Abstract
The enzyme α-isopropylmalate synthase from Mycobacterium tuberculosis (MtIPMS) has been identified as a possible target for design of new anti-tubercular therapeutics. Recently it was shown that MtIPMS is subject to slow-onset, feedback inhibition by L-leucine, the first instance of an allosteric regulator utilizing this mechanism. Structural studies are inconsistent with allosteric mechanisms including changes to the quarternary structure or large, rigid-body conformational changes to the enzyme upon L-leucine binding. Thus, the allosteric regulation may result from a discrete inhibitory signal transmitted to the active site upon L-leucine binding in the regulatory domain, a distance of over 50 Å In order to test this mechanism, site-directed mutagenesis was employed to construct enzymes with substitutions at phylogenetically conserved active site residues near the interface of the catalytic and linker domains. The substitutions had wide ranging effects on the kinetics of L-leucine inhibition, with some modest effects on the kinetic parameters of catalysis. The most dramatic result was the finding that the Y410F mutant form of MtIPMS is insensitive to L-leucine inhibition, suggesting that this residue has completely uncoupled the inhibitory signal to the active site. Overall, the data are consistent with a mechanism of allosteric regulation described by the inter-domain communication of the inhibitory signal from the regulatory to catalytic domain and implicate the interaction between the linker and catalytic domains as critical determinants of inhibitory signal transmission.
Keywords: allosteric regulation, α-isopropylmalate synthase, Mycobacterium tuberculosis, slow-onset inhibition, feedback inhibition, L-leucine, substrate activation
Understanding the mechanism of allosteric regulation in proteins, where an effector molecule exerts influence at a distal site, has been a central pursuit of biochemistry for over forty years (1). Cases where the binding of an effector molecule causes a change in the quarternary structure of a protein or alters the equilibrium between and active and inactive conformers through rigid body motion have been well documented due to their distinct structural and physical properties (2, 3). However, there a numerous reports of proteins that are allosterically regulated which are not consistent with either of these two models (4–7). In these cases, the mechanism of allostery is loosely described by invoking changes in local protein dynamics or equlibria transmitted through a type of contact network between the binding site of the effector molecule and the active site.
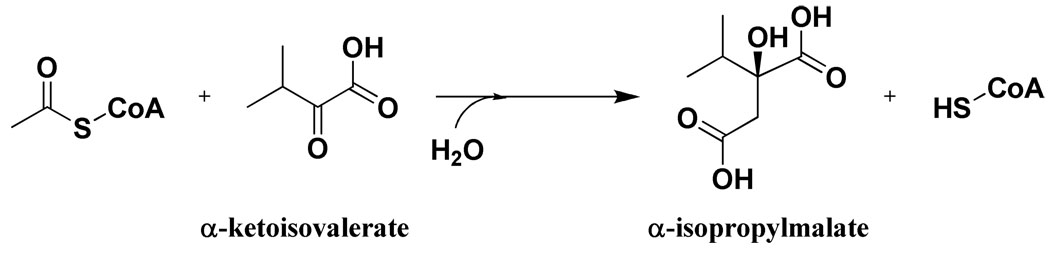
A nearly ubiquitous type of allosteric regulation is described as feedback inhibition. This type of inhibition refers to the regulation of enzymes in biosynthetic pathways by the downstream end-product of the pathway. Recently, we described the feedback inhibition of the enzyme α-isopropylmalate synthase from Mycobacterium tuberculosis (MtIPMS) by L-leucine (8). IPMS catalyzes the acetyl-CoA-dependent carboxymethylation of α-ketoisovalerate (α-KIV) to form α-isopropylmalate (Scheme 1). This reaction is the first committed step in the biosynthesis of L-leucine and as such is subject to feedback inhibition (9). However, closer inspection of the initial velocity progress curves revealed biphasic kinetics. Further kinetic studies established that for the M. tuberculosis enzyme L-leucine acts as a reversible, slow-onset inhibitor (a two-step process where the initial Enz-Leucine complex undergoes a slow equilibration with a more inhibited form). To the best of our knowledge, this is the first reported case of an allosteric inhibitor acting through a slow-onset mechanism for any enzyme system.
Scheme 1.
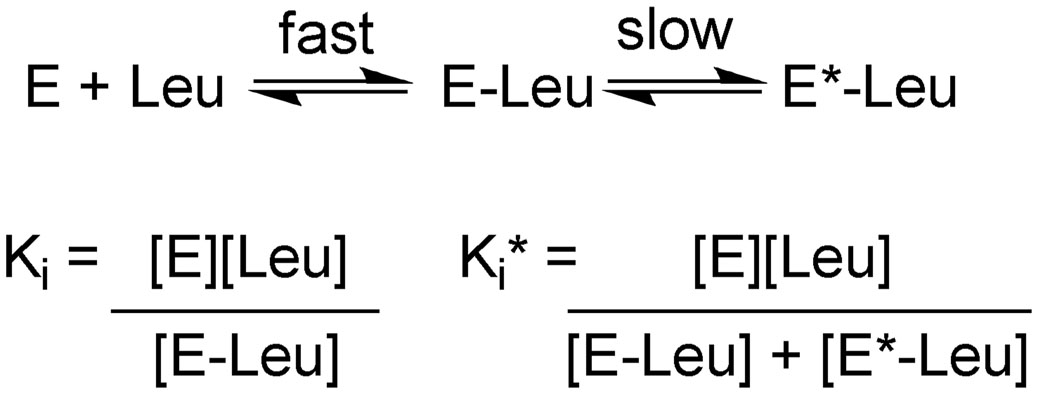
The slow-onset inhibition by L-leucine was consistent with the mechanism shown in Scheme 2, a minimal kinetic mechanism without regard to substrate binding (10). Binding of L-leucine to the free enzyme rapidly forms the inhibited binary complex. The binary complex then undergoes a slow equilibration to a second form of the enzyme which displays a further decrease in activity. The characterization of L-leucine as a slow-onset inhibitor is unique in the fact that all other reported slow-onset inhibitors have been characterized as competitive inhibitors (i.e. at saturating concentrations of substrate, inhibition is relieved), while kinetic studies with L-leucine are consistent with noncompetitive inhibition versus α-KIV. Slow-onset inhibition is traditionally exhibited by transition state or bisubstrate analog inhibitors. The mechanism describing slow-onset inhibition by these types of inhibitors is based on the conversion of the transition state stabilization energy into binding energy in the conversion from the EI to EI* complex. In the case of a slow-onset allosteric inhibitor, it is difficult to rationalize the involvement of the transition state stabilization energy in the mechanism of inhibition. As this type of inhibition is particularly useful in developing therapeutics due to the dramatic increase in residence time of the inhibitor, it is important to develop a better understanding of the mechanism of slow-onset inhibition in an allosteric background.
Scheme 2.
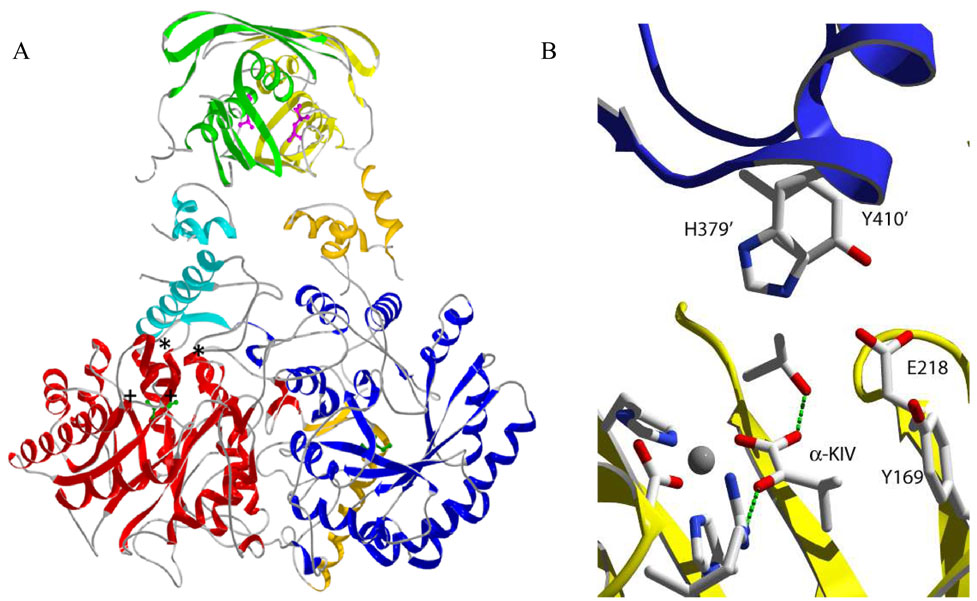
The three-dimensional structure of the MtIPMS-αKIV-M2+ ternary complex has been solved in the presence and absence of L-leucine (11). The enzyme is a homodimer with each monomer consisting of three domains: an N-terminal TIM-barrel catalytic domain (residues 51–368), a subdivided linker domain (residues 369–424 and 434–490), and a C-terminal regulatory domain (residues 491–644) (Figure 1A). L-leucine can be found bound to the regulatory domain (1 molecule per monomer) some 50 Å away from the active site. However, a comparison of the crystallographically-determined structures in the absence and presence of L-leucine shows virtually no changes in the architecture of the active site. Additionally, studies have shown no effect of L-leucine binding on the quarternary structure of MtIPMS, ruling out a mechanism of allosteric regulation involving changes to the quarternary structure of the enzyme (12). Thus, in the absence of structural evidence consistent with a large, rigid-body structural rearrangement or changes to the quarternary structure upon L-leucine binding, a mechanism involving inter-domain communication between the regulatory domain and the active site seems like the most plausible mechanism to describe the allosteric inhibition.
Figure 1.
Ribbon drawing of the three-dimensional structure of α-isopropylmalate synthase from M. tuberculosis. A. Domains colored red/blue, orange/cyan, and yellow/green correspond to the catalytic, linking, and regulatory domain, respectively, of each monomer. L-leucine is shown in purple in the regulatory domain. The substrate (α-KIV) is shown in green to denote the active site. The general location of substituted residues in this study are denoted by (*) for residues in the linker domain and by (+) for residues in the catalytic domain. B. Detailed rendering of residues on monomer A (yellow) and monomer B (purple and prime numbering) selected for mutational analysis. Residues involved in metal binding and α-KIV binding are shown as well. All three-dimensional structures were created with SwissPDB Viewer and rendered with POVRAY 3.5.
In order to test this theory, we have constructed a selected number of mutant forms of MtIPMS in the region of the protein where the linker subdomain I (residues 369–424) interacts with the active site (Figure 1B). In the MtIPMS dimer, each monomer forms a distinct catalytic domain, but the polypeptide chains then extend such that the linker subdomain I of one chain interacts with the active site of the adjacent monomer before forming the remaining portions of the linker and regulatory domains. Based on a sequence alignment, this region contains several phylogenetically conserved residues which we hypothesize may be important for allosteric control. Here, we describe the effects of substitutions at these conserved residues in MtIPMS on the kinetics of the enzyme reaction and inhibition by L-leucine.
Materials and Methods
Materials
Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs and Pfu turbo DNA polymerase was from Stratagene. Oligonucleotides used to amplify the leuA gene were obtained from Invitrogen. All substrates, buffers, and chemicals were obtained from Sigma and Aldrich. The Zero-Blunt Kit was obtained from Invitrogen. The plasmid, pET-28(a)+, and E. coli BL21(DE3) pLysS cells were from Novagen. Chromatographic columns and resins were obtained from Amersham-Pharmacia Biotech and Ni-NTA resin was from Novagen.
Cloning and Expression of Mutant IPMS
Point mutations in the pET28a(+)::leuA plasmid were made using QuikChange Site Directed Mutagenesis Kit (Stratagene). The integrity of the gene and presence of the desired point mutations were confirmed by DNA sequencing. The mutant plasmids were transformed into BL21(DE3)pLysS cells. BL21(DE3)pLysS cells harboring pET-28a(+)::leuA and variants were inoculated into 2×LB media and grown at 37 °C until they reached an OD600 of 0.4, then induced with 1 mM IPTG and incubated at 25 °C for 4 hours. Protein expression and purity was analyzed by SDS-PAGE.
Protein Purification
All steps were performed at 4 °C. Cells were thawed on ice for 30 min with Complete® protease inhibitors (Roche), lysozyme and DNAse I in 20 mM TEA, pH 7.8, containing 300 mM NaCl, and 50 mM imidazole. Cells were lysed by sonication and the soluble extract recovered after two centrifugations at 20,000 rpm. The clear soluble fraction was loaded onto a Nickel-NTA column (100 mL), and eluted with a linear 50 – 500 mM imidazole gradient in 20 mM TEA, pH 7.8, containing 300 mM NaCl. The peak fractions were pooled and dialyzed against 10 L of 20 mM TEA, pH 7.8. The dialyzate was loaded onto a Mono-Q column, and eluted with a linear 0 – 500 mM NaCl gradient in 20 mM TEA, pH 7.8. The peak fractions were pooled and dialyzed against 10 L of 20 mM TEA, pH 7.8. The dialysate was made 1 M in (NH4)2SO4 and loaded onto a Phenyl Sepharose column, and eluted with a linear 1-0 M (NH4)2SO4 gradient in 20 mM TEA, pH 7.8. Fractions containing pure IPMS were pooled and concentrated. After concentration, the IPMS solution was dialyzed against 10 L of 20 mM TEA, pH 7.8, containing 2 mM EDTA and after that, dialyzed versus 10 L of 20 mM TEA, pH 7.8. The protein was stored at −20 °C in 50% glycerol.
Measurement of Enzymatic Activity
Initial velocities for the forward reaction of α-isopropylmalate synthase were determined using 4,4’-dithiodipyridine (DTP) to detect the formation of CoA at 324 nm (ε = 19,800 M−1 cm−1) at 25 °C. A typical reaction mix contained 50 mM potassium phosphate, pH 7.5, 12 mM MgCl2, 50 µM DTP, 300 µM acetyl-CoA (AcCoA), and 300 µM α-ketoisovalerate (α-KIV). Values for the kinetic constants were determined using either the Michaelis-Menten equation (Eq. 1) or, in enzymes exhibiting substrate activation, equation 2, where V1 and V2 are the maximal velocities and K1 and K2 are the Michaelis constants for each phase of the kinetics, respectively.
| (1) |
| (2) |
Inhibition by L-leucine
For inhibition assays, reaction mixtures contained 50 mM potassium phosphate buffer, pH 7.5, 12 mM MgCl2, 100 µM DTP, 300 µM AcCoA, 300 µM α-KIV and variable concentrations of L-leucine. Reactions were initiated by the addition of MtIPMS.
Data Analysis
Progress curves at various L-leucine concentrations were fit to equation 3, where [P]t is the total product produced, [E]t is the total enzyme, vss is the steady state velocity, t is time, vi is the initial velocity, kb is the exponential rate constant for equilibration, and C is a finite intercept (10). The C term is included to allow for any product displacement at t = 0 since a minimal mixing time of two seconds is required prior to measurement (10). The terms vi and vss were then plotted versus L-leucine concentration. The dependence of vi and vss on L-leucine concentration were then analyzed as described below.
| (3) |
Theory
The feedback inhibition of MtIPMS by L-leucine is accurately defined as non-cooperative allosteric inhibition; in situations where the binding of inhibitor and substrate are independent of each other, this mechanism is identical to non-competitive inhibition (13). 2 The velocity equation for this type of allosteric mechanism is shown in equation 4 where V is the velocity in the absence of inhibitor, Ki is the dissociation constant for the inhibitor, β is the fractional activity relative to V at saturating inhibitor concentrations, and KS is the dissociation constant for the substrate. Under saturating substrate conditions, this equation can be reduced to equation 5. The initial velocities determined using equation 1 were plotted versus L-leucine concentration and fit to Equation 5 to determine values for V, β, and Ki.
| (4) |
| (5) |
In the cases of complete inhibition, plots of initial velocities versus L-leucine concentration were fit by equation 5 with β = 0. By analogy with the many descriptions of competitive slow-onset inhibitors, when the steady state velocities are used in place of the initial velocities, the Ki term is replaced with the term Ki*, which describes the overall dissociation constant for L-leucine from the enzyme (10). In the simplest case, the values for the other terms in the equation should be identical regardless of whether the initial or steady state velocities are analyzed.
Sedimentation Velocity Experiments
Sedimentation velocity measurements were conducted at 30,000 rpm in a Beckman XL-I analytical ultracentrifuge using double sector cells and the Ti-60 rotor at 22 °C in pH 7.5 buffer containing 20 mM potassium phosphate and 20 mM MgCl2. Boundary movement was followed at 280 nm using the centrifuge’s absorption optics. Each sample was adjusted to A280 = 1.0 data to facilitate their direct comparison. g(s*) distributions were determined using DCDT+ version 2.0.9 (14). A value of ν̄= 0.7276 was calculated from the amino acid composition using Sednterp v1.06 (15) as were the density (1.01356 g/mL) and viscosity (1.0445 cp) from the buffer composition. The latter values were used to correct the sedimentation coefficients to S20,w.
Isothermal Titration Calorimetry Experiments
Calorimetric titrations of WT, Y410F and E218A IPMS with L-leucine were performed on a VP-ITC microcalorimeter (MicroCal). Protein samples were dialyzed against 50 mM Tris, 25 mM KCl, 12 mM MgCl2, pH 7.5. The ligand solution was prepared by dissolving in protein dialysate buffer, and both ligand and protein solutions were degassed for 5 mins under vacuum prior to each titration. The concentration of protein was determined using Bradford reagent, and the concentration of L-leucine was determined by weight. The 1.46 mL sample cell was filled with 100 µM protein solution and the 250 µL injection syringe with 1 mM L-leucine solution which was injected at a rate of 10 µL every 4 minutes into the sample cell. The titrations were performed at 25 °C and the data was fit using the One Set of Sites Model provided in the Origin 7.0 software to determine Ka,ΔH and N (the stoichiometry for binding).
Product analysis by 1H NMR
The reaction of Y410F IPMS with AcCoA and α-KIV was followed by 1H NMR. The reaction mixture consisted of 50 mM potassium phosphate, pH 7.5, 12 mM MgCl2, 1.1 mM α-KIV, 1 mM AcCoA, and 100 µg of Y410F IPMS. After enzyme addition, the reaction was allowed to proceed at room temperature for 16 hours.
Results
Multiple Sequence Alignment
Multiple alignment of 12 α-IPMS’s from eubacteria, archea, plants, and fungi were performed using ClustalW (Figure S1) (16). The overall identity is 5.6% (equivalent to 35 amino acids), with an additional 4.6% of residues showing strong similarity. The majority of conserved residues map to the active site and subdomain I of the linker domain. Four residues (three strictly conserved, one strongly similar) that reside at the interface of the catalytic domain and linker subdomain I of the adjacent monomer were selected for mutational analysis (Figure 1). Residues Y169 and Y410 were mutated to phenylalanine, and residues E218 and H379 were mutated to alanine.
Steady-State Kinetic Analysis
The kinetic parameters determined for the mutant enzymes are shown in Table 1. Two of the mutant enzymes, E218A and Y410F IPMS, exhibit kcat values that are reduced 35-fold relative to wild-type MtIPMS. Excluding Y410F IPMS, none of the mutations have caused a drastic change in the Km values for either substrate. In the case of Y410F IPMS, the Km values are actually lowered about 10-fold relative to the wild-type enzyme. Two of the substitutions, E218A and H379A, resulted in enzymes which display substrate activation toward α-KIV (Figure S2), suggesting that these mutations have created non-symmetrical active sites in the dimer. In addition, E218A IPMS also displays substrate activation kinetics towards AcCoA, but with a significantly higher Km for the second binding site (10 µM and 700 µM).
Table 1.
Steady-State Kinetic Parameters for MtIPMS and Mutant Enzymesa
| α-KIVb | Acetyl-CoAb | |||||||
|---|---|---|---|---|---|---|---|---|
| Enzyme |
kcat1c (min−1) |
K1c (µM) |
kcat2c (min−1) |
K2c (µM) |
kcat1 (min−1) |
K1 (µM) |
kcat2 (min−1) |
K2 (µM) |
| WT | 152 ± 3 | 7.1 ± 0.5 | - | - | 163 ± 3 | 24 ± 2 | - | - |
| Y169F | 285 ± 16 | 13.4 ± 2.6 | - | - | 340 ± 13 | 40 ± 6 | - | - |
| E218A | 2.2 ± 0.1 | 1.6 ± 0.2 | 1.3 ± 0.1 | 39± 11 | 3.5 ± 0.2 | 9.5 ± 1.1 | 5.6 ± 2.8 | 697 ± 615 |
| H379A | 42 ± 18 | 1.6 ± 1.1 | 89 ± 17 | 19 ± 6 | 153 ± 2 | 23 ± 1.5 | - | - |
| Y410F | 4.5 ± 0.2 | 0.5 ± 0.1 | - | - | 5.6 ± 0.2 | 1.1 ± 0.2 | - | - |
All reactions were performed as described in Materials and Methods.
Varied substrate
For enzymes displaying substrate activation, kinetic parameters with subscripts 1 and 2 represent the kinetic parameters for the unactivated and activated forms of the enzyme, respectively. For enzymes without substrate activation, kcat1 and K1 are kcat and Km, respectively.
Effect of Mutations on Inhibition by L-Leucine
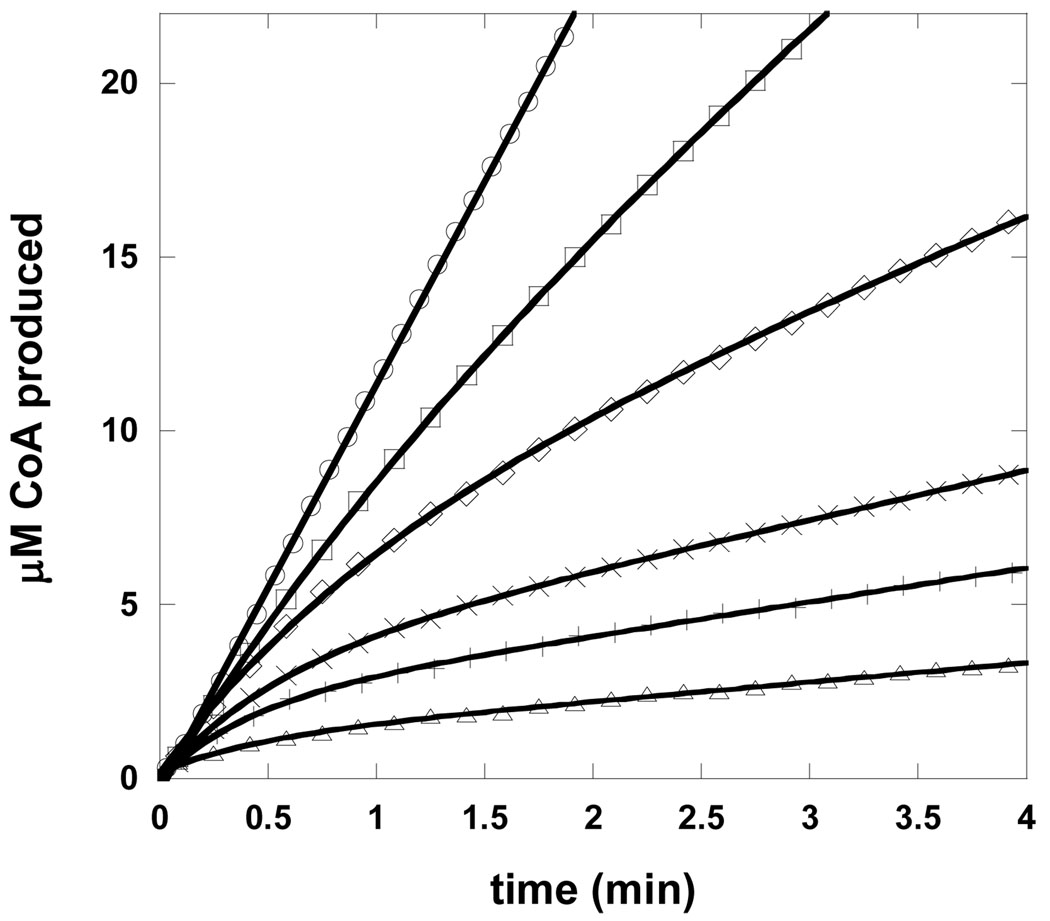
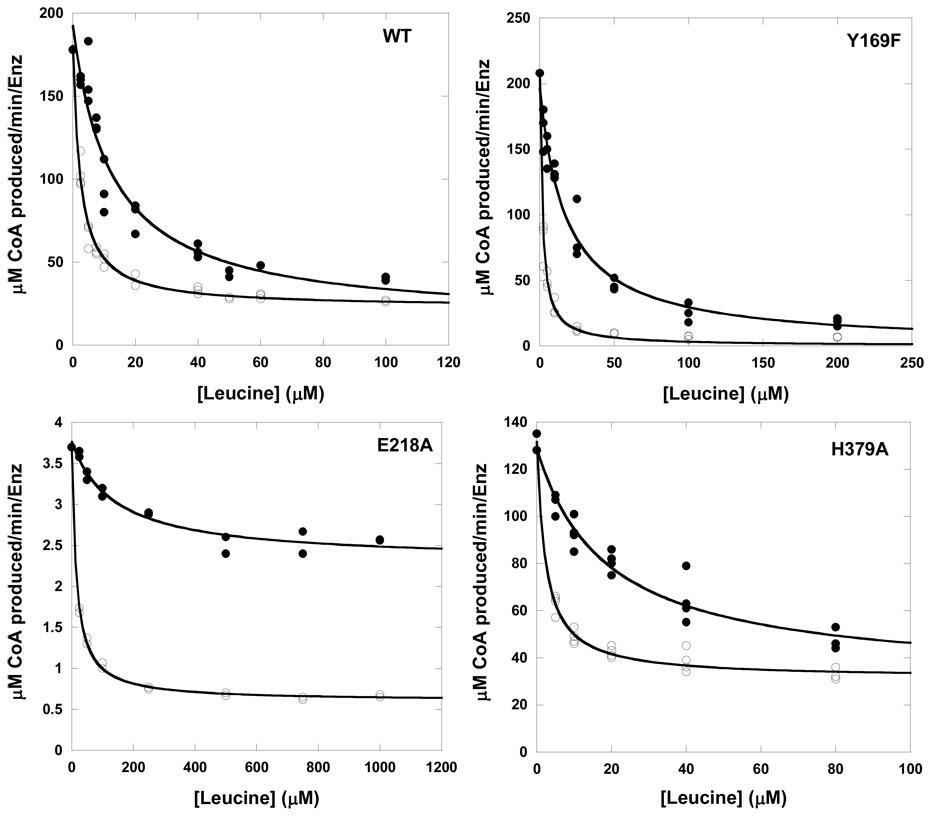
With the exception of Y410F IPMS, all of the variant enzymes displayed slow-onset inhibition kinetics (i.e. biphasic progress curves) in the presence of L-leucine (Figure 2 for Y169F IPMS). In the absence of L-leucine, all enzymes exhibited linear kinetics over the time-frame of the inhibition experiments suggesting that the biphasic kinetics were not due to enzyme instability or tighter product inhibition (data not shown). Inhibition by L-leucine was characterized by evaluating the initial and steady-state velocities of the enzymes at various concentrations of inhibitor as described in Materials and Methods (Figure 3). The inhibition parameters determined for wild-type and the mutant enzymes are shown in Table 2. Closer inspection of the kinetic data for the wild-type enzyme revealed that at saturating concentrations of L-leucine (200 µM or 17-times the Ki value) the enzyme retained approximately 10% activity. The values determined for Ki and Ki* for the wild-type enzyme are identical to those determined previously (8). The Y169F substitution resulted in an enzyme that was fully inhibited in the presence of L-leucine, while the H379A substitution retained 24% activity, without affecting the Ki or Ki* values relative to the wild-type values. This suggests that these residues have not perturbed the binding of L-leucine but have altered the environment of the active site in the E*-leu complex resulting in differing residual activities.
Figure 2.
Kinetics of slow-onset inhibition of Y169F MtIPMS by L-leucine. Reactions were initiated with enzyme and the following concentrations of L-leucine: 0 µM (○), 2.5 µM (□), 5 µM (◊), 10 µM (X), 15 µM (+), 50 µM (Δ). Solid black lines are fits to either a linear equation (0 µM leucine) or equation 3.
Figure 3.
Plots of vi (●) and vss (○) versus L-leucine concentration for wild-type and the Y196F, E218A, and H379A enzymes. Solid lines are from fits to equation 5. Results are shown in Table 2.
Table 2.
L-Leucine Inhibition Parameters for MtIPMS and Mutant Enzymesa
| Enzyme | Ki(µM) | Ki* (µM) | β |
|---|---|---|---|
| WT | 12 ± 3 | 2.3 ± 0.2 | 0.1 |
| Y169F | 18 ± 2 | 1.6 ± 0.1 | -b |
| E218A | 139 ± 42 | 15 ± 1 | 0.61/0.16c |
| H379A | 19 ± 4 | 2.3 ± 0.2 | 0.24 |
| Y410F | -d | - | - |
Performed as described in Materials and Methods.
Data for Y169F IPMS was fit to equation 5 with β = 0.
Limiting values for vi and vss, respectively.
Inhibition not observed.
Two of the mutant enzymes displayed much more dramatic alterations in inhibition kinetics. The results from the E218A enzyme suggest that this substitution has caused a change in the affinity of the free enzyme for L-leucine due to the 10-fold increase in the value for Ki (a true dissociation constant) relative to the value for the wild-type enzyme. Since the Ki* value (in Scheme 2) consists of an arrangement of the rate constants involved in the second step multiplied by the Ki value, the 10-fold change in Ki could account for the 6.5-fold change in the Ki* value. E218A IPMS also gave different values for the residual activity depending on the analysis of the vi or vss data, retaining 61% and 16% activity, respectively. This result suggests that the E218A substitution has also caused a change in the active site architecture of the E-leu complex resulting in significantly higher residual activity. The most interesting result was seen for the Y410F substitution. This mutant enzyme was insensitive to L-leucine at concentrations up to 500 µM.
Effect of L-Leucine on Quarternary Strucutre
It has previously been determined that WT MtIPMS is a stable dimer in solution and addition of L-leucine has no effect on the quarternary structure (12). However, it has been reported that the mechanism of allosteric inhibition by L-leucine of α-IPMS from Salmonella enterica serovar typhimurium is consistent with the dissociation of a tetramer to inactive monomers (17, 18). In order to determine if the differences in inhibition kinetics seen with the mutant enzymes were due to a change in quarternary structure, sedimentation velocity experiments were run in the presence and absence of 1 mM L-leucine. The sedimentation coefficients determined for each enzyme (except Y169F IPMS) in the absence of L-leucine were consistent with a dimer for all of the enzymes (Table S1). When L-leucine was added, no significant changes were seen in the sedimentation coefficients. This result suggests that the mutations have not altered the quaternary structure of the enzyme, and that the altered inhibition kinetics are not due to a dissociation of the active dimer complex in the wild-type or mutant enzymes.
Further Characterization of Y410F IPMS
Due to the unanticipated and dramatic effect of the Y410F substitution, this enzyme form was investigated in greater detail. The spectroscopic assay described for IPMS measures production of CoA. In the wild-type enzyme, the production of CoA has been shown to be strictly coupled to the formation of α-IPM, with no evidence for uncoupled AcCoA hydrolysis (12). To ensure that the activity associated with Y410F IPMS is due to α-IPM production, 1H-NMR was used to monitor the reaction products. There was no evidence for the Y410F catalyzed hydrolysis of AcCoA in the presence of α-KIV (Figure S3).
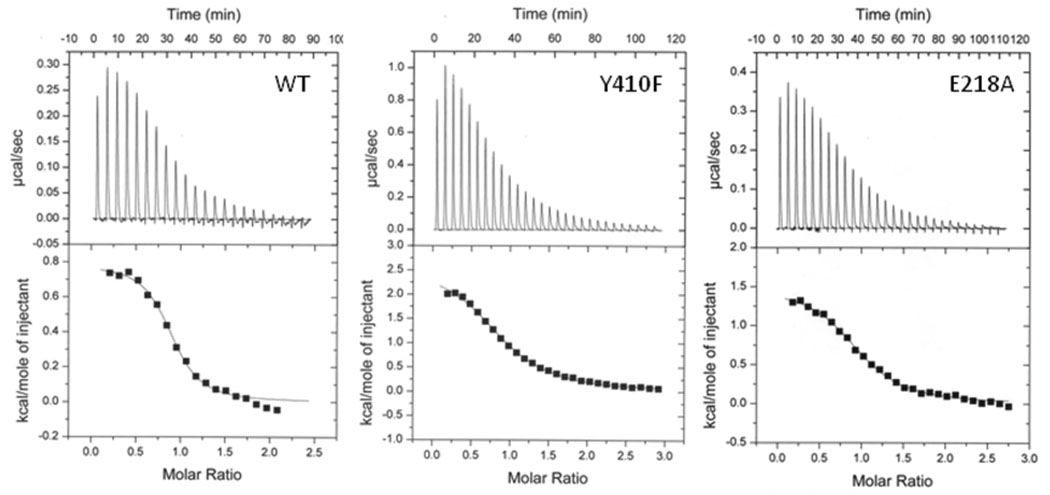
In order to determine if the loss of sensitivity to L-leucine in the Y410F enzyme is due to the inability to bind L-leucine, isothermal titration calorimetry (ITC) was employed to directly determine the dissociation constant for L-leucine for the wild-type and Y410F enzymes (Figure 4). Binding of L-leucine to the wild-type enzyme was endothermic and could be fit to a one-site model consistent with the non-cooperative kinetics of L-leucine inhibition. A Kd value of 3.0 ± 0.6 µM was determined for the wild-type enzyme, consistent with the value for Ki*. Y410F IPMS gave ITC data which could be fit to the same equation, and a value of 21 ± 1 µM was determined for the dissociation constant for L-leucine, suggesting that the substitution has not drastically altered the affinity of the enzyme for L-leucine.
Figure 4.
Isothermal titration calorimetry plots for wildtype, Y410F and E218A MtIPMS. Experiments were performed as described in Materials and Methods. The results from a fit of the data to a One Set of Sites model produced the following values for the binding of L-leucine to the wild-type (N = 0.86 ± 0.02, ΔH = 774 ± 20 cal/mol, ΔS = 27.8 cal/mol/K), Y410F (N = 0.91 ± 0.01, ΔH = 2598 ± 48 cal/mol, `S = 30.1 cal/mol/K) and E218A (N = 0.97 ± 0.01, ΔH = 1486 ± 31 cal/mol, ΔS = 28.5 cal/mol/K) enzymes.
Discussion
The enzyme α-IPMS from M. tuberculosis catalyzes the first step in the biosynthesis of the amino acid L-leucine. We have previously demonstrated the MtIPMS is feedback inhibited by L-leucine through a slow-onset mechanism (8). We believe that this is the first reported case of an allosteric effector that acts through a slow-onset mechanism; all previous inhibitors displaying slow-onset kinetics have been characterized as competitive inhibitors. In the wild-type enzyme, there is no evidence for L-leucine causing a large, rigid-body conformational change or altering the quarternary structure of the enzyme, ruling out either of these allosteric mechanisms. Thus, we hypothesized that there must be some type of inter-domain communication between the regulatory domain and the active site to achieve inhibition. This type of inter-domain communication has also been proposed in the more prevalent ACT regulatory domain-containing enzymes that are responsible for the regulation of a number of metabolic systems involving amino acid metabolism (19). Studies of the enzyme D-3-phosphoglycerate dehydrogenase (PGDH), which catalyzes the first step in the biosynthesis of D-serine, demonstrate that PGDH is subject to feedback regulation by D-serine as a V-type inhibitor similar to that seen in MtIPMS (20). Recently, structures of both the apo- and serine-bound enzyme from M. tuberculosis have been reported which demonstrate that serine binding produces only subtle movements of residues involved in binding the inhibitor and no domain movement, consistent with a allosteric mechanism involving inter-domain communication (21, 22).
According to the three-dimensional structure of MtIPMS, a probable intersection for this type of communication would occur where the linker subdomain I of one monomer interacts with the catalytic domain of the adjacent monomer (11). In order to probe this interface, mutagenesis of specific residues at the interface was undertaken in an attempt to disrupt the allosteric communication. With respect to the roles of the selected residues in catalysis, rather than regulation, an analysis of the kinetics of the substituted enzymes was surprising rather than enlightening. MtIPMS belongs to a family of enzymes that catalyze the Claisen-condensation reaction between an α-keto acid and acetyl-CoA. Within this family, the enzyme homocitrate synthase catalyzes the carboxymethylation of α-ketoglutarate by acetyl-CoA. Recently, mutational studies on homocitrate synthase from Saccharomyces cerevisiae have concluded that the residues equivalent to E218 and H379 act as a catalytic dyad in the chemical mechanism (23). In fact, no detectable activity was seen when the histidine residue was mutated to alanine and the substitution of alanine for the glutamate residue reduced the kcat value 1000-fold, in sharp contrast to the results with MtIPMS. However, the mutation equivalent to Y410F in homocitrate synthase had a modest effect on activity, lowering the kcat value only 25-fold similar to the effect we report. In light of the comparison with homocitrate synthase, it is clear that none of these residues are critical for catalysis, as evidenced by the maximal drop of about 40-fold in the kcat value. In addition, none of the mutations adversely affects the Michaelis constants for either substrate. However, it is known that for wild-type MtIPMS that chemistry is not rate-limiting, making interpretation of the Michaelis constant difficult (12). Consistent with a proposed rate-limiting product release, the Y169F substitution results in an enzyme that exhibits a two-fold increase in the value for kcat relative to the wild-type enzyme. This residue is located between the α-KIV binding site and the opening of the active-site cleft, suggesting that loss of the hydroxyl group allows for more facile release of the product from the active site.
Two of the substitutions appear to have created kinetic cooperativity between the two active sites. E218A and H379A IPMS have initial velocity kinetics consistent with substrate activation towards α-KIV. In the three-dimensional structure, these two residues (one from each monomer, thus E218A and H379’A) are located 4.5 Å from each other and within 5–6 Å of α-KIV. The simplest explanation for these results is that removal of these charged residues at the active site interface of the two monomers has resulted in non-symmetrical active site architecture in the presence of α-KIV. Another possible explanation for substrate activation kinetics which cannot be explicitly ruled out is the presence of a contaminating enzyme. However, neither of these variant enzymes produces CoA in the absence of α-KIV under the experimental conditions, suggesting that any contaminating enzyme must also be dependent on α-KIV, making this explanation unlikely. E218A IPMS also displays substrate activation towards AcCoA, however this result is less interpretable because of the inability to identify the binding site of AcCoA in structural studies.
The effects of the mutations on L-leucine inhibition of IPMS were determined by analysis of the initial and steady-state velocities of the biphasic reaction. Surprisingly, all of the mutant enzymes (except Y410F IPMS) displayed slow-onset inhibition kinetics. This result suggests that the E-Leu to E*-Leu transition is not governed through this domain interface and may be the result of a more global change in conformation. Initial attempts to fit the initial velocity data for the L-leucine inhibition of the wild-type enzyme to an equation derived for competitive slow-onset inhibitors quickly led to the realization that the wild-type enzyme retains approximately 10% activity at saturating L-leucine concentrations. However, in a slow-onset mechanism where there are two distinct enzyme-inhibitor forms (E-Leu and E*-Leu) it is difficult to determine conclusively whether the residual activity is due to a single form, or a combination of both forms.
The activity of Y169F IPMS, however, is completely inhibited in the presence of saturating concentrations of L-leucine. This substitution has not affected the values for the Ki or Ki* for L-leucine, but it has altered the architecture of the active site in the presence of L-leucine relative to the wild-type enzyme. H379A IPMS retains 24% activity in the presence of saturating concentrations of L-leucine, more than twice the relative residual activity of the wild-type enzyme. The results seen with these substitutions demonstrate that changes in the linker domain/active site interface can affect the kinetics of inhibition without altering the binding of L-leucine.
More drastic effects are seen with the E218A enzyme. The inhibition constants for L-leucine determined with E218A IPMS are elevated (6.5- to 10-fold) relative to the values determined with the wild-type enzyme. This suggests that the substitution has moderately affected the affinity of the enzyme for L-leucine, despite the fact that the residue is 50 Å from the inhibitor binding site. A Kd value of 7.1 ± 0.8 μM for L-leucine binding to the E218A enzyme was determined using ITC experiments (Figure 4). This value is elevated 2.3-fold relative to the Kd value determined with the wild-type enzyme. The E218A substitution has also significantly altered the residual activity of the enzyme-Leu complexes. The initial velocity retains nearly 60% of the uninhibited activity, while the final, steady-state velocity only retains 16% activity. This suggests that changes at the linker domain/active site interface can influence the architecture of the L-leucine binding site and the active site when L-leucine is bound.
Kinetic experiments with Y410F IPMS suggest that this substitution has rendered the enzyme insensitive to L-leucine. Control experiments confirmed that Y410F IPMS has not abolished L-leucine binding, is chemically competent, and exists as a stable dimer, ruling out these alternate explanations for the absence of inhibition. The results from ITC experiments are consistent with the Y410F mutation altering the affinity of the enzyme for L-leucine seven-fold, relative to the value determined with the wild-type enzyme. The ability of the Y410F and E218A substitutions to alter the affinity of the enzyme for L-leucine is consistent with the presence of a network of residues connecting the active site and the regulatory domain. In addition, the Y410F substitution appears to have severed the network as evidence by the uncoupling of the binding of L-leucine in the regulatory domain from changes to the active site architecture.
From the crystal structure, it was straightforward to propose that the Y410-containing loop plays a role in transmitting the signal from the linker subdomain I to the active site (11); however, the remainder of the network lacks similar definition. Mutations that resulted in resistance to inhibition by trifluoroleucine produced by genetic studies on IPMS from Saccharomyces cerevisiae all mapped to the regulatory domain near the L-leucine binding site (24). Only one of the residues implicated in trifluoroleucine resistance, A568, is positioned at the interface of the regulatory domain and subdomain II of the linker domain. This interface is made up of three beta sheets on the regulatory domain and the three helixes of subdomain II of the linker domain. There are numerous possible interactions consisting of ionic and aromatic residues, making identification of important interactions difficult. Tracing the proposed network between the linker subdomains becomes even more challenging due to asymmetry and missing electron density in crystal structure.
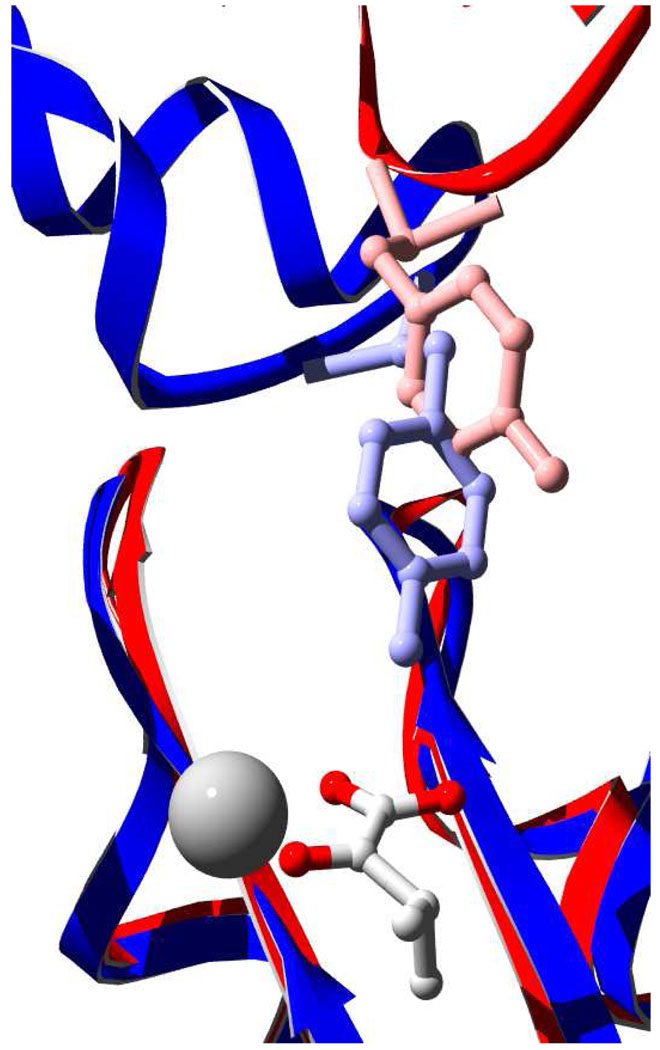
Additional support for the inclusion of the Y410-containing loop in an inter-domain allosteric network comes from the three-dimensional structure of the bifunctional enzyme 4-hydroxy-2-ketovalerate aldolase (DmpG)/acylating acetaldehyde dehydrogenase (DmpF) (25). DmpG and IPMS share the basic TIM barrel catalytic domain and have nearly identical active site architecture resulting in the DmpG structure as the best match to the catalytic domain using the secondary structure matching algorithm SSM (11). In addition, DmpG has a linker domain with an α-helical structure similar to that of IPMS. DmpG catalyzes the formation of acetaldehyde and pyruvate from 4-hydroxy-2-ketovalerate. The acetaldehyde is proposed to move through a 29 Å tunnel to the DmpF active site. Access to the tunnel is controlled by the presence of NAD+, a substrate for DmpF, at the DmpF active site coupled to the motion of residue Y291 on the linker domain of DmpG. When the structures of DmpG and MtIPMS are superimposed, Y291 DmpG is found to be in a position similar to Y410 IPMS (Figure 5).
Figure 5.
Superimposition of MtIPMS and DmpG. Swiss-PDB Viewer was used to align the E chain of DmpG (1NMV) (blue) with MtIPMS (1SR9) (red). The alignment included 808 atoms and had an RMS of 1.4Å. α-KIV and a Mg2+ atom are shown to indicate the active site of MtIPMS. MtIPMS Y410 (red) and DmpG Y291 (blue) are depicted as ball and stick representations.
The results of kinetic and thermodynamic studies presented here are consistent with an allosteric network linking the catalytic and regulatory domains in MtIPMS. The perturbation of the affinity of the enzyme for L-leucine caused by the E218A and Y410F substitutions, located 50 Å from the L-leucine binding site, implicate these residues as belonging to that network and broadly suggest that the interface between the linker and catalytic domains contains important contacts in the network. More specifically, as the Y410F substitution has uncoupled the binding of L-leucine from changes at the active site, it is clear that the Y410F-containing loop is an integral part of the allosteric network in MtIPMS. While the upstream connections of the allosteric network remain unidentified, Y410F IPMS now provides an excellent negative control for future structural studies aimed at more precisely defining the mechanism of inter-domain communication in this biologically important enzyme.
Supplementary Material
Information regarding the results of the ultracentrifugation experiments, the multiple sequence alignment, Eadie-Hofstee plots, and the 1H-NMR spectrum of the reaction product. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements
We thank the College of Medicine for their support of the analytical ultracentrifuge facility and Michael Brenowitz for conducting the experiments. We also thank Achelle Edwards for assistance in performing the isothermal titration calorimetry experiments.
Abbreviations
- MtIPMS
M. tuberculosis α-isopropylmalate synthase
- α-KIV
α-ketoisovalerate
- AcCoA
acetyl-coenzyme A
- α-IPM
α-isopropylmalate
- DTP
4,4’-dithiodipyridine
Footnotes
This work was supported by NIH grant AI33696 (JSB) and a postdoctoral fellowship from the Charles H. Revson Foundation (PAF)
An alternate kinetic analysis of allosteric effectors has previously been discussed. (Reinhart, .D. (1983) Archives of Biochemistry and Biophysics. 224, pp. 389–401) In the absence of ooperativity, the equations in this reference are essentially identical to those presented by Segel (13).
References
- 1.Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 2.Kantrowitz ER, Lipscomb WN. Escherichia coli aspartate transcarbamoylase: the molecular basis for a concerted allosteric transition. Trends Biochem Sci. 1990;15:53–59. doi: 10.1016/0968-0004(90)90176-c. [DOI] [PubMed] [Google Scholar]
- 3.Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 4.Daily MD, Upadhyaya TJ, Gray JJ. Contact rearrangements form coupled networks from local motions in allosteric proteins. Proteins. 2008;71:455–466. doi: 10.1002/prot.21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandhi PS, Chen Z, Mathews FS, Di Cera E. Structural identification of the pathway of long-range communication in an allosteric enzyme. Proc Natl Acad Sci U S A. 2008;105:1832–1837. doi: 10.1073/pnas.0710894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai CJ, del Sol A, Nussinov R. Allostery: absence of a change in shape does not imply that allostery is not at play. J Mol Biol. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenton AW, Paricharttanakul NM, Reinhart GD. Disentangling the web of allosteric communication in a homotetramer: heterotropic activation in phosphofructokinase from Escherichia coli. Biochemistry. 2004;43:14104–14110. doi: 10.1021/bi048569q. [DOI] [PubMed] [Google Scholar]
- 8.de Carvalho LP, Argyrou A, Blanchard JS. Slow-onset feedback inhibition: inhibition of Mycobacterium tuberculosis alpha-isopropylmalate synthase by L-leucine. J Am Chem Soc. 2005;127:10004–10005. doi: 10.1021/ja052513h. [DOI] [PubMed] [Google Scholar]
- 9.Stieglitz BI, Calvo JM. Distribution of the isopropylmalate pathway to leucine among diverse bacteria. J Bacteriol. 1974;118:935–941. doi: 10.1128/jb.118.3.935-941.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison JF, Walsh CT. The behavior and significance of slow-binding enzyme inhibitors. Adv Enzymol Relat Areas Mol Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 11.Koon N, Squire CJ, Baker EN. Crystal structure of LeuA from Mycobacterium tuberculosis, a key enzyme in leucine biosynthesis. Proc Natl Acad Sci U S A. 2004;101:8295–8300. doi: 10.1073/pnas.0400820101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Carvalho LP, Blanchard JS. Kinetic and chemical mechanism of alpha-isopropylmalate synthase from Mycobacterium tuberculosis. Biochemistry. 2006;45:8988–8999. doi: 10.1021/bi0606602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segel IH. Enzyme kinetics : behavior and analysis of rapid equilibrium and steady state enzyme systems. New York: Wiley; 1975. [Google Scholar]
- 14.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 15.Hayes B, Laue T, Philo J. Sedimentation Interpretation Program. University of New Hampshire; 2003. [Google Scholar]
- 16.de Carvalho LP. Department of Biochemistry. New York: Albert Einstein College of Medicine; 2006. Investigations of Mycobacterium tuberculosis α-Isopropylmalate Synthase. [Google Scholar]
- 17.Leary TR, Kohlhaw GB. -isopropylmalate synthase from Salmonella typhimurium. Analysis of the guaternary structure and its relation to function. J Biol Chem. 1972;247:1089–1095. [PubMed] [Google Scholar]
- 18.Leary TR, Kohlhaw G. Dissociation of alpha-isopropylmalate synthase from Salmonella typhimurium by its feedback inhibitor leucine. Biochem Biophys Res Commun. 1970;39:494–501. doi: 10.1016/0006-291x(70)90605-4. [DOI] [PubMed] [Google Scholar]
- 19.Chipman DM, Shaanan B. The ACT domain family. Curr Opin Struct Biol. 2001;11:694–700. doi: 10.1016/s0959-440x(01)00272-x. [DOI] [PubMed] [Google Scholar]
- 20.Grant GA, Schuller DJ, Banaszak LJ. A model for the regulation of D-3-phosphoglycerate dehydrogenase, a Vmax-type allosteric enzyme. Protein Sci. 1996;5:34–41. doi: 10.1002/pro.5560050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dey S, Burton RL, Grant GA, Sacchettini JC. Structural analysis of substrate and effector binding in Mycobacterium tuberculosis D-3-phosphoglycerate dehydrogenase. Biochemistry. 2008;47:8271–8282. doi: 10.1021/bi800212b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dey S, Grant GA, Sacchettini JC. Crystal structure of Mycobacterium tuberculosis D-3-phosphoglycerate dehydrogenase: extreme asymmetry in a tetramer of identical subunits. J Biol Chem. 2005;280:14892–14899. doi: 10.1074/jbc.M414489200. [DOI] [PubMed] [Google Scholar]
- 23.Qian J, Khandogin J, West AH, Cook PF. Evidence for a catalytic dyad in the active site of homocitrate synthase from Saccharomyces cerevisiae. Biochemistry. 2008;47:6851–6858. doi: 10.1021/bi800087k. [DOI] [PubMed] [Google Scholar]
- 24.Cavalieri D, Casalone E, Bendoni B, Fia G, Polsinelli M, Barberio C. Trifluoroleucine resistance and regulation of alpha-isopropyl malate synthase in Saccharomyces cerevisiae. Mol Gen Genet. 1999;261:152–160. doi: 10.1007/s004380050952. [DOI] [PubMed] [Google Scholar]
- 25.Manjasetty BA, Powlowski J, Vrielink A. Crystal structure of a bifunctional aldolase-dehydrogenase: sequestering a reactive and volatile intermediate. Proc Natl Acad Sci U S A. 2003;100:6992–6997. doi: 10.1073/pnas.1236794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information regarding the results of the ultracentrifugation experiments, the multiple sequence alignment, Eadie-Hofstee plots, and the 1H-NMR spectrum of the reaction product. This material is available free of charge via the Internet at http://pubs.acs.org.