Abstract
Background
The clinical relevance of the amount of human papillomavirus type 18 (HPV18) DNA in cervical tissue (ie, HPV18 DNA load) is unknown.
Methods
Study subjects were 303 women who were HPV18 positive at enrollment into the Atypical Squamous Cells of Undetermined Significance (ASC-US) and Low-Grade Squamous Intraepithelial Lesion (LSIL) Triage Study. HPV18 DNA load, expressed as copies of HPV18 per nanogram of cellular DNA, at enrollment was quantitatively measured. Subjects were followed up semiannually for a period of 2 years for detection of cervical intraepithelial neoplasia 2–3 (CIN2–3). A linear regression model was used to examine associations of CIN2–3 with HPV18 DNA load. All statistical tests were two-sided.
Results
CIN2–3 was confirmed in 92 of 303 (30.4%) HPV18-positive women. Among women without CIN2–3, HPV18 DNA load was positively associated with increasing severity of cervical cytology at enrollment (Ptrend < .001). However, among those with CIN2–3, HPV18 DNA load was not associated with severity of cervical cytology at enrollment (Ptrend = .33). The ratios of geometric means of HPV18 DNA load at enrollment among women with CIN2–3, relative to those without, were 6.06 (95% confidence interval [CI] = 0.31 to 117.92) for those with normal cytology at enrollment, 0.50 (95% CI = 0.10 to 2.44) for those with ASC-US, 0.11 (95% CI = 0.03 to 0.46) for those with LSIL, and 0.07 (95% CI = 0.01 to 0.80) for those with high-grade squamous intraepithelial lesion (HSIL). After adjusting for age and coinfection with other high-risk HPVs, a statistically significant association of lower HPV18 DNA load with CIN2–3 was observed among women with LSIL or HSIL at enrollment (P = .02). Within the 2-year period, HPV18 DNA load was unrelated to the timing of CIN2–3 diagnosis. Overall results were similar when the outcome was CIN3.
Conclusions
HPV18 DNA load was higher for women with LSIL or HSIL at enrollment with no evidence of CIN2–3 during the 2-year follow-up period than it was for women with CIN2–3. Thus, testing for high levels of HPV18 DNA does not appear to be clinically useful.
CONTEXT AND CAVEATS
Prior knowledge
Human papillomavirus type 18 (HPV18) is often detected in invasive cervical cancer, but it is unknown whether high amounts of HPV18 DNA in cervical tissue are associated with cervical intraepithelial neoplasia (CIN) grades 2–3, the precursor of cervical cancer.
Study design
Women (n = 303) who were HPV18 positive at enrollment in the Atypical Squamous Cells of Undetermined Significance and Low-Grade Squamous Intraepithelial Lesion Triage Study were monitored semiannually for 2 years.
Contributions
CIN2–3 was diagnosed in 92 women. The amount of HPV18 DNA was positively associated with severity of cervical cytology at enrollment among women without CIN2-3 during the 2-year study period, and among those with low- or high-grade sequamous intraepithelial lesions at enrollment, it was lower in women with CIN2-3 than those without CIN2-3.
Implications
Testing for high amounts of HPV18 DNA in cervical tissue samples may not be clinically useful.
Limitations
The study sample size is small. Lesions would have likely been detected earlier in the study than in the general population. HPV18 DNA levels may fluctuate during the course of infection.
From the Editors
Although human papillomavirus type 18 (HPV18) is the second most common type of HPV detected in invasive cervical cancers (1) and is more prevalent in cervical adenocarcinoma/adenosquamous carcinoma than in squamous cell carcinoma (2), it is infrequently detected in cross-sectional studies of women with cervical intraepithelial neoplasia grades 2–3 (CIN2–3), the precursor of cervical cancer (3–6). Possible explanations for the limited detection of HPV18-related CIN2–3 include the following: HPV18-associated CIN2–3 may progress rapidly to invasive disease (5) and HPV18-associated CIN2–3 may be underdiagnosed, perhaps because HPV18 has a tropism to glandular cells in which the infection is less accessible to sampling and the lesion is difficult to detect or because HPV18 is less likely to cause a cytological high-grade squamous intraepithelial lesion (HSIL) than HPV16 (7), thereby leading to underrepresentation of HPV18 in CIN2–3 patients. Another possibility for the underdiagnosis of HPV18-related CIN2–3 is that replication of HPV18 DNA may be below a detectable threshold during neoplastic transformation. This possibility can be indirectly evaluated by comparing the HPV18 DNA concentrations in cervical samples (ie, HPV DNA load) of women with CIN2–3 and without CIN2–3 who have a detectable HPV18 infection.
Previous attempts to examine the clinical relevance of HPV DNA load have focused mainly on HPV16 (8–19) and have shown that the elevated viral load is associated with risk of cervical cancer and its precursor. Data from quantitative analyses of viral load of other HPV types are rare. The reported findings regarding HPV18 DNA load and risk of high-grade lesions are controversial—some studies (12,19–23) found a positive association; others (7,8,24) did not. Most of the previous studies are of cross-sectional observations with only a single measurement of outcome of interest. Although few studies with longitudinal settings have been reported (13,18,25), the findings are generally limited by the small sample size. It remains largely undetermined whether, and to what extent, the relationship between HPV18 DNA load and CIN2–3 differs by cervical cytology, the factor known to reflect productive HPV infections and is associated with underlying histopathology.
To better understand the clinical implications of HPV18 DNA load, we examined associations of HPV18 DNA load with severity of cervical cytology at enrollment into the Atypical Squamous Cells of Undetermined Significance (ASC-US) and Low-Grade Squamous Intraepithelial Lesion (LSIL) Triage Study (ALTS), a clinical trial that was designed to evaluate management strategies for women with mild or equivocally abnormal community-read Pap smears. We compared cytology-stratified HPV18 DNA load of women with CIN2–3 and without CIN2–3 during a 2-year follow-up period and between CIN2–3 patients who were initially diagnosed at the time concurrent with and subsequent to the visit at which HPV18 DNA load was measured.
Study Subjects and Methods
Subjects and Study Design
Study subjects were women who participated in ALTS. A full description of the ALTS design and study population is available elsewhere (26,27). Briefly, between January 1, 1997, and December 31, 1998, a total of 5060 women who had a Pap smear showing ASC-US or LSIL within 6 months (an average of 2 months) were enrolled. Women were randomly assigned to one of three management arms (ie, immediate colposcopy, HPV triage, or conservative management). These arms differed only in referral for colposcopy at enrollment: An entry colposcopic examination with biopsy of any visible lesions was referred for all women in the immediate colposcopy arm, women who had an entry cytology result of HSIL in the conservative management arm, and women who had either an entry HPV test result of positive for high-risk types or an entry cytology result of HSIL in the HPV triage arm. Participants in all study arms were scheduled to return for Pap smear and HPV testing every 6 months for 2 years. During follow-up, women were rereferred for colposcopy and biopsy if HSIL was identified. At the end of the study, participants were required to undergo an exit procedure including cytology, HPV testing, and colposcopic examination with biopsy of any visible lesions.
An ALTS participant was eligible for this study if HPV18 DNA was detected by polymerase chain reaction (PCR)–based reverse-line blot assay in her cervical swab sample at enrollment (28,29). The ALTS protocol was approved by the institutional review boards at the National Cancer Institute and at each of the four clinical centers involved in the trial, and written informed consent was obtained from all ALTS participants. The protocol for this ancillary study was also approved by the institutional review board at the University of Washington. Of 312 eligible women, 8 were excluded because their enrollment samples were not available for viral load quantification. We also excluded one woman whose enrollment sample was positive for HPV18 but was negative for cellular DNA, leaving 303 women in the analysis.
In ALTS, cervical cytology and histology were initially diagnosed by clinical center pathologists and then reviewed by a panel of expert pathologists. Histological diagnosis was made on tissues that were obtained by biopsy, endocervical curettage, and/or excision procedure. The most severe diagnosis was assigned if there was more than one tissue block examined at a single visit. The clinical endpoint used for this study was based on diagnoses by the panel of expert pathologists. The results did not change appreciably when the diagnosis by the clinical center pathologists was used (data not shown).
Quantification of HPV18 DNA Load
HPV18 DNA load in enrollment cervical swab sample was measured by multiplex real-time PCR. DNA was extracted and purified as previously described (30). The assay was set up in a reaction volume of 25 μL with the TaqMan Universal PCR Master Mix kit (Applied Biosystems, Foster City, CA). Sequences of primers and probe for the HPV18 E7 gene were as follows: forward primer, nucleotide position 686–710, 5′-GACTCAGAGGAAGAAAACGATGAAA; reverse primer, nucleotide position 766–748, 5′-GTGACGTTGTGGTTCGGCT; and probe, nucleotide position 715–739, 5′-TGGAGTTAATCATCAACATTTACCA. The probe was labeled with a fluorescence reporter dye (VIC, a proprietary fluorescent dye; Applied Biosystems) at the 5′ end and a quencher dye (6-carboxy-tetramethyl-rhodamine) at the 3′ end. The primers and probe for the β-actin gene (cellular DNA) were commercially available (Applied Biosystems). PCR amplification was carried out on Applied Biosystems 7900 HT Sequence Detection System with a cycling program of holding at 50°C for 2 minutes and then at 95°C for 10 minutes followed by a two-step cycle of 10 seconds at 95°C and 1 minute at 60°C for 40 cycles.
Two logarithmic-phase five-point standard curves were implemented in each set of real-time PCR assays—one for HPV18 DNA and the other for cellular DNA. The viral load was normalized to the input amount of cellular DNA and expressed as HPV18 E7 copy number per nanogram of cellular DNA. Each sample was assayed in triplicate, and the mean value from the three triplicate measurements was used for analysis. The estimated reliability of the mean of the triplicate measures was 0.96 with an intraclass correlation coefficient, r = .89 (95% confidence interval [CI] = 0.87 to 0.91), as assessed by one-way ANOVA with random effects.
HPV18 E7 DNA was not detected by real-time PCR in 22 cervical samples that tested positive by initial PCR-based reverse-line blot assay. The negative result was not explained by a lack of sufficient sample DNA input or presence of inhibitors because the amount of cellular DNA in samples with and without detectable HPV18 E7 DNA was similar (data not shown). Considering that the result might be due to a tiny amount of HPV18 DNA in the sample, a value of one viral copy per nanogram of cellular DNA was arbitrarily assigned to each of these samples. Similar results were obtained when these samples were excluded from the analysis (data not shown).
Statistical Analyses
The quantity of HPV18 DNA at enrollment was treated as a continuous variable, and log10-transformed values were used for statistical analysis. Because hierarchically HPV16 confers a higher risk of CIN2–3 than does HPV18, the results were presented separately for all HPV18-positive women and for those without HPV16 coinfection at enrollment.
The cross-sectional associations of HPV18 DNA load with risk of abnormal cervical cytology at enrollment were measured by use of risk ratios (RRs), with normal cervical cytology as the reference group. The risk ratios and 95% confidence intervals were estimated using log regression analysis (31) and adjusted for age at enrollment (18–24 vs ≥25 years) and coinfection with other high-risk HPV types, which include types 16, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 (yes or no).
A linear regression model (32) was used to compare cytology-stratified HPV18 DNA load between women with CIN2–3 and those without CIN2–3 and between women with CIN3 and those without CIN2–3, while controlling for age at enrollment and coinfection with other high-risk HPV types. To minimize the possibility of verification bias being introduced by missing and/or delayed diagnoses at enrollment, the 2-year cumulative diagnosis of CIN2–3 (or CIN3) was used as the endpoint. For women with CIN2–3 (or CIN3) that was detected at more than one time point, only the first diagnosis was counted.
Among CIN2–3 patients, we examined HPV18 DNA load at enrollment when the initial CIN2–3 diagnosis occurred (at enrollment vs during follow-up). To minimize a possible bias being introduced by the study arm–related referral of entry colposcopy and biopsy, this analysis was restricted to women who were enrolled in the immediate colposcopy or HPV triage arm. Per ALTS protocol, women in these two arms who tested positive for high-risk HPV types at enrollment were required to undergo an entry colposcopic examination.
To examine the association between HPV18 DNA load at enrollment and risk of concurrent CIN2–3 in this subset of women, we estimated risk ratios and 95% confidence intervals using log regression analyses. A Cox proportional hazards regression model (33) was used to estimate hazard ratios (HRs) of CIN2–3 initially diagnosed during follow-up associated with HPV18 DNA load at enrollment. The latter analysis was restricted to women without histologically confirmed CIN2–3 at enrollment. In this analysis, time to the event was measured from the date of study entry to the onset of CIN2–3. The time of onset was defined as the midpoint between the visit at which CIN2–3 was initially diagnosed and the most recent previous visit. Women who did not develop CIN2–3 were censored at the date of their last visit. The proportional hazards assumption of the Cox regression was examined by the rank test; no violation was found.
We used Student t test or one-way ANOVA, whichever was appropriate, to compare HPV18 DNA load by age at enrollment, study arm, race (white or nonwhite), current smoking status (yes or no), current use of hormonal contraceptives (yes or no), lifetime number of sex partners (0–5 or ≥6), HPV18 variant (European, African, or Asian American), and coinfection with other high-risk HPV types (yes or no). A linear trend for increase in HPV18 DNA load with increasing severity of cervical cytology at enrollment was tested by assigning scores to cytological diagnoses and treating this scored factor as a continuous variable. P values were two-sided; comparisons for which P was less than .05 were considered statistically significantly different.
Results
Among 303 women who tested positive for HPV18 at enrollment, the geometric mean of HPV18 DNA load (expressed as HPV18 E7 copy number per nanogram of cellular DNA) was 7.37 × 103 (95% CI = 4.78 × 103 to 1.13 × 104). Abnormal cytology at enrollment was detected in 243 (80.2%, 95% CI = 75.3% to 84.5%) of 303 women, including 82 (27.1%) with ASC-US, 127 (41.9%) with LSIL, and 34 (11.2%) with HSIL (Table 1). Among women who had normal cytology at enrollment and no evidence of CIN2–3 during the 2-year follow-up interval, the mean log10-transformed HPV18 DNA load at enrollment differed statistically significantly between those with and without coinfection with other high-risk HPV types (2.03 vs 3.36, difference = 1.33, 95% CI = 0.38 to 2.27; P = .007). No appreciable difference in HPV18 DNA load was observed by age at enrollment, race, lifetime number of sex partners, current smoking status, current use of hormonal contraceptives, HPV18 variant, and study arm (data not shown).
Table 1.
Associations between abnormal cervical cytology and a 1-unit increase in log10-transformed HPV18 DNA load at enrollment*
| Cytology at enrollment | No. | Mean (SD) of log10 HPV18 E7 copy number per nanogram of cellular DNA | Crude RR (95% CI) | Adjusted RR (95% CI)† |
| All women | ||||
| Within normal limits | 60 | 2.82 (±1.70) | 1.00 (Referent) | 1.00 (Referent) |
| ASC-US | 82 | 3.87 (±1.48) | 1.51 (1.20 to 1.91) | 1.61 (1.26 to 2.06) |
| LSIL | 127 | 4.19 (±1.59) | 1.64 (1.33 to 2.01) | 1.85 (1.46 to 2.33) |
| HSIL | 34 | 4.49 (±1.51) | 2.01 (1.39 to 2.90) | 2.49 (1.64 to 3.78) |
| Women without HPV16 coinfection | ||||
| Within normal limits | 52 | 3.05 (±1.57) | 1.00 (Referent) | 1.00 (Referent) |
| ASC-US | 72 | 3.95 (±1.44) | 1.50 (1.16 to 1.95) | 1.57 (1.19 to 2.07) |
| LSIL | 107 | 4.26 (±1.61) | 1.59 (1.26 to 1.99) | 1.78 (1.38 to 2.29) |
| HSIL | 23 | 4.79 (±1.47) | 2.34 (1.46 to 3.77) | 2.67 (1.61 to 4.45) |
HPV = human papillomavirus; SD = standard deviation; RR = risk ratio (a ratio of the probability of having abnormal cervical cytology associated with a 1-unit increase in log10-transformed HPV18 DNA load); CI = confidence interval; ASC-US = atypical squamous cells of undetermined significance; LSIL = low-grade squamous intraepithelial lesion; HSIL = high-grade squamous intraepithelial lesion.
Adjusted for age at enrollment and coinfection with other high-risk HPV types (ie, HPV16, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 for all women and HPV31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 for those without coinfection with HPV16).
After adjusting for age at enrollment and coinfection with other high-risk HPV types, the risk ratios of cytological interpretations of ASC-US, LSIL, and HSIL (as compared with within normal limits) per 1-unit increase of log10-transformed HPV18 DNA load at enrollment were 1.61 (95% CI = 1.26 to 2.06), 1.85 (95% CI = 1.46 to 2.33), and 2.49 (95% CI = 1.64 to 3.78), respectively (Table 1). The risk ratios remained similar when the analysis was restricted to women without HPV16 coinfection at enrollment. Among 76 women who were infected with HPV18 as the only HPV type at enrollment, the viral load remained higher in women with abnormal cytology than for those with normal enrollment cytology (RR = 1.37, 95% CI = 1.00 to 1.89).
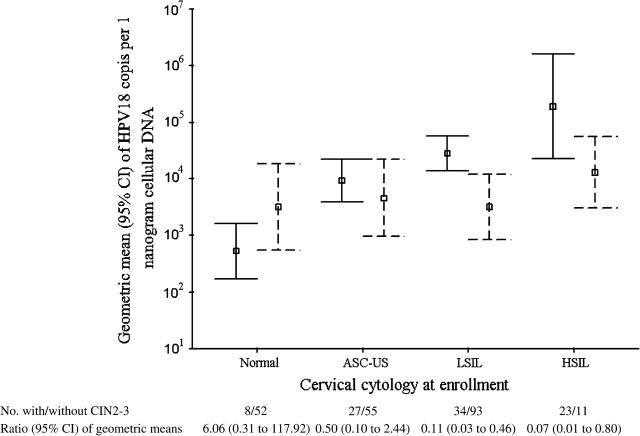
During the 2-year study period, CIN2–3 was histologically confirmed in 92 (30.4%, 95% CI = 25.2% to 35.9%) of the 303 HPV18-positive women: eight of 60 (13.3%) with normal cytology at enrollment, 27 of 82 (32.9%) with ASC-US, 34 of 127 (26.8%) with LSIL, and 23 of 34 (67.6%) with HSIL. We calculated the geometric means of cytology-stratified HPV18 DNA load between women with and without CIN2–3 during the 2-year study period (Figure 1). In contrast to a trend of increasing HPV18 DNA load with increasing severity of cervical cytology at enrollment among women without a diagnosis of CIN2–3 (Ptrend < .001), the viral load across cytological diagnoses at enrollment was comparable among those with CIN2–3 (Ptrend = .33). The ratios of geometric means of HPV18 DNA load at enrollment among women with CIN2–3 relative to those without were 6.06 (95% CI = 0.31 to 117.92) for those with normal, 0.50 (95% CI = 0.10 to 2.44) with ASC-US, 0.11 (95% CI = 0.03 to 0.46) for those with LSIL, and 0.07 (95% CI = 0.01 to 0.80) for those with HSIL cytology at enrollment, respectively. Among women with no evidence of CIN2–3 during the 2-year follow-up period, HPV18 DNA load at enrollment was not appreciably associated with a likelihood of becoming HPV18 negative at month 12 or month 24 visit (data not shown).
Figure 1.
Geometric means (squares) and 95% confidence intervals (CIs; upper and lower bound) of human papillomavirus (HPV) type 18 DNA load at enrollment between women with (dashed line) and without (solid line) cervical intraepithelial neoplasia grades 2–3 (CIN2–3) during the 2-year study period. Among women without CIN2–3, the geometric mean of HPV18 E7 copy number per nanogram of cellular DNA was 522 (95% CI = 172 to 1598) for those with normal cytology, 9309 (95% CI = 4078 to 21 251) for those with atypical squamous cells of undetermined significance (ASC-US), 28 045 (95% CI = 13 900 to 56 621) for those with low-grade squamous intraepithelial lesion (LSIL), and 189 409 (95% CI = 28 976 to 1 238 112) for those with high-grade squamous intraepithelial lesion (HSIL) at enrollment (Ptrend < .001); the corresponding values among women with CIN2–3 were 3166 (95% CI = 741 to 13,534), 4623 (95% CI = 1050 to 20 350), 3151 (95% CI = 881 to 11 277), and 12 891 (95% CI = 3280 to 50 672), respectively (Ptrend < .33). P values (two-sided) were calculated with a trend test.
HPV18 DNA load at enrollment was statistically significantly lower among women with CIN2–3 than those without CIN2–3 who had either LSIL (Pcrude = .002) or HSIL cytology (Pcrude = .04) at enrollment (Table 2). After adjusting for age at enrollment and coinfection with other high-risk HPV types, the difference remained statistically significant among women with LSIL at enrollment (Padjusted = .009). We observed no difference in HPV18 DNA load between women with and without CIN2–3 who had normal cytology or ASC-US at enrollment. Similar patterns of HPV18 DNA load were observed when the patient group was restricted to 53 women with a diagnosis of CIN3. When the analysis was restricted to women without HPV16 coinfection at enrollment, the results did not change considerably, except that the difference between women with CIN3 and those without CIN2–3 was statistically significant among those with HSIL at enrollment (Padjusted = .03) and was close to being statistically significant among those with LSIL at enrollment (Padjusted = .06). When women with a cytological diagnosis of LSIL and HSIL were grouped, the association of lower HPV18 DNA load with underlying CIN2–3 was statistically significant overall in women with squamous intraepithelial lesion (SIL) cytology at enrollment (Padjusted = .02) or in those without HPV16 coinfection at enrollment (Padjusted = .04). Among those infected with HPV18 as the only HPV type at enrollment, the HPV18 DNA load (expressed as the mean log10-transformed HPV18 DNA copy number per nanogram of cellular DNA) was similar between the 10 women with and the 34 women without a diagnosis of 2-year cumulative CIN2–3 (4.11 vs 3.77, difference = 0.34, 95% CI = –0.83 to 1.51; P = .56) who had normal cytology or ASC-US at enrollment, but the HPV18 DNA load was lower for the three women with than the 29 women without a diagnosis of 2-year cumulative CIN2–3 (4.39 vs 5.28, difference = –0.89, 95% CI = –2.58 to 0.81; P = .29) who had SIL at enrollment.
Table 2.
HPV18 DNA load at enrollment of women with and without CIN2–3 during the 2-year study period: results stratified by cervical cytology at enrollment*
| Cytology at enrollment | Women without CIN2–3 | Women with CIN2–3 | Women with CIN3 | |||||||
| No. | Mean load (±SD) | No. | Mean load (±SD) | Pcrude | Padjusted | No. | Mean load (±SD) | Pcrude | Padjusted | |
| All women | ||||||||||
| Overall | 211 | 3.94 (±1.70) | 92 | 3.70 (±1.57) | .08 | .21 | 53 | 3.69 (1.35) | .10 | .24 |
| Within normal limits | 52 | 2.72 (±1.78) | 8 | 3.50 (±0.91) | .18 | .26 | 3 | 3.23 (0.44) | .56 | .58 |
| ASC-US | 55 | 3.97 (±1.36) | 27 | 3.66 (±1.71) | .40 | .47 | 14 | 3.58 (1.78) | .39 | .53 |
| LSIL | 93 | 4.45 (±1.50) | 34 | 3.50 (±1.65) | .002 | .009 | 19 | 3.66 (1.36) | .04 | .06 |
| HSIL | 11 | 5.28 (±1.38) | 23 | 4.11 (±1.45) | .04 | .18 | 17 | 3.90 (1.07) | .02 | .11 |
| Women without HPV16 coinfection | ||||||||||
| Overall | 190 | 4.03 (±1.65) | 63 | 3.82 (±1.54) | .23 | .29 | 31 | 3.74 (1.21) | .24 | .28 |
| Within normal limits | 45 | 2.96 (±1.64) | 7 | 3.61 (±0.92) | .29 | .40 | 2 | 3.48 (0.06) | .63 | .72 |
| ASC-US | 52 | 3.95 (±1.39) | 19 | 3.95 (±1.61) | 1.00 | .93 | 7 | 4.33 (1.34) | .53 | .56 |
| LSIL | 82 | 4.49 (±1.54) | 25 | 3.53 (±1.65) | .006 | .01 | 14 | 3.63 (1.43) | .05 | .06 |
| HSIL | 11 | 5.28 (±1.38) | 12 | 4.33 (±1.45) | .14 | .27 | 8 | 3.50 (0.72) | .01 | .03 |
HPV18 DNA load was expressed as log10 (HPV18 E7 copies per nanogram of cellular DNA). Data were adjusted for age at enrollment and coinfection with other high-risk HPV types (ie, HPV16, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 for all women and HPV31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 for those without coinfection with HPV16). CIN = cervical intraepithelial neoplasia; SD = standard deviation; ASC-US = atypical squamous cells of undetermined significance; LSIL = low-grade squamous intraepithelial lesion; HSIL = high-grade squamous intraepithelial lesion; HPV = human papillomavirus. P values (two-sided) were calculated using linear regression analyses.
Fifty of 303 (17%) women exited the trial before the scheduled last visit. The number of women examined at month 6, 12, 18, and 24 follow-up visits was 257, 245, 235, and 253, respectively. SIL was detected at enrollment in 66% (62% with LSIL and 4% with HSIL) of the 50 women who did not complete their last visit and in 50.5% (38% with LSIL and 13% with HSIL) of the 253 women who did (P = .05). The rate of 2-year cumulative CIN2–3 was similar among 125 women who did and 17 women who did not complete their last visit (25% vs 24%, P = .91) who had normal cytology or ASC-US at enrollment but was higher for 128 women who did than for 33 women who did not complete their last visit (42% vs 9%, difference = 33%, 95% CI = 20% to 46%; P < .001) who had SIL at enrollment. Among those with SIL at enrollment, the HPV18 DNA load (expressed as the mean value of log10-transformed HPV18 DNA load) was comparable between women who completed their last visit and those who did not (4.24 vs 4.33, P = .75). Additional adjustment for number of follow-up visits or restricting analysis to women without CIN2–3 to those who had completed their last visit did not change the results appreciably (data not shown). HPV18 DNA became undetectable in seven of 92 CIN2–3 patients at the time concurrent with and one visit before the one in which the lesion was diagnosed. Considering that their CIN2–3 lesion might be HPV18 unrelated, these seven patients were excluded from the analysis. The exclusion did not appreciably change our findings (data not shown).
The analysis of HPV18 DNA load at enrollment by the timing of CIN2–3 diagnosis was restricted to women who were enrolled in the immediate colposcopy or HPV triage arm. CIN2–3 was histologically confirmed in 63 of 188 (33.5%) women during the 2-year study period. Because few women were diagnosed with CIN2–3 during follow-up and the patterns of HPV18 DNA load by histopathology were similar among women with LSIL and HSIL cytology at enrollment, these patient groups were combined (SIL) for statistical analysis. HPV18 DNA load was similar among CIN2–3 patients who were diagnosed at enrollment and during follow-up, regardless of cytology results at enrollment (Table 3). The results remained similar when the analysis was restricted to CIN3 patients (data not shown). In this subset of women, HPV18 DNA load at enrollment continued to be lower in those with CIN2–3 than without CIN2–3 who had SIL cytology at enrollment (P = .003 for all women; P = .02 for those without HPV16 coinfection).
Table 3.
HPV18 DNA load at enrollment between patients who were initially diagnosed with CIN2–3 at enrollment and those diagnosed during follow-up who were enrolled in the immediate colposcopy or HPV triage arm*
| Cytology at enrollment | Women without CIN2–3 | Women with entry CIN2–3 | Women with follow-up CIN2–3 | P† | P‡ | |||
| No. | Mean load (±SD) | No. | Mean load (±SD) | No. | Mean load (±SD) | |||
| All women | ||||||||
| Overall | 125 | 3.95 (±1.80) | 46 | 3.59 (±1.66) | 17 | 3.89 (±1.09) | .53 | .30 |
| Within normal limits | 33 | 2.66 (±1.82) | 2 | 3.51 (±0.95) | 5 | 3.51 (±1.11) | 1.00 | .24 |
| ASC-US | 33 | 4.06 (±1.64) | 16 | 3.61 (±2.01) | 5 | 4.38 (±1.27) | .44 | .58 |
| LSIL | 51 | 4.51 (±1.50) | 18 | 3.41 (±1.61) | 6 | 3.89 (±1.05) | .73 | .003 |
| HSIL | 8 | 5.17 (±1.42) | 10 | 3.91 (±1.38) | 1 | 3.34 | — | — |
| Women without HPV16 coinfection | ||||||||
| Overall | 110 | 4.01 (±1.77) | 32 | 3.78 (±1.58) | 14 | 4.00 (±1.15) | .71 | .58 |
| Within normal limits | 29 | 2.88 (±1.69) | 2 | 3.51 (±0.95) | 4 | 3.70 (±1.18) | .86 | .30 |
| ASC-US | 30 | 4.04 (±1.72) | 11 | 3.91 (±1.90) | 5 | 4.38 (±1.27) | .62 | .99 |
| LSIL | 43 | 4.53 (±1.55) | 12 | 3.57 (±1.45) | 5 | 3.87 (±1.17) | .86 | .02 |
| HSIL | 8 | 5.17 (±1.42) | 7 | 4.01 (±1.63) | 0 | — | — | — |
HPV18 DNA load was expressed as log10 (HPV18 E7 copies per nanogram of cellular DNA). A category of LSIL was combined with HSIL for statistical testing. CIN2–3 = cervical intraepithelial neoplasia grades 2–3; SD = standard deviation; ASC-US = atypical squamous cells of undetermined significance; LSIL = low-grade squamous intraepithelial lesion; HSIL = high-grade squamous intraepithelial lesion; — = values missing because they were not available or not applicable; HPV = human papillomavirus.
Comparison of HPV18 DNA load at enrollment between CIN2–3 patients who were initially diagnosed at enrollment and those diagnosed during follow-up. P values (two-sided) were calculated using a Student t test.
Comparison of HPV18 DNA load at enrollment between women with and without CIN2–3 during the 2-year study period. P values (two-sided) were calculated using a Student t test.
We further examined the associations of HPV18 DNA load at enrollment with concurrent or subsequent diagnoses of CIN2–3 among women who were enrolled in the immediate colposcopy or HPV triage arm. The risk ratio of having a CIN2–3 diagnosis at enrollment per 1-unit increase in log10-transformed HPV18 DNA load was 0.79 (95% CI = 0.68 to 0.92) for women with SIL cytology at enrollment (Table 4). For the evaluation of risk for progression to CIN2–3, we excluded 46 women who had CIN2–3 at enrollment and eight women who did not complete a follow-up visit, leaving 134 in the analysis. The inverse association between HPV18 DNA load at enrollment and risk of CIN2–3 that was diagnosed during follow-up for women with SIL cytology at enrollment was marginally statistically significant (per 1-unit increase in log10-transformed HPV18 DNA load, HR = 0.48, 95% CI = 0.21 to 1.06). Among women who had normal cytology or ASC-US at enrollment, neither concurrent nor subsequent risk of CIN2–3 was associated with HPV18 DNA load at enrollment. The associations remained similar when the analysis was restricted to women without HPV16 coinfection at enrollment.
Table 4.
Association of per 1-unit increase in log10-transformed HPV18 DNA load at enrollment with risk of concurrent or subsequent diagnoses of CIN2–3 among women who were enrolled in the immediate colposcopy or HPV triage arm*
| Cytology at enrollment | No. with CIN2–3 at enrollment/No. of subjects | RR (95% CI) | No. with follow-up CIN2–3/person-years at risk | HR (95% CI) |
| All women | ||||
| Within normal limits or ASC-US | 18/94 | 1.04 (0.82 to 1.32) | 10/153.4 | 1.10 (0.74 to 1.64) |
| LSIL or HSIL | 28/94 | 0.79 (0.68 to 0.92) | 7/114.9 | 0.48 (0.21 to 1.06) |
| Women without HPV16 coinfection | ||||
| Within normal limits or ASC-US | 13/81 | 1.09 (0.81 to 1.48) | 9/138.4 | 1.16 (0.75 to 1.78) |
| LSIL or HSIL | 19/75 | 0.80 (0.65 to 0.99) | 5/98.2 | 0.53 (0.22 to 1.23) |
CIN2–3 = cervical intraepithelial neoplasia grades 2–3; RR = risk ratio (a ratio of the probability of having CIN2–3 at enrollment associated with per 1-unit increase in log10-transformed HPV18 DNA load); CI = confidence interval; HR = hazard ratio; ASC-US = atypical squamous cells of undetermined significance; LSIL = low-grade squamous intraepithelial lesion; HSIL = high-grade squamous intraepithelial lesion; HPV = human papillomavirus.
Discussion
We found that among ALTS participants with HPV18 infection at enrollment, viral load was associated with the severity of concurrent cervical cytology, a result that is consistent with previous reports (21,22,34). Because the measurement of HPV18 DNA load was performed without knowledge of any clinical or epidemiological information and the diagnosis of cervical disease was provided by a panel of expert pathologists, potential biases in ascertainments of HPV18 DNA load and cervical lesion were minimized. The association of HPV18 DNA load with abnormal cervical cytology was not surprising. As shown in a study of women with CIN3 (35), HPV DNA load was closely associated with the number of ASC-US and LSIL cells present in exfoliative cervical samples. This study further demonstrates that the associations between HPV18 DNA load and cytological abnormalities differ by underlying histopathology: In contrast to a statistically significant stepwise increase in HPV18 DNA load at enrollment with increasing severity of cervical cytology among women without CIN2–3, the viral load was relatively constant across cytological diagnoses among those with CIN2–3.
The most striking finding of this study was that among women with LSIL or HSIL cytology at enrollment, HPV18 DNA load was statistically significantly lower in those with than those without a diagnosis of CIN2–3 during the 2-year follow-up period. This difference was not explained by age at enrollment or coinfection with HPV16 or other high-risk HPV types. It was not explained by incomplete outcome ascertainment or by misclassification of non–HPV18-relevant to HPV18-relevant CIN2–3 because the results remained similar when the analysis was performed with additional adjustment for number of follow-up visits, was restricted to women without CIN2–3 who had completed the scheduled last visit, or was restricted to CIN2–3 patients who continued to be HPV18 positive at the time concurrent with or one visit before the one in which CIN2–3 was diagnosed. We noticed that among those with SIL cytology at enrollment, the rate of CIN2–3 diagnosis during the 2-year follow-up period was higher for women who completed the last visit than for those who did not. If a high viral load had been related to missing diagnoses of CIN2–3, the association of CIN2–3 with lower HPV18 DNA load among women with SIL cytology at enrollment would have been biased. Arguing against the possibility of higher viral load being associated with missing diagnoses of CIN2–3 is the fact that the overall HPV18 DNA load was similar between women with and without completion of their last visit who had SIL cytology at enrollment.
A possible interpretation of our findings is that we observed fluctuations of HPV18 DNA load that were based on the natural course of infection and the cervical location of HPV18-related neoplasia. The group of women with normal enrollment cytology and no evidence of CIN2–3 during follow-up might have had a lower average level of HPV18 DNA than the other groups of women because their cervical infections might have been resolving or might have been from a noncervical transformation zone site (eg, the vaginal wall). The group of women with SIL cytology at enrollment and no CIN2–3 during follow-up had a higher average level of HPV18 DNA than all other groups of women because they might have been more likely to have had acute and productive HPV18 infection. Finally, the group of women with CIN2–3 (regardless of enrollment cytology) had an average level of HPV18 DNA that was lower than that for those with SIL cytology and no histological evidence of CIN2–3. This lower level of HPV18 DNA might have occurred because HPV18-related precancerous lesions and cancers tend to develop in columnar epithelial cells that can be difficult to sample with swabs, brushes, brooms, or spatulas. Such cells, which are often glandular in origin, are found in the endocervical canal and the clefts and crypts of the transformation zone. As shown in previous studies (36–39), the frequency of positive detection is usually higher for HPV18 than for HPV16 in adenocarcinoma in situ of the cervix, which is histologically characterized by epitheliomatous transformation of endocervical glands.
Another possible interpretation for the lower average HPV18 DNA load in the group of women with CIN2–3 than the group with cytological evidence of a lesion that was not confirmed as CIN2–3 is that factors associated with underlying CIN2–3, such as cell type–related and integration-related reduction in viral replication, might somewhat limit the ability of the virus to replicate. Unlike the multilayers of squamous epithelial cells where the HPV lifecycle is closely linked to epithelial differentiation (from basal layers up to superficial layers), the columnar epithelial cells have only a single layer. The HPV lifecycle in the single layer of epithelium is virtually unknown. If the virus is less capable of productive replication in the columnar epithelial cells and HPV18 preferentially induces neoplastic transformation of these cells, our observation might be somewhat explained by the behavior of the virus in these cells. Integration of viral DNA into the host genome occurs preferentially with disruption of the coding sequence for the E2 protein. The E2 protein behaves not only as a transcriptional regulator to modulate viral transcription (40–45) but also as an activator of viral DNA replication by interacting cooperatively with the viral helicase E1 at the origin of replication (46,47). Thus, although an integration-induced disruption of the E2 gene plays a role in the development of precancerous lesion, it may lead to a decrease in replication of viral DNA. However, the published data (48–50) usually suggest that most cases of HPV18-positive CIN2–3 do not have a detectable integration of viral DNA, although a high frequency of integration in women with CIN3 or carcinoma in situ was also reported (51,52). Whether the comparable HPV18 DNA load across cytological diagnoses at enrollment among women with underlying CIN2–3 is a result of the balance between the abnormal cytology-related increase in viral load and the integration-related loss of viral DNA replication deserves further investigation.
The finding that women who were diagnosed with CIN2–3 at enrollment and those diagnosed during the 2-year follow-up had similar HPV18 DNA load at enrollment further suggests that suppression of viral DNA replication (either due to HPV18 infection of progenitor glandular cells that are difficult to sample and weakly permissive for viral replication or to increased integration) might explain the lower than expected risk of CIN2–3 than cervical cancer associated with detectable HPV18 infection (3). This clinical phenomenon has been poorly understood so far.
In our previous study of ALTS participants with HPV16 infection at enrollment, we found that HPV16 DNA levels were higher in women with CIN2–3 than those without CIN2–3 (53), suggesting that the pattern of viral load by histopathology is related to HPV type. Although the underlying mechanism for this difference in HPV16 and HPV18 DNA levels and detection of CIN2–3 is not clear, it does suggest that extensive examination of precancerous and benign lesions for DNA levels of all high-risk HPV types must be undertaken before the clinical usefulness of measuring HPV level can be determined.
Several limitations of this study should be addressed. First, women who were eligible for participation in ALTS were required to have a Pap smear showing ASC-US or LSIL within 6 months of screening. Thus, a cytological interpretation of within normal limits, ASC-US or LSIL, and HSIL at enrollment when the viral load was measured can also be viewed as cytological regression, persistent mild abnormality, and cytological progression, respectively. Also, due to intensive procedures for follow-up and clinical examination implemented in ALTS, the lesions identified in this study were expected to be smaller than those seen in the general population. Therefore, HPV18 DNA load detected in a particular cytological category or among CIN2–3 patients may not be generalizable to that present in the general population. However, this lack of generalizability does not affect the validity of relative comparisons of cytology-stratified HPV18 DNA load between women with and without underlying CIN2–3. Second, HPV18 DNA load was assessed on enrollment samples from women with prevalent HPV18 infection. The viral load may naturally fluctuate during the course of the infection. Although the analysis of consecutive samples may provide additional insights into the dynamics of HPV18 DNA load, the comparison of groups at the same time point is still valid. Third, the cervical samples that were used for this study tested HPV18 positive by PCR-based reverse-line blot assay. Thus, we were not measuring very low viral loads that would be missed by this assay. Exclusion of these infections may lead to overestimates of the mean value of viral load. However, this overestimation is likely to occur in each group of women and therefore does not affect the validity of relative comparisons. Finally, approximately 17% of women who tested HPV18 positive withdrew from the trial before the scheduled last visit. Biases could have been introduced had a loss to follow-up been differentially related to HPV18 DNA load. However, there was no appreciable difference in HPV18 DNA load between women who completed the last visit and those who did not, and additional adjustment for number of follow-up visits did not change the estimates appreciably.
In summary, our data indicated that HPV18 DNA levels were highest among women with evidence of a benign squamous intraepithelial lesion, intermediate among those with CIN2–3, and lowest among those with normal cytological findings. Thus, testing for high levels of HPV18 DNA does not appear to be clinically useful. Further studies to examine possible mechanisms involving lower HPV18 DNA levels in women with CIN2–3 than those with benign squamous intraepithelial lesions are of interest.
Funding
The work was supported by a Public Health Service grant (CA 84396 to LFX). ALTS was supported by the National Cancer Institute, National Institutes of Health Department of Health and Human Services contracts CN-55153, CN-55154, CN-55155, CN-55156, CN-55157, CN-55158, CN-55159, and CN-55105 and by the Intramural Research Program of the National Cancer Institute. In conducting ALTS, some of the equipment and supplies used in these studies were donated or provided at reduced cost by Digene Corporation (Gaithersburg, MD); Cytyc Corporation (Boxborough, MA); National Testing Laboratories (Fenton, MO); DenVu (Tucson, AZ); TriPath Imaging, Inc. (Burlington, NC); and Roche Molecular Systems, Inc. (Alameda, CA). C. M. Wheeler received reagents from Roche Molecular Systems, Inc. and funds to conduct HPV vaccine trial from Merck and GlaxoSmith Kline. L. A. Koutsky received research funds from Merck Research Laboratories.
Footnotes
This study was part of the project ancillary to the ALTS clinical trial but does not represent the views of the ALTS Group. We would like to thank the ALTS Group for providing the biological specimens and HPV typing results and Dr Kathrin Jansen for providing the HPV18 DNA standard. We thank the ALTS Group Investigators for their help in planning and in conducting the trial. We thank Information Management Services, Inc. (Rockville, MD), for data management and programming support.
The sponsors had no role in the study design, the data collection and analysis, the interpretation of the results, the preparation of the manuscript, or the decision to submit the manuscript for publication.
References
- 1.Munoz N, Bosch FX, de Sanjose S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348(6):518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 2.Smith JS, Lindsay L, Hoots B, et al. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: a meta-analysis update. Int J Cancer. 2007;121(3):621–632. doi: 10.1002/ijc.22527. [DOI] [PubMed] [Google Scholar]
- 3.Bulk S, Berkhof J, Rozendaal L, et al. The contribution of HPV18 to cervical cancer is underestimated using high-grade CIN as a measure of screening efficiency. Br J Cancer. 2007;96(8):1234–1236. doi: 10.1038/sj.bjc.6603693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woodman CB, Collins S, Rollason TP, et al. Human papillomavirus type 18 and rapidly progressing cervical intraepithelial neoplasia. Lancet. 2003;361(9351):40–43. doi: 10.1016/S0140-6736(03)12120-4. [DOI] [PubMed] [Google Scholar]
- 5.Kurman RJ, Schiffman MH, Lancaster WD, et al. Analysis of individual human papillomavirus types in cervical neoplasia: a possible role for type 18 in rapid progression. Am J Obstet Gynecol. 1988;159(2):293–296. doi: 10.1016/s0002-9378(88)80070-x. [DOI] [PubMed] [Google Scholar]
- 6.Bergeron C, Barrasso R, Beaudenon S, Flamant P, Croissant O, Orth G. Human papillomaviruses associated with cervical intraepithelial neoplasia. Great diversity and distinct distribution in low- and high-grade lesions. Am J Surg Pathol. 1992;16(7):641–649. doi: 10.1097/00000478-199207000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Kovacic MB, Castle PE, Herrero R, et al. Relationships of human papillomavirus type, qualitative viral load, and age with cytologic abnormality. Cancer Res. 2006;66(20):10112–10119. doi: 10.1158/0008-5472.CAN-06-1812. [DOI] [PubMed] [Google Scholar]
- 8.Swan DC, Tucker RA, Tortolero-Luna G, et al. Human papillomavirus (HPV) DNA copy number is dependent on grade of cervical disease and HPV type. J Clin Microbiol. 1999;37(4):1030–1034. doi: 10.1128/jcm.37.4.1030-1034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ylitalo N, Sorensen P, Josefsson AM, et al. Consistent high viral load of human papillomavirus 16 and risk of cervical carcinoma in situ: a nested case-control study. Lancet. 2000;355(9222):2194–2198. doi: 10.1016/S0140-6736(00)02402-8. [DOI] [PubMed] [Google Scholar]
- 10.Josefsson AM, Magnusson PK, Ylitalo N, et al. Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet. 2000;355(9222):2189–2193. doi: 10.1016/S0140-6736(00)02401-6. [DOI] [PubMed] [Google Scholar]
- 11.van Duin M, Snijders PJ, Schrijnemakers HF, et al. Human papillomavirus 16 load in normal and abnormal cervical scrapes: an indicator of CIN II/III and viral clearance. Int J Cancer. 2002;98(4):590–595. doi: 10.1002/ijc.10232. [DOI] [PubMed] [Google Scholar]
- 12.Moberg M, Gustavsson I, Gyllensten U. Type-specific associations of human papillomavirus load with risk of developing cervical carcinoma in situ. Int J Cancer. 2004;112(5):854–859. doi: 10.1002/ijc.20480. [DOI] [PubMed] [Google Scholar]
- 13.Monnier-Benoit S, Dalstein V, Riethmuller D, Lalaoui N, Mougin C, Pretet JL. Dynamics of HPV16 DNA load reflect the natural history of cervical HPV-associated lesions. J Clin Virol. 2006;35(3):270–277. doi: 10.1016/j.jcv.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Fiander AN, Hart KW, Hibbitts SJ, et al. Variation in human papillomavirus type-16 viral load within different histological grades of cervical neoplasia. J Med Virol. 2007;79(9):1366–1369. doi: 10.1002/jmv.20875. [DOI] [PubMed] [Google Scholar]
- 15.Cricca M, Morselli-Labate AM, Venturoli S, et al. Viral DNA load, physical status and E2/E6 ratio as markers to grade HPV16 positive women for high-grade cervical lesions. Gynecol Oncol. 2007;106(3):549–557. doi: 10.1016/j.ygyno.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Bavin PJ, Giles JA, Deery A, et al. Use of semi-quantitative PCR for human papillomavirus DNA type 16 to identify women with high grade cervical disease in a population presenting with a mildly dyskaryotic smear report. Br J Cancer. 1993;67(3):602–605. doi: 10.1038/bjc.1993.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuzick J, Terry G, Ho L, Hollingworth T, Anderson M. Type-specific human papillomavirus DNA in abnormal smears as a predictor of high-grade cervical intraepithelial neoplasia. Br J Cancer. 1994;69(1):167–171. doi: 10.1038/bjc.1994.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalstein V, Riethmuller D, Pretet JL, et al. Persistence and load of high-risk HPV are predictors for development of high-grade cervical lesions: a longitudinal French cohort study. Int J Cancer. 2003;106(3):396–403. doi: 10.1002/ijc.11222. [DOI] [PubMed] [Google Scholar]
- 19.Moberg M, Gustavsson I, Wilander E, Gyllensten U. High viral loads of human papillomavirus predict risk of invasive cervical carcinoma. Br J Cancer. 2005;92(5):891–894. doi: 10.1038/sj.bjc.6602436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho CM, Chien TY, Huang SH, Lee BH, Chang SF. Integrated human papillomavirus types 52 and 58 are infrequently found in cervical cancer, and high viral loads predict risk of cervical cancer. Gynecol Oncol. 2006;102(1):54–60. doi: 10.1016/j.ygyno.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 21.Carcopino X, Henry M, Benmoura D, et al. Determination of HPV type 16 and 18 viral load in cervical smears of women referred to colposcopy. J Med Virol. 2006;78(8):1131–1140. doi: 10.1002/jmv.20673. [DOI] [PubMed] [Google Scholar]
- 22.Flores R, Papenfuss M, Klimecki WT, Giuliano AR. Cross-sectional analysis of oncogenic HPV viral load and cervical intraepithelial neoplasia. Int J Cancer. 2006;118(5):1187–1193. doi: 10.1002/ijc.21477. [DOI] [PubMed] [Google Scholar]
- 23.Snijders PJ, Hogewoning CJ, Hesselink AT, et al. Determination of viral load thresholds in cervical scrapings to rule out CIN 3 in HPV 16, 18, 31 and 33-positive women with normal cytology. Int J Cancer. 2006;119(5):1102–1107. doi: 10.1002/ijc.21956. [DOI] [PubMed] [Google Scholar]
- 24.Ho CM, Cheng WF, Chu TY, et al. Human papillomaviral load changes in low-grade squamous intraepithelial lesions of the uterine cervix. Br J Cancer. 2006;95(10):1384–1389. doi: 10.1038/sj.bjc.6603430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlecht NF, Trevisan A, Duarte-Franco E, et al. Viral load as a predictor of the risk of cervical intraepithelial neoplasia. Int J Cancer. 2003;103(4):519–524. doi: 10.1002/ijc.10846. [DOI] [PubMed] [Google Scholar]
- 26.Schiffman M, Adrianza ME. ASCUS-LSIL Triage Study. Design, methods and characteristics of trial participants. Acta Cytol. 2000;44(5):726–742. doi: 10.1159/000328554. [DOI] [PubMed] [Google Scholar]
- 27.The ASCUS-LSIL Traige Study (ALTS) Group. Results of a randomized trial on the management of cytology interpretations of atypical squamous cells of undetermined significance. Am J Obstet Gynecol. 2003;188(6):1383–1392. doi: 10.1067/mob.2003.457. [DOI] [PubMed] [Google Scholar]
- 28.Gravitt PE, Peyton CL, Alessi TQ, et al. Improved amplification of genital human papillomaviruses. J Clin Microbiol. 2000;38(1):357–361. doi: 10.1128/jcm.38.1.357-361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peyton CL, Gravitt PE, Hunt WC, et al. Determinants of genital human papillomavirus detection in a US population. J Infect Dis. 2001;183(11):1554–1564. doi: 10.1086/320696. [DOI] [PubMed] [Google Scholar]
- 30.Xi LF, Kiviat NB, Hildesheim A, et al. Human papillomavirus type 16 and 18 variants: race-related distribution and persistence. J Natl Cancer Inst. 2006;98(15):1045–1052. doi: 10.1093/jnci/djj297. [DOI] [PubMed] [Google Scholar]
- 31.Blizzard L, Hosmer DW. The log multinomial regression model for nominal outcomes with more than two attributes. Biom J. 2007;49(6):889–902. doi: 10.1002/bimj.200610377. [DOI] [PubMed] [Google Scholar]
- 32.Slinker BK, Glantz SA. Multiple linear regression is a useful alternative to traditional analyses of variance. Am J Physiol. 1988;255(3 pt 2):R353–R367. doi: 10.1152/ajpregu.1988.255.3.R353. [DOI] [PubMed] [Google Scholar]
- 33.Cox DR. Regression models and life tables. J R Stat Soc (Ser B) 1972;34:187–220. [Google Scholar]
- 34.Gravitt PE, Burk RD, Lorincz A, et al. A comparison between real-time polymerase chain reaction and hybrid capture 2 for human papillomavirus DNA quantitation. Cancer Epidemiol Biomarkers Prev. 2003;12(6):477–484. [PubMed] [Google Scholar]
- 35.Sherman ME, Wang SS, Wheeler CM, et al. Determinants of human papillomavirus load among women with histological cervical intraepithelial neoplasia 3: dominant impact of surrounding low-grade lesions. Cancer Epidemiol Biomarkers Prev. 2003;12(10):1038–1044. [PubMed] [Google Scholar]
- 36.Bekkers RL, Bulten J, Wiersma-van Tilburg A, et al. Coexisting high-grade glandular and squamous cervical lesions and human papillomavirus infections. Br J Cancer. 2003;89(5):886–890. doi: 10.1038/sj.bjc.6601204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hadzisejdc I, Krasevic M, Haller H, Grahovac B. Distribution of human papillomavirus types in different histological subtypes of cervical adenocarcinoma. Coll Antropol. 2007;31(suppl 2):97–102. [PubMed] [Google Scholar]
- 38.Zielinski GD, Snijders PJ, Rozendaal L, et al. The presence of high-risk HPV combined with specific p53 and p16INK4a expression patterns points to high-risk HPV as the main causative agent for adenocarcinoma in situ and adenocarcinoma of the cervix. J Pathol. 2003;201(4):535–543. doi: 10.1002/path.1480. [DOI] [PubMed] [Google Scholar]
- 39.Duggan MA, Benoit JL, McGregor SE, Inoue M, Nation JG, Stuart GC. Adenocarcinoma in situ of the endocervix: human papillomavirus determination by dot blot hybridization and polymerase chain reaction amplification. Int J Gynecol Pathol. 1994;13(2):143–149. [PubMed] [Google Scholar]
- 40.Thierry F, Heard JM, Dartmann K, Yaniv M. Characterization of a transcriptional promoter of human papillomavirus 18 and modulation of its expression by simian virus 40 and adenovirus early antigens. J Virol. 1987;61(1):134–142. doi: 10.1128/jvi.61.1.134-142.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demeret C, Yaniv M, Thierry F. The E2 transcriptional repressor can compensate for Sp1 activation of the human papillomavirus type 18 early promoter. J Virol. 1994;68(11):7075–7082. doi: 10.1128/jvi.68.11.7075-7082.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong G, Broker TR, Chow LT. Human papillomavirus type 11 E2 proteins repress the homologous E6 promoter by interfering with the binding of host transcription factors to adjacent elements. J Virol. 1994;68(2):1115–1127. doi: 10.1128/jvi.68.2.1115-1127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan SH, Leong LE, Walker PA, Bernard HU. The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E6 promoter through displacement of Sp1 and TFIID. J Virol. 1994;68(10):6411–6420. doi: 10.1128/jvi.68.10.6411-6420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thierry F, Howley PM. Functional analysis of E2-mediated repression of the HPV18 P105 promoter. New Biol. 1991;3(1):90–100. [PubMed] [Google Scholar]
- 45.Romanczuk H, Thierry F, Howley PM. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J Virol. 1990;64(6):2849–2859. doi: 10.1128/jvi.64.6.2849-2859.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desaintes C, Demeret C. Control of papillomavirus DNA replication and transcription. Semin Cancer Biol. 1996;7(6):339–347. doi: 10.1006/scbi.1996.0043. [DOI] [PubMed] [Google Scholar]
- 47.Yasugi T, Benson JD, Sakai H, Vidal M, Howley PM. Mapping and characterization of the interaction domains of human papillomavirus type 16 E1 and E2 proteins. J Virol. 1997;71(2):891–899. doi: 10.1128/jvi.71.2.891-899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vinokurova S, Wentzensen N, Kraus I, et al. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 2008;68(1):307–313. doi: 10.1158/0008-5472.CAN-07-2754. [DOI] [PubMed] [Google Scholar]
- 49.Cullen AP, Reid R, Campion M, Lorincz AT. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasm. J Virol. 1991;65(2):606–612. doi: 10.1128/jvi.65.2.606-612.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hopman AH, Kamps MA, Smedts F, Speel EJ, Herrington CS, Ramaekers FC. HPV in situ hybridization: impact of different protocols on the detection of integrated HPV. Int J Cancer. 2005;115(3):419–428. doi: 10.1002/ijc.20862. [DOI] [PubMed] [Google Scholar]
- 51.Hudelist G, Manavi M, Pischinger KI, et al. Physical state and expression of HPV DNA in benign and dysplastic cervical tissue: different levels of viral integration are correlated with lesion grade. Gynecol Oncol. 2004;92(3):873–880. doi: 10.1016/j.ygyno.2003.11.035. [DOI] [PubMed] [Google Scholar]
- 52.Manavi M, Hudelist G, Fink-Retter A, Gschwantler-Kaulich D, Pischinger K, Czerwenka K. Human papillomavirus DNA integration and messenger RNA transcription in cervical low- and high-risk squamous intraepithelial lesions in Austrian women. Int J Gynecol Cancer. 2008;18(2):285–294. doi: 10.1111/j.1525-1438.2007.01017.x. [DOI] [PubMed] [Google Scholar]
- 53.Xi LF, Kiviat NB, Galloway DA, Zhou XH, Ho J, Koutsky LA. Effect of cervical cytologic status on the association between human papillomavirus type 16 DNA load and the risk of cervical intraepithelial neoplasia grade 3. J Infect Dis. 2008;198(3):324–331. doi: 10.1086/589715. [DOI] [PMC free article] [PubMed] [Google Scholar]