S., Wilkins C. L., O'Brien R. M. 2006. Characterizing the phospholipid profiles in mammalian tissues by MALDI FTMS. Anal. Chem. 78: 3062–3071. [DOI] [PubMed] [Google Scholar]
- 58.McLean J. A., Ridenour W. B., Caprioli R. M. 2007. Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J. Mass Spectrom. 42: 1099–1105. [DOI] [PubMed] [Google Scholar]
- 59.Jackson S. N., Ugarov M., Egan T., Post J. D., Langlais D., Albert Schultz J., Woods A. S. 2007. MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J. Mass Spectrom. 42: 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayasaka T., Goto-Inoue N., Sugiura Y., Zaima N., Nakanishi H., Ohishi K., Nakanishi S., Naito T., Taguchi R., Setou M. 2008. Matrix-assisted laser desorption/ionization quadrupole ion trap time-of-flight (MALDI-QIT-TOF)-based imaging mass spectrometry reveals a layered distribution of phospholipid molecular species in the mouse retina. Rapid Commun. Mass Spectrom. 22: 3415–3426. [DOI] [PubMed] [Google Scholar]
- 61.Pulfer M., Murphy R. C. 2003. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 22: 332–364. [DOI] [PubMed] [Google Scholar]
- 62.Norris J. L., Cornett D. S., Mobley J. A., Andersson M., Seeley E. H., Chaurand P., Caprioli R. M. 2007. Processing MALDI mass spectra to improve mass spectral direct tissue analysis. Int. J. Mass Spectrom. 260: 212–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hitzemann R. J., Johnson D. A. 1983. Developmental changes in synaptic membrane lipid composition and fluidity. Neurochem. Res. 8: 121–131. [DOI] [PubMed] [Google Scholar]
- 64.Zerouga M., Jenski L. J., Stillwell W. 1995. Comparison of phosphatidylcholines containing one or two docosahexaenoic acyl chains on properties of phospholipid monolayers and bilayers. Biochim. Biophys. Acta. 1236: 266–272. [DOI] [PubMed] [Google Scholar]
- 65.Salem N., Jr., Litman B., Kim H. Y., Gawrisch K. 2001. Mechanisms of action of docosahexaenoic acid in the nervous system. Lipids. 36: 945–959. [DOI] [PubMed] [Google Scholar]
- 66.Kim H. Y. 2007. Novel metabolism of docosahexaenoic acid in neural cells. J. Biol. Chem. 282: 18661–18665. [DOI] [PubMed] [Google Scholar]
- 67.Stubbs C. D., Smith A. D. 1984. The modification of mammalian membrane polyunsaturated fatty acid composition in relation to membrane fluidity and function. Biochim. Biophys. Acta. 779: 89–137. [DOI] [PubMed] [Google Scholar]
- 68.Surette M. E., Chilton F. H. 1998. The distribution and metabolism of arachidonate-containing phospholipids in cellular nuclei. Biochem. J. 330: 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hishikawa D., Shindou H., Kobayashi S., Nakanishi H., Taguchi R., Shimizu T. 2008. Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity. Proc. Natl. Acad. Sci. USA. 105: 2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.