Abstract
Aims: To develop a panel of markers able to extract full haplotype information for candidate genes in alcoholism, other addictions and disorders of mood and anxiety. Methods: A total of 130 genes were haplotype tagged and genotyped in 7 case/control populations and 51 reference populations using Illumina GoldenGate SNP genotyping technology, determining haplotype coverage. We also constructed and determined the efficacy of a panel of 186 ancestry informative markers. Results: An average of 1465 loci were genotyped at an average completion rate of 91.3%, with an average call rate of 98.3% and replication rate of 99.7%. Completion and call rates were lowered by the performance of two datasets, highlighting the importance of the DNA quality in high throughput assays. A comparison of haplotypes captured by the Addictions Array tagging SNPs and commercially available whole-genome arrays from Illumina and Affymetrix shows comparable performance of the tag SNPs to the best whole-genome array in all populations for which data are available. Conclusions: Arrays of haplotype-tagged candidate genes, such as this addictions-focused array, represent a cost-effective approach to generate high-quality SNP genotyping data useful for the haplotype-based analysis of panels of genes such as these 130 genes of interest to alcohol and addictions researchers. The inclusion of the 186 ancestry informative markers allows for the detection and correction for admixture and further enhances the utility of the array.
Introduction
Unraveling the underlying mechanisms behind genetically complex traits remains one of the principal goals in psychiatric neurogenetics. The challenges associated with identifying the underlying causes of complex diseases are well illustrated by alcoholism, addictions and other psychiatric diseases. These are complex disorders with moderate to high heritability (approximate range 0.4–0.6) (Goldman et al., 2005). The high incidence and complex inheritance patterns suggest that the elucidation of the roles of common genetic variations in vulnerability might be critical for a better understanding of the pathophysiologies and for the improvement in diagnostic specificity. Whilst several functional loci have been identified (e.g. ADH1B His47Arg and ALDH2 Glu487 in alcoholism (Quertemont, 2004), the MAOA VNTR in dyscontrol behaviors (Popova, 2006; Craig, 2007) and HTTLPR in anxiety/dysphoria (Heinz et al., 2001)), the underlying origins of the genetic variance in vulnerability to addictions and other major psychiatric diseases remain largely unknown.
Analysis of markers throughout the genome has shown that alleles of single nucleotide polymorphisms (SNPs) are often linked to each other in stretches that can range in size from <5 Kb up to >100 Kb (Gabriel et al., 2002). These combinations of linked alleles (haplotypes) allow the entire genome (or portions thereof) to be analyzed using a relatively small number of SNPs. Disease causing SNPs will therefore be linked to other markers and can be identified through their association with other markers even if the causative SNP itself is not assayed (Risch and Merikangas, 1996; reviewed in Kruglyak, 2008).
Until recently researchers were limited in their options for genetic analysis by the limited number of available markers, coupled with comparatively high cost for each genotype obtained. Classical genetic linkage approaches could only be applied when families could be recruited. With the rapid increase in marker information from the HapMap (http://www.hapmap.org/) and GenBank (www.ncbi.nlm.nih.gov/Genbank/) data- bases and the availability of high-density SNP genotyping platforms, researchers now have the possibility of comprehensively interrogating candidate genes and entire biosynthetic/physiological pathways (Perlis et al., 2008) for their genetic contribution to a disorder or phenotype, as well as of performing genome wide scans to identify new candidate genes.
Whole-genome association studies have shown promise in the identification of causative genes in disease (Wellcome Trust Case Control Consortium, 2007; Easton et al., 2007; Hunter et al., 2007; Frayling et al., 2007; Rioux et al., 2007). However, several problems remain with widespread use of this technology. Published whole-genome association studies have demonstrated that common vulnerability alleles often lead to odds ratios of less than 2, and due to the genome-wide nature of these analyses, and the need for statistical correction (Risch and Merikangas, 1996; Hirschhorn and Daly, 2005) (although the required degree of correction for multiple correction remains uncertain), large sample sizes in excess of several thousand cases and controls are needed to detect loci influencing risk (Wang et al., 2005). Furthermore, in the case of bipolar disorder a recent whole-genome association study that compared 2000 cases to 3000 controls identified only a single association signal that survived criteria for genome-wide significance, and this locus accounts for only a small part of the variance in vulnerability attributable to genetic factors. The relatively high per sample cost and the requirement for large numbers of cases and control subjects to identify alleles of modest effect size with associations that are able to withstand correction for multiple testing, make the widespread use of this approach impractical and financially burdensome for many research groups unless pooling approaches are adopted (Shifman et al., 2002; Liu et al., 2006; Johnson et al., 2006).
The complexity of neuropsychiatric and behavioral disorders coupled with the fact that phenoptype can be modulated by environmental factors and that clinical diagnostic criteria likely miss possible etiological heterogeneity only detectible by biologic measures has promoted researchers to use so-called endophenotypes as surrogates for disease states. These endophenotypes are heritable quantitative measurable traits that are inherited in a stable manner and that are more frequently observed in both cases and their first degree relatives and potentially confer vulnerability to a disorder (Gottesman and Gould, 2003; Flint and Munafo, 2007; Frederick and Iacono, 2006; Enoch et al., 2003). Often these endophenotypes are measured by the use of imaging technologies (MRI and PET) (Martinez et al., 2001; Meyer-Lindenberg and Weinberger, 2006) or by EEG measures (Yoon et al., 2006), techniques which due to their cost, invasive nature, requirement for expensive, specialized equipment and length of time required for data acquisition are impractical to use on large cohorts. The practicality of the whole-genome association approach to the study of quantitative imaging traits is being assessed, and although no studies are currently published, the data appear to be promising.
Although candidate gene studies have their own inherent limitations (reviewed in Tabor et al., 2002), the use of smaller focused arrays possibly represents a more practical approach for many studies. These focused arrays are able to overcome the issues of inadequate gene coverage and ethnic stratification by providing full coverage for a limited number of candidate genes and by the inclusion of ancestry informative markers (AIMs). Such focused arrays offer the advantages of lower cost and lower false discovery rate, especially in situations where a dataset may have inadequate power for WGA either because of size or other reasons. In the future it also appears likely that such arrays will be required for follow-up on genomic regions identified by linkage and association studies. Studies on individual candidate genes or small groups of such genes have led to the discovery of functional loci such as the ones cited earlier, but on the other hand these studies have been hampered in other ways. Many linkage and association studies on the role of candidate genes in complex disorders have used single non-functional markers that do not capture sufficient information or do not evaluate all genes in the functional domain of interest. In many instances different markers are selected by groups to interrogate a single gene, making the comparison of data difficult. An additional confound in these single gene studies has been the general failure to control for unrecognized ethnic stratification within the cohort that can lead to the generation of both false positive and false negative signals (Schork et al., 2001; Rosenberg and Nordborg, 2006). Such unrecognized stratification is problematic for genetic studies and can also confound studies relating phenotype to phenotype or risk variable to outcome. In such instances ethnicity can represent a hidden variable.
Recent advances in the neurobiology of addiction, mood disorders and psychoses have established the importance of several mechanisms, including reward, stress resiliency and executive cognitive control (reviewed in Goldman et al., 2005). These studies thereby implicate several molecular networks that are integral to those processes and genes necessary for their function. These molecular pathways include signaling networks, stress/endocrine genes, key neurotransmitter systems including dopamine, serotonin, glutamate, GABA and acetylcholine. In several instances, particular genes and molecules have also been specifically implicated in addiction liability or in addictions-related phenotypes by whole-genome or candidate-gene-focused linkage results.
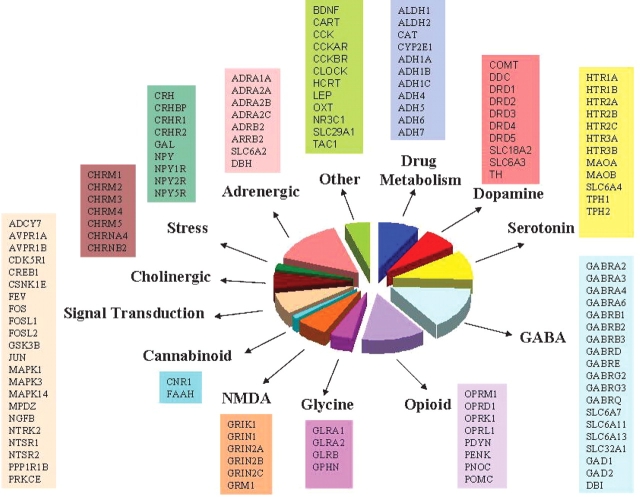
We have designed a 1536 SNP array, implemented on the Illumina Goldengate assay platform. This array includes 1350 SNPs selected for 130 genes and 186 markers that are highly informative for AIMs. The 130 candidate genes were selected on the basis of their roles in functional domains important in the addictions and in the related phenotypes of anxiety and depression. Figure 1 lists the 130 candidate genes organized into one somewhat arbitrary scheme of functional categorization. The candidate genes included a limited number involved in the pharmacokinetic domain (e.g. several genes in the ADH gene cluster, and ALDH genes). The majority of the genes represent the domains of vulnerability to drug use and pharmacodynamic response. These include dopamine, serotonin, glutamine, GABA, and opioid neurotransmitter genes, signaling genes, and genes modulating stress resiliency and behavioral dyscontrol domains. There is a high degree of overlap between functional gene categories because of pleiotropic actions of molecules on behavior.
Fig. 1.
Gene content of the Addictions Array.
Methods
Array design
SNPs for candidate genes
A total of 1350 SNPs (Table 1) from 130 candidate genes (Fig. 1) were selected for inclusion on the array. Tagging SNPs were identified for these genes using the following design pipeline:
A genomic region containing sequence 5 kb upstream and 1 kb downstream for each candidate gene was retrieved from NCBI Human Genome Build 35.1.
Genotype data for the African Yoruban population were obtained from HapMap Project Public Release #18. Haplotype structures for each gene were generated using SNPHAP. The Yoruban data were used since Africans generally show the greatest haplotype diversity and therefore require a larger number of tag SNPs to capture full haplotype information as compared to any other population for which data are available.
The minimum number of index SNPs that captured haplotypes with a frequency of at least 0.006 was selected using a double classification tree search algorithm (Zhang et al., 2004).
Large genes with complex haplotype structures were split into two or three regions according to the haplotype block structure and each of these regions was tagged separately.
SNPs encoding non-synonymous amino-acid substitutions and SNPs within 10 bp of intron/exon splice sites with the potential to alter splicing efficiency were specifically included by forcing these SNPs into the panels used for haplotype tagging (Table 2).
Table 1.
List of genes included on the array, chromosome and number of tag SNPs
| Gene ID | Selected SNPs | Gene symbol | Chr | Gene product name |
|---|---|---|---|---|
| 553 | 4 | AVPR1B | 1 | Arginine vasopressin receptor 1B |
| 1,131 | 20 | CHRM3 | 1 | Cholinergic receptor, muscarinic 3 |
| 1,141 | 5 | CHRNB2 | 1 | Cholinergic receptor, nicotinic, beta |
| 2,166 | 7 | FAAH | 1 | Fatty acid amide hydrolase |
| 2,563 | 4 | GABRD | 1 | Gamma-aminobutyric acid (GABA) A receptor |
| 3,725 | 4 | JUN | 1 | v-jun avian sarcoma virus 17 oncogene homolog |
| 4,803 | 15 | NGFB | 1 | Nerve growth factor, beta polypeptide precursor |
| 4,985 | 11 | OPRD1 | 1 | Opioid receptor, delta 1 |
| 151 | 4 | ADRA2B | 2 | Alpha-2B-adrenergic receptor |
| 1,385 | 10 | CREB1 | 2 | cAMP responsive element binding protein 1 |
| 1,622 | 5 | DBI | 2 | Diazepam binding inhibitor |
| 54,738 | 4 | FEV | 2 | FEV (ETS oncogene family) |
| 2,355 | 9 | FOSL2 | 2 | FOS-like antigen 2 |
| 2,571 | 12 | GAD1 | 2 | Glutamate decarboxylase 1 |
| 3,357 | 6 | HTR2B | 2 | 5-Hydroxytryptamine (serotonin) receptor 2B |
| 23,620 | 4 | NTSR2 | 2 | Neurotensin receptor 2 |
| 5,443 | 4 | POMC | 2 | Proopiomelanocortin preproprotein |
| 5,581 | 21 | PRKCE | 2 | Protein kinase C, epsilon |
| 885 | 7 | CCK | 3 | Cholecystokinin preproprotein |
| 1,814 | 16 | DRD3 | 3 | Dopamine receptor D3 |
| 2,932 | 16 | GSK3B | 3 | Glycogen synthase kinase 3 beta |
| 6,538 | 19 | SLC6A11 | 3 | Solute carrier family 6 (neurotransmitter |
| 124 | 4 | ADH1A | 4 | Class I alcohol dehydrogenase, alpha subunit |
| 125 | 5 | ADH1B | 4 | Alcohol dehydrogenase 1B (class I), beta |
| 126 | 6 | ADH1C | 4 | Class I alcohol dehydrogenase, gamma subunit |
| 127 | 6 | ADH4 | 4 | Class II alcohol dehydrogenase 4 pi subunit |
| 128 | 8 | ADH5 | 4 | Class III alcohol dehydrogenase 5 chi subunit |
| 130 | 4 | ADH6 | 4 | Class V alcohol dehydrogenase 6 |
| 131 | 9 | ADH7 | 4 | Class IV alcohol dehydrogenase 7 mu or sigma |
| 152 | 4 | ADRA2C | 4 | Alpha-2C-adrenergic receptor |
| 886 | 7 | CCKAR | 4 | Cholecystokinin A receptor |
| 9,575 | 10 | CLOCK | 4 | Clock |
| 1,816 | 6 | DRD5 | 4 | Dopamine receptor D5 |
| 2,555 | 12 | GABRA2 | 4 | Gamma-aminobutyric acid A receptor, alpha 2 |
| 2,557 | 12 | GABRA4 | 4 | Gamma-aminobutyric acid A receptor, alpha 4 |
| 2,560 | 19 | GABRB1 | 4 | Gamma-aminobutyric acid (GABA) A receptor, beta |
| 2,743 | 14 | GLRB | 4 | Glycine receptor, beta |
| 4,886 | 6 | NPY1R | 4 | Neuropeptide Y receptor Y1 |
| 4,887 | 4 | NPY2R | 4 | Neuropeptide Y receptor Y2 |
| 4,889 | 4 | NPY5R | 4 | Neuropeptide Y receptor Y5 |
| 154 | 4 | ADRB2 | 5 | Adrenergic, beta-2-, receptor, surface |
| 9,607 | 6 | CART | 5 | Cocaine- and amphetamine-regulated transcript |
| 1,393 | 8 | CRHBP | 5 | Corticotropin releasing hormone binding protein |
| 1,812 | 8 | DRD1 | 5 | Dopamine receptor D1 |
| 2,559 | 6 | GABRA6 | 5 | Gamma-aminobutyric acid A receptor, alpha 6 |
| 2,561 | 18 | GABRB2 | 5 | Gamma-aminobutyric acid (GABA) A receptor, beta |
| 2,566 | 15 | GABRG2 | 5 | Gamma-aminobutyric acid A receptor, gamma 2 |
| 2,741 | 10 | GLRA1 | 5 | Glycine receptor, alpha 1 |
| 3,350 | 4 | HTR1A | 5 | 5-Hydroxytryptamine (serotonin) receptor 1A |
| 2,908 | 10 | NR3C1 | 5 | Nuclear receptor subfamily 3, group C, member 1 |
| 6,531 | 12 | SLC6A3 | 5 | Solute carrier family 6 (neurotransmitter |
| 6,534 | 7 | SLC6A7 | 5 | Solute carrier family 6, member 7 |
| 1,268 | 8 | CNR1 | 6 | Central cannabinoid receptor |
| 2,911 | 22 | GRM1 | 6 | Glutamate receptor, metabotropic 1 |
| 3,351 | 4 | HTR1B | 6 | 5-Hydroxytryptamine (serotonin) receptor 1B |
| 1,432 | 11 | MAPK14 | 6 | Mitogen-activated protein kinase 14 |
| 4,988 | 24 | OPRM1 | 6 | Opioid receptor, mu 1 |
| 2,030 | 9 | SLC29A1 | 6 | Solute carrier family 29 (nucleoside |
| 1,129 | 20 | CHRM2 | 7 | Cholinergic receptor, muscarinic 2 |
| 1,395 | 13 | CRHR2 | 7 | Corticotropin releasing hormone receptor 2 |
| 1,644 | 22 | DDC | 7 | Dopa decarboxylase (aromatic L-amino acid |
| 3,952 | 4 | LEP | 7 | Leptin precursor |
| 4,852 | 4 | NPY | 7 | Neuropeptide Y |
| 6,863 | 4 | TAC1 | 7 | Tachykinin 1 |
| 148 | 19 | ADRA1A | 8 | Alpha-1A-adrenergic receptor |
| 1,392 | 4 | CRH | 8 | Corticotropin releasing hormone precursor |
| 4,986 | 12 | OPRK1 | 8 | Opioid receptor, kappa 1 |
| 5,179 | 8 | PENK | 8 | Proenkephalin |
| 5,368 | 7 | PNOC | 8 | Prepronociceptin |
| 216 | 26 | ALDH1A1 | 9 | Aldehyde dehydrogenase 1A1 |
| 1,621 | 24 | DBH | 9 | Dopamine beta-hydroxylase precursor |
| 2,902 | 5 | GRIN1 | 9 | NMDA receptor 1 |
| 8,777 | 18 | MPDZ | 9 | Multiple PDZ domain protein |
| 4,915 | 17 | NTRK2 | 9 | Neurotrophic tyrosine kinase, receptor, type 2 |
| 150 | 4 | ADRA2A | 10 | Alpha-2A-adrenergic receptor |
| 1,571 | 6 | CYP2E1 | 10 | Cytochrome P450, family 2, subfamily E |
| 2,572 | 17 | GAD2 | 10 | Glutamate decarboxylase 2 |
| 6,571 | 14 | SLC18A2 | 10 | Solute carrier family 18 (vesicular monoamine) |
| 627 | 9 | BDNF | 11 | Brain-derived neurotrophic factor |
| 847 | 11 | CAT | 11 | Catalase |
| 887 | 5 | CCKBR | 11 | Cholecystokinin B receptor |
| 1,128 | 6 | CHRM1 | 11 | Cholinergic receptor, muscarinic 1 |
| 1,132 | 4 | CHRM4 | 11 | Cholinergic receptor, muscarinic 4 |
| 1,813 | 16 | DRD2 | 11 | Dopamine receptor D2 |
| 1,815 | 4 | DRD4 | 11 | Dopamine receptor D4 |
| 8,061 | 5 | FOSL1 | 11 | FOS-like antigen 1 |
| 51,083 | 4 | GAL | 11 | Galanin preproprotein |
| 3,359 | 7 | HTR3A | 11 | 5-Hydroxytryptamine (serotonin) receptor 3A |
| 9,177 | 9 | HTR3B | 11 | 5-Hydroxytryptamine (serotonin) receptor 3B |
| 7,054 | 4 | TH | 11 | Tyrosine hydroxylase |
| 7,166 | 7 | TPH1 | 11 | Tryptophan hydroxylase 1 |
| 217 | 8 | ALDH2 | 12 | Mitochondrial aldehyde dehydrogenase 2 |
| 552 | 9 | AVPR1A | 12 | Arginine vasopressin receptor 1A |
| 2,904 | 25 | GRIN2B | 12 | N-Methyl-d-aspartate receptor subunit 2B |
| 6,540 | 19 | SLC6A13 | 12 | Solute carrier family 6 (neurotransmitter |
| 121,278 | 22 | TPH2 | 12 | Neuronal tryptophan hydroxylase |
| 3,356 | 19 | HTR2A | 13 | 5-Hydroxytryptamine (serotonin) receptor 2A |
| 2,353 | 4 | FOS | 14 | v-fos FBJ murine osteosarcoma viral oncogene |
| 10,243 | 23 | GPHN | 14 | Gephyrin |
| 1,133 | 17 | CHRM5 | 15 | Cholinergic receptor, muscarinic 5 |
| 2,562 | 21 | GABRB3 | 15 | Gamma-aminobutyric acid (GABA) A receptor, beta |
| 2,567 | 19 | GABRG3 | 15 | Gamma-aminobutyric acid (GABA) A receptor, gamma |
| 113 | 14 | ADCY7 | 16 | Adenylate cyclase 7 |
| 2,903 | 18 | GRIN2A | 16 | N-Methyl-d-aspartate receptor subunit 2A |
| 5,595 | 4 | MAPK3 | 16 | Mitogen-activated protein kinase 3 |
| 6,530 | 22 | SLC6A2 | 16 | Solute carrier family 6 member 2 |
| 409 | 4 | ARRB2 | 17 | Arrestin beta 2 |
| 8,851 | 4 | CDK5R1 | 17 | Cyclin-dependent kinase 5, regulatory subunit 1 |
| 1,394 | 9 | CRHR1 | 17 | Corticotropin releasing hormone receptor 1 |
| 2,905 | 4 | GRIN2C | 17 | N-Methyl-d-aspartate receptor subunit 2C |
| 3,060 | 4 | HCRT | 17 | Orexin precursor |
| 84,152 | 4 | PPP1R1B | 17 | Protein phosphatase 1, regulatory (inhibitor) |
| 6,532 | 15 | SLC6A4 | 17 | Solute carrier family 6 member 4 |
| 1,137 | 4 | CHRNA4 | 20 | Cholinergic receptor, nicotinic, alpha |
| 4,923 | 12 | NTSR1 | 20 | Neurotensin receptor 1 |
| 4,987 | 7 | OPRL1 | 20 | Opiate receptor-like 1 |
| 5,020 | 4 | OXT | 20 | Oxytocin-neurophysin I preproprotein |
| 5,173 | 8 | PDYN | 20 | Beta-neoendorphin-dynorphin preproprotein |
| 140,679 | 4 | SLC32A1 | 20 | Solute carrier family 32, member 1 |
| 2,897 | 16 | GRIK1 | 21 | Glutamate receptor, ionotropic, kainate 1 |
| 1,312 | 16 | COMT | 22 | Catechol-O-methyltransferase |
| 1,454 | 11 | CSNK1E | 22 | Casein kinase 1 epsilon |
| 5,594 | 19 | MAPK1 | 22 | Mitogen-activated protein kinase 1 |
| 2,556 | 16 | GABRA3 | X | Gamma-aminobutyric acid A receptor, alpha 3 |
| 2,564 | 10 | GABRE | X | Gamma-aminobutyric acid (GABA) A receptor |
| 55,879 | 6 | GABRQ | X | Gamma-aminobutyric acid (GABA) receptor, theta |
| 2,742 | 16 | GLRA2 | X | Glycine receptor, alpha 2 |
| 3,358 | 19 | HTR2C | X | 5-Hydroxytryptamine (serotonin) receptor 2C |
| 4,128 | 9 | MAOA | X | Monoamine oxidase A |
| 4,129 | 13 | MAOB | X | Amine oxidase (flavin-containing) |
Table 2.
Table showing SNP content of Addictions Array, including cSNPs and other putative functional SNPs
| Total SNPs for 130 genes | 1350 |
|---|---|
| SNPs in intron within 10 bp of splice site | 37 |
| SNPs in transcripts | 167 |
| SNPs in UTR (5′ or 3′) | 55 |
| SNPs in coding sequence | 122 |
| Non-synonymous | 86 |
| Synonymous | 26 |
| Ancenstry informative markers (AIMs) | 186 |
Ancestry informative markers
A panel of 186 SNPs was selected as genomic controls based on the following criteria: (i) reference allele frequency (RFA) of pairwise SNPs from the HapMap Project was at least 0.75; (ii) the minimum distance between SNPs was 80 kb; (iii) the absolute value of log (RFA1/RFA2) was greater than 1 (i.e. there was a 10-fold difference). The selected SNPs represent a sub-fraction of a larger 204 SNP AIMS panel (Enoch et al., 2006) previously tested on the Illumina platform where failed or uninformative assays have been removed from the assay pool. AIMs data were analyzed using structure 2.1 to generate population assignments for all individuals (Pritchard et al., 2000). For the CEPH (Centre Etudes du Polymorphisme Humain) diversity panel, the run parameters used were 1051 individuals, 179 loci, 51 populations assumed, 100,000 Burn-in period and 200,000 Reps. For the test populations, the same run parameters were used, with 5 populations assumed for the 564 samples and 159 loci. The output was graphically represented using the distruct program (Rosenberg, 2004; http://rosenberglab.bioinformatics.med.umich.edu/distruct.html).
Samples
All samples used were collected under protocols approved by the relevant institutional IRB, with participants providing written informed consent for use of their samples in genetic studies.
Genotyping
Genotyping was performed using the Illumina GoldenGate genotyping protocols on 96-well format Sentrix® arrays. Five hundred nanogram of sample DNA was used per assay. All pre-PCR processing was performed using a TECAN liquid handling robot running Illumina protocols. Arrays were imaged using an Illumina Beadstation GX500 and the data analyzed using GenCall v6.2.0.4 and GTS Reports software v5.1.2.0 (Illumina). Genotype clusters were determined for a test dataset and this template was applied to all subsequent datasets. Data for each dataset were polished by manual adjustment of the clustering for each marker to correct for differences between datasets arising from sample integrity and concentration. Loci for which three distinct clusters could not be resolved were assigned zero scores. Data were further polished as follows: genotypes with low GenCall scores (<0.25) were called as undetermined. The GenCall score is a value between 0 and 1 giving a confidence score for that genotype call (the higher the score the higher the confidence in the call) and is derived from the tightness of the clusters for a given locus and the position of the sample relative to its cluster.
Loci with a call rate >90% were determined to have failed and were excluded. At this point deviation from Hardy–Weinberg equilibrium was not used as an exclusion criterion since all datasets contained both case and control samples and, in general, were of mixed ethnic composition.
A total of 8309 unique samples were genotyped from seven different datasets. DNA samples were excluded using the following criteria. The GenTrain scores for a sample for all loci are used to determine the 10% percentile GenCall score (%10 GC) for that sample. The sample exclusion threshold is based on a single project and is calculated by taking the 90th percentile of %10 GC scores for all samples in the project and multiplying by 0.85. Any sample with the %10 GC value below that threshold was classified as failed and removed from the analysis.
Genotyping accuracy was determined based on genotype concordance between DNA replicates. The level of sample replication varied between datasets averaging 16% across all seven datasets.
Haplotypes were derived using the program Phase 2.0 (Li and Stephens, 2003).
Results
Five of the seven datasets (sets A, B, C, D and G) averaged 1481 passing loci, with an average completion rate of 97.60% for those loci (Table 3). Datasets E and F had fewer passing loci, 1351 and 1387 respectively, and greatly reduced completion rates, 86% and 67%. Once all failing DNAs were removed, the average call rate per sample for the datasets was 99.31%, with all but dataset F having a call rate of 90.4%. The reduced performance of the array for datasets E and F is likely due to issues of DNA concentration and quality since the average replication rate for all seven datasets was 99.7% and datasets E and F recorded replication rates of 99.5% (99.95% if one pair were excluded) and 99.6%, respectively, indicating the high quality of genotyping generated for these two datasets.
Table 3.
Table showing the performance of the Addictions Array assays over seven datasets
| Dataset | Passing loci | Completion rate (%) | Average call rate (%) | Replication rate (%) |
|---|---|---|---|---|
| A | 1487 | 98.1 | 99.7 | 99.9 |
| B | 1484 | 96.5 | 99.2 | 99.7 |
| C | 1492 | 99.7 | 99.9 | 99.8 |
| D | 1479 | 98.9 | 99.9 | 99.8 |
| E | 1351 | 86.2 | 99.6 | 99.5 |
| F | 1387 | 66.9 | 90.6 | 99.6 |
| G | 1463 | 94.8 | 99.6 | 99.4 |
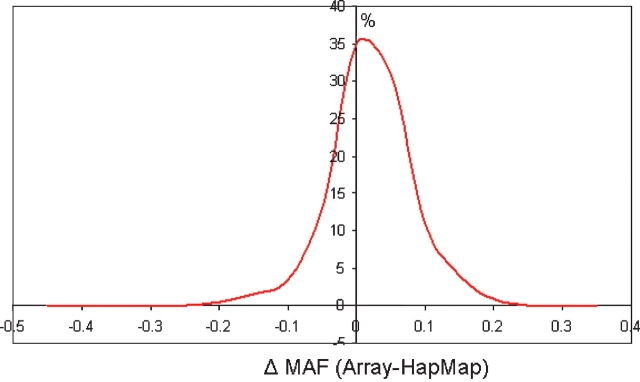
One of the datasets was derived from a Finnish population which allowed us to estimate the genotyping accuracy by the comparison of the minor allele frequency (MAF) for all passing loci in this dataset to the MAF (where known) for the HapMap Caucasian population. This similarity in MAF for the 1440+ loci (Fig. 2) suggests that the genotyping clusters were correctly assigned. Only one marker showed a deviation in MAF >±0.25. This marker rs4824001 is one of the 186 AIMs and was originally selected for its high MAF in the Yoruban population (MAF = 0.833), intermediate frequency in Asian populations (MAF = 0.471) and low MAF (0.017) in Caucasians. The observed MAF (0.498) was confirmed by inspection of the cluster file, which showed clear cluster separation (data not shown). This suggests that this marker, in conjunction with others, may have utility for identifying population stratification in Caucasian populations.
Fig. 2.
Minor allele frequency (MAF) for all array markers was determined for a Finnish Caucasian population and compared to the expected MAF from the HapMap Caucsian data. The difference in experimentally determined and expected MAF was plotted against frequency. Clustering of the data around a MAF of 0 provides confidence that experimentally determined genotype is accurate. Deviation from 0 MAF highlights the possibility of detectable stratification between Caucasian populations.
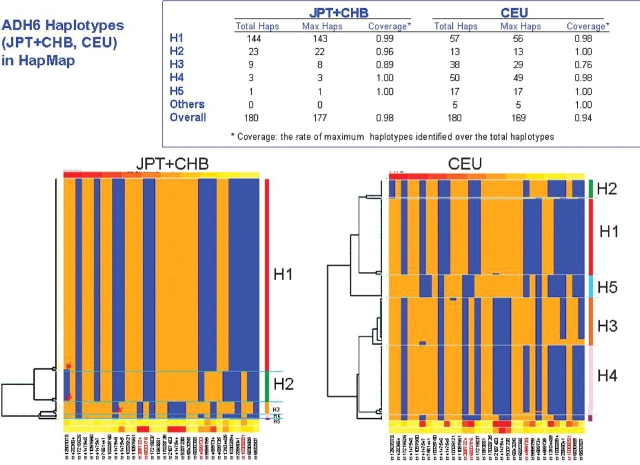
The array was designed to allow haplotype analysis. Tagging SNPs were selected to be able to detect haplotypes present at a haplotype frequency of 0.006 or higher. However, subsequent to the design of the oligo pool additional SNPs have been identified and genotyped in the HapMap populations resulting in an increase in the number of possible haplotypes. The haplotype coverage offered by the tagging SNPs Addictions Array for alcohol dehydrogenase 6 (ADH6) was compared to haplotypes calculated for data from HapMap release 21 (Fig. 3) for the combined Asian and Caucasian samples. To facilitate the analysis haplotypes for Nigeria (YRI) and Utah (CEU) samples in the chromosome region were downloaded from HapMap project release 21 (http://www.hapmap.org/). Based on Manhattan distances weighted by minor allele frequency and marker average LD, haplotypes were clustered hierarchically using R (http://www.r-project.org). Haplotype coverage was determined by dividing the number of haplotypes correctly identified by the tag SNP set divided by the total number of SNPs within the corresponding cluster. As shown in Fig. 3, the majority of all the haplotypes could be correctly called in the combined Asian sample with only three minor haplotypes not being determined by the tag SNP set. Overall in the Asian population haplotype coverage averaged 0.98. In Caucasians the overall haplotype coverage remained at 0.94; however, of the 11 minor haplotypes not detected, the majority (9) were cladistically related, arising in the H3 cluster.
Fig. 3.
Haplotype capture for the ADH6 gene in two HapMap populations. Using genotypes for the HapMap Caucasian and combined Chinese and Japanese populations the ability of the array marker set to capture HapMap release 2 haplotypes was analyzed. Haplotypes were cladistically clustered (see the diagram) and the ability of the panel to correctly identify haplotypes in the HapMap Asian and Caucasian populations was determined (see the table on top of the figure).
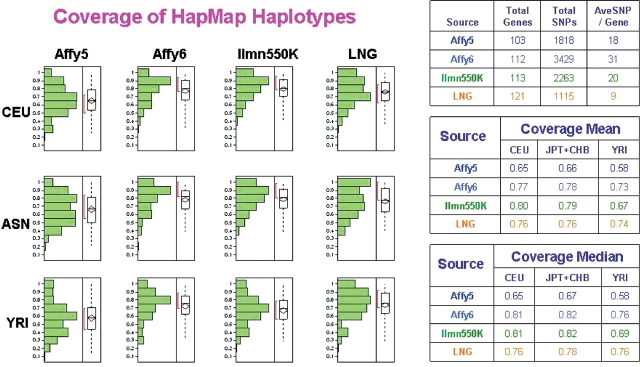
The average haplotype coverage for the genes analyzed by the Addictions Array was compared to the coverage provided by the Illumina HumanHap 550®, the Affymetrix Human-Wide SNP Array 5.0® whole-genome association array and the Affymetrix Human-Wide SNP Array 6.0® (Fig. 4). Only 121 of the 130 genes represented on the Addictions Array were analyzed because X-linked phased haplotypes carried a discrepancy warning from HapMap and because in the case of several smaller genes only two markers had been genotyped in HapMap. The subsets of genes analyzed for the Illumina and Affymetrix arrays were not completely overlapping. Out of 121 Addiction Array genes only 113 were represented on the Illumina array, 112 on the Affymetrix 6.0 array and 103 on the Affymetrix 5.0 array. The whole-genome arrays on average used more than twice the number of SNPs (averaging 18, 31 and 20 SNPs per gene for the Affymetrix 5.0, Affymetrix 6.0 and Illumina 550 arrays, respectively) to cover each gene compared to the Addictions Array (average 9 SNPs per gene). Despite the reduced number of SNPs per gene, the average haplotype coverage (HCM–haplotype coverage mean) for the Addictions Array was consistently higher than that of the Affymetrix 5.0 Array for all three HapMap populations. The superior performance of the Addictions Array over the Affymetrix 5.0 array product was also confirmed by the coverage median values in all three populations. The Addictions Array performed comparably to the Illumina humanhap 550 array, and the Affymetrix 6.0 array for the Caucasian and Asian HapMap populations with an HCM of 0.76 for the Caucasian and Asian groups, compared to the 0.80 and 0.79 values for the Illumina 550 k array, and 0.77 and 0.78 for the Affymetrix 6.0 array. The Addictions Array produced a higher HCM (0.74) and coverage median (0.76) for the Yoruban population than the Illumina array (HCM 0.67, median coverage 0.69) and comparable results to the Affymetrix 6.0 array (HCM 0.73; median coverage 0.76).
Fig. 4.
Comparison of overall haplotype capture in three HapMap populations by the Addictions Array, the Affymetrix Human-Wide SNP 5.0®, the Affymetrix Human-Wide SNP 6.0® and the Illumina HumanHap550® arrays. Overall the Addictions array was able to capture haplotypes with a similar efficiency the Illumina product in all three populations. Due to the SNP selection on the Affymetrix 5.0 array the Addictions array, the Illumina 550 HumanHap550® array and the 1 million SNP Affymetrix 6.0 were able to capture more haplotype diversity.
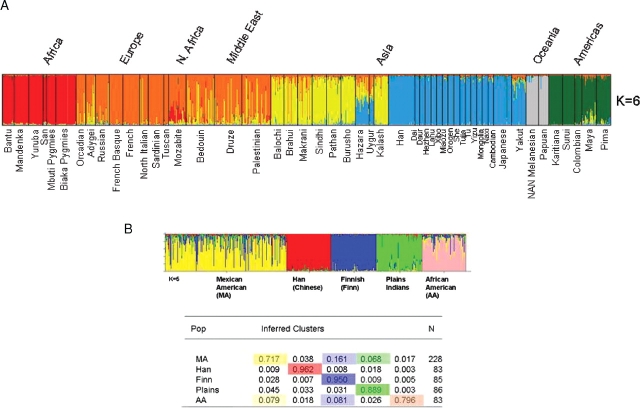
The ability of the AIMs panel to detect differences between populations that were not originally used in the design of the panel was tested by genotyping the CEPH diversity panel (Cann et al., 2002). Genotyping data were analyzed using structure 2.2 for a six-population solution (Fig. 5a). Using the combined global data the AIMs panel is able to distinguish six distinct populations that segregate along continental lines. This solution is similar to that obtained by Rosenberg et al. using a panel of 377 micro-satellite markers. Additionally the two samples previously shown to be misidentified (Rosenberg et al., 2002), as members of the Biaka pygmy and Japanese cohorts, were correctly assigned by this AIMs panel to their correct continental groups (Europe/Middle East and the Americas, respectively).The ability of AIMs panel to detect admixture was then tested by analyzing the combined data from five populations, Finnish Caucasians (n = 85), African Americans from New Jersey (n = 83), Native Americans from the Midwest (n = 86), Han Chinese (n = 83) and Mexican Americans from California (n = 228). The analysis was performed using the assumption of five populations using data for 159 loci and the results are shown in Fig. 5b. All individuals were correctly assigned to their ethnic cluster, although individuals can be seen to vary in their degree of admixture. The admixture contribution of a cluster to each population is shown as a percentage of the inferred clusters (Fig. 5). As expected the African American and Mexican American populations showed higher degrees of admixture than the Finnish and Han Chinese samples, both of which had been previously shown to be relatively homogenous groups (Enoch et al., 2006).
Fig. 5.
(A) Analysis of the AIMs data for the CEPH diversity panel. The AIMs panel is able to identify six distinct groups from the global population, based upon geographically relationships. (B) Analysis of five test samples including Mexican Americans (MA), African Americans (AA), Finnish Caucasians, Han Chinese and Midwestern Native Americans (Plains) using the AIMs panel. All individuals were correctly assigned to their respective ethnic groupings. Five distinct groups were determined. The table shows the calculated contribution of each ethnic factor to the other populations. As anticipated the Mexican American, African American and to a lesser extent the Native American samples showed a higher degree of admixture than the Finnish and Han Chinese samples.
Discussion
Technologies for genotyping have increased genotyping throughput whilst at the same time decreasing the cost per genotype. At present up to 1 million SNPs can be interrogated simultaneously in an individual allowing for whole-genome association studies. Such studies have successfully identified susceptibility loci for obesity (Freyling et al., 2007) and breast cancer (Hunter et al., 2007; Easton et al., 2007) as well as for bipolar disorder, coronary heart disease, Crohns disease, rheumatoid arthritis, and type 1 and type 2 diabetes (Wellcome trust Case Control Consortium, 2007; Rioux et al., 2007). The cost of this approach remains prohibitively high for generalized use and requires large datasets to obtain the necessary power to detect association to a phenotype. This is particularly problematic for those datasets where PET or MRI imaging is performed since the high cost of the scans coupled with the time required to acquire the data makes the collection of large datasets impractical. Pooling of samples has been successfully used to reduce the overall number of arrays required for a study; however, this approach has not gained widespread acceptance or use due to the practical issues of sample normalization, statistical testing and the loss of individual haplotype information needed amongst other reasons to validate the homogeneity of the phenotypic groups. Although the cost per sample of the whole-genome arrays is constantly falling, and the data could be used for haplotype-based analysis of individual genes, these arrays are likely to remain inappropriate for candidate gene analysis due to issues of sample throughput. Certainly the use of these arrays would allow for more fine-tuned control and correction of population stratification due to the higher number of markers. Currently, however, the use of more focused arrays represents a more appropriate approach for many studies where the number of subjects is limited, and where the investigators wish to study a specific hypothesis where candidate genes are selected on the basis of function or where individual SNPs are known to alter the expression or biological activity of the gene product. Additionally in future, once a number of large whole-genome association studies have been completed, it may be more appropriate to use focused arrays to interrogate genomic regions identified as potential candidate regions in a large number of smaller datasets. In this context where there are convergences of whole-genome association data to previously identified candidate genes, the two approaches act synergistically as cross-validation of each positive association finding.
The SNP tagging pipeline for this array used the HapMap data for the Yoruban population as its basis. Whilst it would be preferred that a tag set was used for each unique population it has been shown that tagging SNP sets have high portability across populations (deBakker et al., 2006; Conrad et al., 2006; Gonzalez-Niera et al., 2006). The discovery of additional SNPs subsequent to the array design has resulted in a reduction in the haplotype capture or coverage, but it remains at levels comparable to the high-density arrays available. Use of clustering on a cladistic basis allows the grouping of related haplotypes, particularly those with low frequency. It might be considered desirable to generate an array capable of universally high haplotype capture in all populations; however, such a goal is unlikely to be achieved. For complete haplotype capture in all three HapMap populations the number of tagged SNPs for each gene would have to be increased, with a concomitant reduction in the number of genes that can be interrogated. That reduction in the number of genes is likely to make any array less attractive to researchers as it increases the likelihood that one or more genes of interest will be absent from the array.
In this array we have focused on genes of particular interest to alcohol researchers, which are also of interest to the general neuropsychiatric community. The use of SNP-tagging allows the reduction in the number of SNPs required to successfully interrogate each gene, and maximizes the utility of any array design by increasing the number of candidates that can be incorporated in to the design. This is an important consideration for custom designs, the cost of which falls as the number of samples screened increases. In addition we have been able to include a large panel of AIMs for the detection and correction for population stratification. To genotype such a large SNP panel on a SNP-by-SNP basis would be uneconomic and take a considerable time to accomplish. Since one of the possible confounds in association studies is false positive (and negative) finding arising from differences in the makeup of the control and case groups the detection of any stratification is of the highest importance. Usually this problem has been handled by careful selection and matching of the case and control groups, often resulting in increased costs and the time of study participant recruitment. Such selection of participants usually results in the exclusion of minorities and represents a contributory factor in racial disparities in healthcare and is obviously undesirable both scientifically and socially. Even when the issue of population stratification was addressed by using genotypes from markers unlinked to each other and to the gene of interest, it was rarely demonstrated that the markers used were in fact capable of detecting it. By genotyping the AIMs in the CEPH reference populations a canonical dataset was created enabling the computation of ethnic factor scores anchored against worldwide genetic diversity and allowing direct dataset-to-dataset comparisons. Fixed solutions for admixture correction can be performed using individual ethic factor scores as covariates, or alternatively association data can be corrected using programs such as STRAT (Pritchard et al., 2000) that directly use the output of the STRUCTURE 2.0 to correct for any detected population stratification.
Comparisons between the results from published candidate gene studies have been hampered in the past by the use of different sets of markers for the interrogation of the same gene. Whilst often this results from studies being performed contemporaneously or due to constraints of a particular genotyping platform, clearly it is desirable to be able to easily correlate data from different studies. This issue has clearly been seen in the study of DISC1 as a candidate gene for schizophrenia where multiple groups have performed association studies using many different markers for their analysis (Hwu et al., 2003; Hennah et al., 2003; Hodgkinson et al., 2004; Thompson et al., 2005). Although the studies have provided supportive evidence for each other, the identification of the functional loci has been hampered. Similarly the general region in which the GABAA subunit gene cluster is located on chromosome 4p was implicated in alcohol dependence by family linkage scans and a series of more recent studies (beginning Long et al., 1998; Porjesz et al., 2002; Song et al., 2003; Edenberg et al., 2004; Lappalainen et al., 2005; Prescott et al., 2006; Drgon et al., 2006) have now demonstrated linkage disequilibrium within the GABAA subunit gene cluster itself including the same alleles and haplotypes as determined by analysis of the data from the partially overlapping loci evaluated in these studies. Frequently, a SNP or multilocus haplotype can be used to impute a different SNP (Wellcome Trust); however, the ability to compare across studies is made considerably more challenging by the genotyping of different markers in different studies. In this context and others, the use of genotyping tools, including commercially available arrays that access common sets of markers, is highly advantageous. Although information from the International HapMap Project provides valuable information about linkage disequilibrium between markers can assist in cross-study comparisons, the process is clearly inefficient, time-consuming and not without error as only four populations are currently represented in the database.
Widespread use of haplotype capture arrays such as this addictions array would greatly facilitate cross-study comparisons and use of large panels of AIMs might permit the data from different studies to be combined and analyzed by allowing for population admixture to be controlled for in the analysis.
Acknowledgments
We would like to thank Amy Doebber and Rema Paudel for technical assistance. This work was supported by 1RO1 AA13640, 5P50 AA11998, 1RO1 NS43762, The Blanche F. Ittleson Endowment Fund (RDT), NIH Grants R01 DA 12422, K02 DA 15766 (JFC) and an unrestricted research grant from Glaxo-Smith-Kline (EBB), grants AA06420 and AA10201 to CLE and NIH grants MH062185, MH048514 and MH056390 to J.J.M. This project was also supported by the National Institute on Alcohol Abuse and Alcoholism Intramural Research Program.
References
- Cann HM, de Toma C, Cazes L, et al. A human genome diversity cell line panel. Science. 2002;296:261–2. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Jakobsson M, Coop G, et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genet. 2006;38:1251–60. doi: 10.1038/ng1911. [DOI] [PubMed] [Google Scholar]
- Craig IW. The importance of stress and genetic variation in human aggression. Bioessays. 2007;29:227–36. doi: 10.1002/bies.20538. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Burtt NP, Graham RR, et al. Transferability of tag SNPs in genetic association studies in multiple populations. Nat Genet. 2006;38:1298–303. doi: 10.1038/ng1899. [DOI] [PubMed] [Google Scholar]
- Drgon T, D’Addario C, Uhl GR. Linkage disequilibrium, haplotype and association studies of a chromosome 4 GABA receptor gene cluster: candidate gene variants for addictions (Comparative Study. Journal Article. Research Support, N.I.H., Intramural) Am J Med Genet Part B, Neuropsychiatr Genet. 2006;141:854–60. doi: 10.1002/ajmg.b.30349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, et al. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–14. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch M-A, Shen P-H, Xu K, et al. Using ancestry-informative markers to define populations and detect population stratification. J Psychopharmacol. 2006;20(Suppl):19–26. doi: 10.1177/1359786806066041. [DOI] [PubMed] [Google Scholar]
- Enoch MA, Schuckit MA, Johnson BA, et al. Genetics of alcoholism using intermediate phenotypes. Alcohol: Clin Exp Res. 2003;27:169–76. doi: 10.1097/01.ALC.0000052702.77807.8C. [DOI] [PubMed] [Google Scholar]
- Flint J, Munafo MR. The endophenotype concept in psychiatric genetics. Psychol Med. 2007;37:163–80. doi: 10.1017/S0033291706008750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick JA, Iacono WG. Beyond the DSM: defining endophenotypes for genetic studies of substance abuse. Curr Psychiatry Rep. 2006;8:144–50. doi: 10.1007/s11920-006-0014-2. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human enome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6:521–32. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Niera A, Ke X, Lao O, et al. The portability of tagSNPs across populations: a worldwide survey. Genome Res. 2006;16:323–30. doi: 10.1101/gr.4138406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–45. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Heinz A, Mann K, Weinberger DR, et al. Serotonergic dysfunction, negative mood states, and response to alcohol. Alcohol: Clin Exp Res. 2001;25:487–95. [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennah W, Varilo T, Kestilä M, et al. Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Hum Mol Genet. 2003;12:3151–9. doi: 10.1093/hmg/ddg341. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Goldman D, Jaeger J, et al. Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am J Hum Genet. 2004;75:862–72. doi: 10.1086/425586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwu HG, Liu CM, Fann CS, et al. Linkage of schizophrenia with chromosome 1q loci in Taiwanese families. Mol Psychiatry. 2003;8:445–52. doi: 10.1038/sj.mp.4001235. [DOI] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Liu QR, et al. Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am J Med Genet Neuropsychiatr Genet. 2006;141 B:844–53. doi: 10.1002/ajmg.b.30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L. The road to genome-wide association studies. Nat Rev Genet. 2008;9:314–18. doi: 10.1038/nrg2316. [DOI] [PubMed] [Google Scholar]
- Lappalainen J, Krupitsky E, Remizov M, et al. Association between alcoholism and gamma-amino butyric acid alpha2 receptor subtype in a Russian population. Alcohol: Clin Exp Res. 2005;29:493–8. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- Li N, Stephens M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 2003;165:2213–33. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QR, Drgon T, Johnson C, et al. Addiction molecular genetics: 639,401 SNP whole genome association identifies many “cell adhesion” genes. Am J Med Genet Neuropsychiatr Genet. 2006;141:918–25. doi: 10.1002/ajmg.b.30436. [DOI] [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, et al. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–21. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Martinez D, Broft A, Laruelle M. Imaging neurochemical endophenotypes: promises and pitfalls. Pharmacogenomics. 2001;2:223–37. doi: 10.1517/14622416.2.3.223. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7:818–27. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- Perlis RH, Purcell S, Fagerness J, et al. Family-based association study of lithium-related and other candidate genes in bipolar disorder. Arch Gen Psychiatry. 2008;65:53–61. doi: 10.1001/archgenpsychiatry.2007.15. [DOI] [PubMed] [Google Scholar]
- Popova NK. From genes to aggressive behavior: the role of serotonergic system. Bioessays. 2006;28:495–503. doi: 10.1002/bies.20412. [DOI] [PubMed] [Google Scholar]
- Porjesz B, Begleiter H, Wang K, et al. Linkage and linkage disequilibrium mapping of ERP and EEG phenotypes. Biol Psychol. 2002;61:229–48. doi: 10.1016/s0301-0511(02)00060-1. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg N, Stephens M, et al. Association mapping in structured populations. Am J Hum Genet. 2000;67:170–81. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quertemont E. Genetic polymorphism in ethanol metabolism: acetaldehyde contribution to alcohol abuse and alcoholism. Mol Psychiatry. 2004;9:570–81. doi: 10.1038/sj.mp.4001497. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Sullivan PF, Kuo PH, et al. Genomewide linkage study in the Irish affected sib pair study of alcohol dependence: evidence for a susceptibility region for symptoms of alcohol dependence on chromosome 4. Mol Psychiatry. 2006;11:603–11. doi: 10.1038/sj.mp.4001811. [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–7. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and omplicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Nordborg M. A general population-genetic model for the production by population structure of spurious genotype-phenotype associations in discrete, admixed or spatially distributed populations. Genetics. 2006;173:1665–78. doi: 10.1534/genetics.105.055335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. Distruct: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–8. [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298:2381–85. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Schork NJ, Fallin D, Thiel B, et al. The future of genetic case-control studies. Adv Genet. 2001;42:191–212. doi: 10.1016/s0065-2660(01)42023-2. [DOI] [PubMed] [Google Scholar]
- Shifman S, Pisante-Shalom A, Yakir B, et al. Quantitative technologies for allele frequency estimation of SNPs in DNA pools. Mol Cell Probes. 2002;16:429–34. doi: 10.1006/mcpr.2002.0440. [DOI] [PubMed] [Google Scholar]
- Song J, Koller DL, Foroud T, et al. Association of GABA(A) receptors and alcohol dependence and the effects of genetic imprinting. Am J Med Genet Part B, Neuropsychiatr Genet. 2003;117:39–45. doi: 10.1002/ajmg.b.10022. [DOI] [PubMed] [Google Scholar]
- Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet. 2002;3:1–7. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- Thompson PA, Wray NR, Millar JK, et al. Association between the TRAX/DISC locus and both bipolar disorder and schizophrenia in the Scottish population. Mol Psychiatry. 2005;616:657–68. doi: 10.1038/sj.mp.4001669. [DOI] [PubMed] [Google Scholar]
- Wang WY, Barratt BJ, Clayton DG, et al. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6:109–18. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HH, Iacono WG, Malone SM, et al. Using the brain P300 response to identify novel phenotypes reflecting genetic vulnerability for adolescent substance misuse. Addict Behavs. 2006;31:1067–87. doi: 10.1016/j.addbeh.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Zhang P, Sheng H, Uehara R. A double classification tree search algorithm for index SNP selection. BMC Bioinformatics. 2004;5:89. doi: 10.1186/1471-2105-5-89. [DOI] [PMC free article] [PubMed] [Google Scholar]