Abstract
AU-rich element-binding proteins (ARE-BPs) regulate the stability and/or translational efficiency of mRNAs containing cognate binding sites. Many targeted transcripts encode factors that control processes like cell division, apoptosis, and angiogenesis, suggesting that disregulated ARE-BP expression could dramatically influence oncogenic phenotypes. Using several approaches, we evaluated the expression of four well characterized ARE-BPs across a variety of human neoplastic syndromes. AUF1, TIA-1, and HuR mRNAs were not systematically disregulated in cancers; however, tristetraprolin (TTP) mRNA levels were significantly decreased across many tumor types, including advanced cancers of the breast and prostate. Restoring TTP expression in an aggressive tumor cell line suppressed three key tumorgenic phenotypes: cell proliferation, resistance to pro-apoptotic stimuli, and expression of VEGF mRNA. However, the cellular consequences of TTP expression varied across different cell models. Analyses of gene array datasets revealed that suppression of TTP expression is a negative prognostic indicator in breast cancer, since patients with low tumor TTP mRNA levels were more likely to present increased pathological tumor grade, VEGF expression, and mortality from recurrent disease. Collectively, these data establish that TTP expression is frequently suppressed in human cancers, which in turn can alter tumorigenic phenotypes that influence patient outcomes.
Keywords: mRNA turnover, RNA-binding protein, prognostic indicator, tumor progression, VEGF
Introduction
The initiation and progression of cancer are exacerbated by disregulated expression of proteins controlling diverse cellular phenotypes including differentiation, the cell cycle, apoptosis, angiogenesis, and cell invasiveness (1). Synthesis of these proteins is strongly influenced by the cytoplasmic concentrations of their respective mRNAs, which in turn depend on the kinetics of both mRNA synthesis and degradation. The cytoplasmic stability of many mRNAs encoding oncoproteins, cytokines, and inflammatory mediators is controlled by AU-rich elements (AREs), a family of RNA sequences located within the 3’UTR of each transcript (2, 3). ARE-directed control of mRNA decay is mediated, in part, through interactions with specific ARE-binding proteins (ARE-BPs). For example, association of tristetraprolin (TTP), KH-type splicing regulatory protein, and some isoforms of AUF1 accelerate decay of targeted transcripts, while some other proteins, like HuR, protect mRNAs from degradation. By contrast, TIA-1 and TIAR do not appear to influence mRNA decay directly, but rather suppress gene expression by inhibiting translation of associated mRNAs (reviewed in Refs. 4, 5).
The presence of AREs in many mRNAs encoding cancer-related proteins, coupled with the role of ARE-BPs in regulating these transcripts, raises the possibility of links between the expression and/or activity of selected ARE-BPs and the initiation or progression of neoplasia. Recent studies have provided data supporting this hypothesis in selected experimental systems. For example, mice that constitutively overexpress the p37 isoform of AUF1 (p37AUF1) develop sarcomas (6). By contrast, prostaglandin A2-dependent induction of p45AUF1 in non-small-cell lung cancer cells suggested an antiproliferative role for this protein, since cyclin D1 mRNA was destabilized concomitant with enhanced p45AUF1 binding to the cyclin D1 mRNA 3’UTR (7). Elevated HuR levels are associated with higher tumor grade in breast carcinomas (8), while ectopic overexpression of TTP in Ras-dependent tumor cells delayed tumor growth (9) and vascularization (10) in murine xenografts concomitant with destabilization of select target mRNAs. Finally, the translational regulatory protein TIA-1 was first identified as a pro-apoptotic factor in lymphocytes (11). Together, these examples suggest close and potentially complex relationships between ARE-BPs and cellular proliferation, angiogenesis, apoptosis, and even the stage of tumor development.
Although many mRNAs that encode tumorigenic products may be regulated by AREBPs, the prevalence of disregulated ARE-BP expression and/or activity in the development of human cancers is largely unknown. Furthermore, mechanisms linking altered expression of specific ARE-BPs to neoplastic phenotypes remain poorly defined. To address these questions, we surveyed the expression of four well-characterized ARE-BPs across a wide range of tumor types using cDNA arrays and bioinformatic approaches. Although levels of each ARE-BP mRNA varied in selected cancers, expression of TTP was significantly repressed in the majority of solid tumors from diverse tissue sources. TTP (also known as TIS11, ZFP36, and Nup475) is the prototype of a small family of RNA-binding proteins containing tandem CCCH zinc finger domains, and recognizes ARE sequences through adjacent UAUU binding sites (12, 13). Given our observations that TTP expression is diminished in many cancers, we predicted that decreasing TTP levels may enhance one or more tumorigenic phenotypes, thus giving cells that weakly express TTP a competitive growth or survival advantage. To test this model, we assayed the influence of TTP expression on three key tumorigenic phenotypes: cell proliferation, sensitivity to apoptotic stimuli, and expression of VEGF, in two different cultured cell models. Finally, we surveyed gene array datasets to determine whether loss of TTP correlated with tumor grade, enhanced expression of a known TTP substrate mRNA, and patient mortality. Together, these data show that TTP expression is widely suppressed in human cancers and that limiting cellular TTP levels can modulate several neoplastic phenotypes in a cell type-specific manner, which may contribute to increases in tumor grade and negative patient outcome in some tissues.
Materials and Methods
Cancer profiling cDNA array
A Cancer Profiling Array II (BD Biosciences) was probed for the expression AUF1, TTP, HuR, and TIA-1 mRNAs using radiolabeled cDNA fragments in ExpressHyb solution (Clontech) according to the manufacturer’s instructions. Hybridized 32P-labeled DNA probes were quantified using a Phosphorimager (GE Biosciences). Sample loading on the array was standardized using a ubiquitin cDNA probe (BD Biosciences).
Cell culture and transfections
HeLa/Tet-Off cells (Clontech) were maintained at 37 °C and 5% CO2 in DMEM (Invitrogen) supplemented with 10% fetal calf serum (FCS; Atlanta Biologicals) and 100 µg/ml G148 (Cellgro). Plasmids pT2hyg-FLAG-TTPwt and pT2hyg-FLAG-TTP C147R were transfected using Superfect (Qiagen), and stably transformed HeLa cell clones isolated by selection in 100 µg/ml hygromycin B (Roche). Doxycycline (Dox; Sigma) was maintained (2 µg/ml) during selection and subsequent clonal expansion to prevent any TTP-dependent effects on cell viability. Several dozen independent hygromycin-resistant lines were screened for Dox-regulated expression of FLAG-TTPwt (or C147R) by Western blot using anti-FLAG antibodies.
Murine embryonic fibroblast (MEF) cultures were derived from E14.5 embryos of TTP knockout mice (Zfp36−/−) and wild type littermates (Zfp36+/+) as described previously (14) and were maintained in DMEM supplemented with 10% FCS, 100 U/ml penicillin, 100 µg/ml streptomycin, and 2 mM L-glutamine (Cellgro). All experiments involving MEFs in this study were performed prior to the 12th cell passage.
Proliferation and apoptosis sensitivity assays
HeLa or MEF cells were seeded in 96-well plates at 1000 cells/well, and then returned to the tissue culture incubator. When measuring proliferation rates, cells were counted using the DHL Cell Viability and Proliferation Assay Kit (Anaspec) according to the manufacturer’s instructions. Cell numbers were determined by comparison of background-corrected fluorescence to standard curves of fluorescence versus cell number for each cell type, and were consistent with data obtained from Trypan Blue exclusion assays (data not shown). To measure the sensitivity of HeLa and MEF lines to proapoptotic stimuli, cells were similarly seeded in 96-well plates and allowed to grow for 24 hours prior to adding varying concentrations of staurosporine or cisplatin. Twenty-four hours afterwards, surviving cells were counted as described above. The IC50 for each apoptotic stimulus was resolved using a four parameter logistic equation (PRISM v3.03).
Measurement of VEGF mRNA decay kinetics
Cellular VEGF mRNA decay rates were measured using actinomycin D (actD) time course assays. Briefly, transcription was inhibited by addition of actD (5 µg/ml; Calbiochem) to the culture medium, and total RNA purified at selected times thereafter. Time courses were limited to 4 hours to avoid complicating cellular mRNA decay pathways by actD-enhanced apoptosis (15). VEGF mRNA levels were measured at each time point by quantitative real-time reverse transcription-PCR (qRT-PCR) and normalized to GAPDH mRNA as described in Supplementary Information. First-order decay constants (k) were solved by nonlinear regression (PRISM) of the percentage of VEGF mRNA remaining versus time of actD treatment. Resolved VEGF mRNA half-lives (t1/2 = ln2/k) are based on the mean ± SD of n independent time course experiments where n ≥ 3, or the mean ± spread where n = 2. Ribonucleoprotein-immunoprecipitations (RNP-IPs) used to detect interactions between FLAG-TTP and cellular VEGF mRNA were adapted from previously described methods (16).
Data analysis and statistics
Comparisons of mRNA levels and decay kinetics, drug IC50 values, and cell proliferation rates were performed using the unpaired t test. Differences yielding P < 0.05 were considered significant, with the exception of gene array comparisons, where a threshold of P < 0.001 was employed. Correlation analyses used the Spearman non-parametric test while Kaplan-Meier comparisons were performed using the log-rank test with events limited to death from recurrent disease. For correlation and survival analyses, differences yielding P < 0.05 were considered significant.
Results
TTP expression is significantly repressed in many human tumors and tumor cell lines
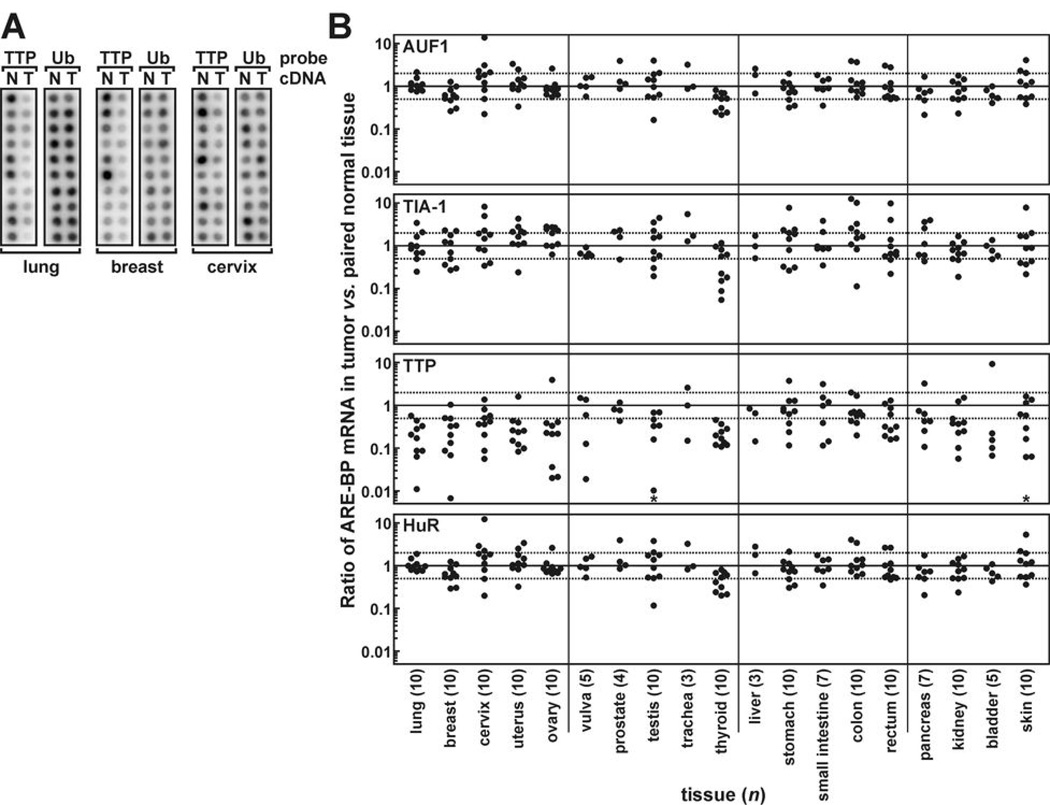
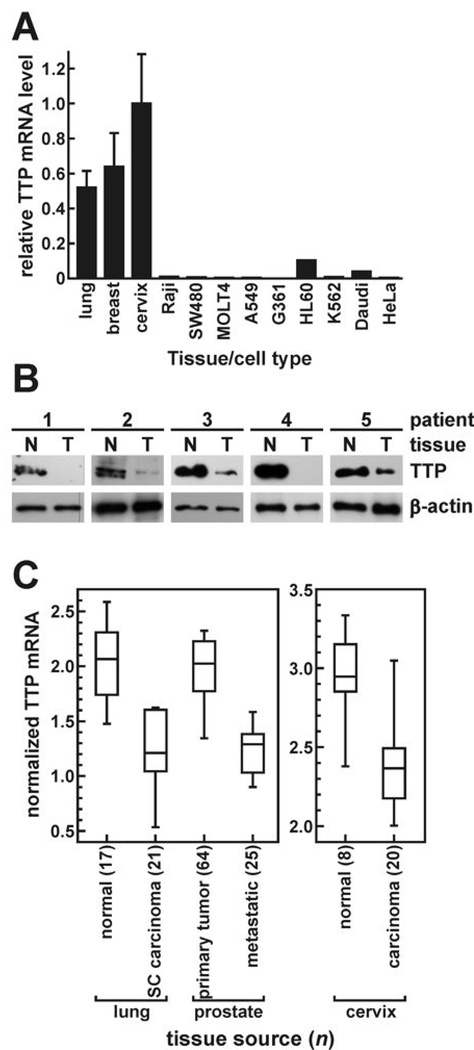
To determine whether disregulated ARE-BP expression might contribute to human tumor development, we first probed a Cancer Profiling Array to compare levels of AUF1, TIA-1, TTP, and HuR mRNAs between tumors and peripheral non-transformed tissues of 154 patients representing 19 different tissues. A change of one log2 unit (100% increase or 50% decrease) in ARE-BP expression between tumor and patient-matched normal tissue was considered substantial. By this criterion, AUF1 and HuR expression were altered in only a small subset of tumors (Fig. 1 and Supplementary Table S1). TIA-1 mRNA was more variable, substantially increasing or decreasing in half of all tumors. However, TTP mRNA levels were substantially decreased in tumors relative to patient-matched non-transformed tissues in 65% of patients tested, and particularly frequently in tumors of the thyroid (10/10), lung (9/10), ovary (9/10), uterus (9/10), and breast (8/10). Consistent with suppression of TTP expression in many tumor types, nine human cancer cell lines included on the array also displayed very low constitutive TTP mRNA levels. For example, TTP mRNA was barely detectable in the cervical adenocarcinoma cell line HeLa or the lung adenocarcinoma cell model A549, yet was abundantly expressed in corresponding non-transformed tissues (Fig. 2A and Ref. 17). Finally, Western blot analyses of extracts from five human breast tumors versus patient-matched non-transformed tissue verified that TTP is also suppressed in tumors at the protein level (Fig. 2B).
Figure 1.
ARE-BP mRNA levels in tumors versus peripheral non-transformed tissues. A cDNA Cancer Profiling Array was probed for selected ARE-BP mRNAs. A, array hybridization signals from lung, breast, and cervical cDNA samples probed for TTP and ubiquitin (ub) expression in tumors (T) and patient-matched, non-transformed peripheral tissue (N). B, scatter plots showing ubiquitin-normalized ratios of AUF1, TIA-1, TTP, and HuR cDNAs derived from tumors versus patient-matched normal tissues. Solid lines (ratio = 1) indicate equivalent ARE-BP expression in tumors and normal tissues, dotted lines show a 100% increase or 50% decrease in tumor versus normal tissues, and n is the number of matched patient sample pairs for each tissue type (bottom). Asterisks in TTP panels denote selected tumors (3 testicular, 1 skin) where TTP cDNA was undetectable above background.
Figure 2.
Repression of TTP expression in cancer cell lines and human tumors. A, TTP mRNA was measured in nine human cancer cell lines concomitantly with tissue samples on the Cancer Profiling Array. Bars labeled lung, breast, and cervix are the mean TTP hybridization signals (± SD) from ten non-transformed tissues normalized to ubiquitin. B, Western blots probed for TTP and β-actin proteins in whole cell extracts from breast tumors (T) and patient-matched non-transformed tissue (N) from five patients: 1, invasive lobular, undefined grade; 2, invasive ductal and ductal carcinoma in situ (DCIS), grade 3 (Nottingham); 3, poorly differentiated invasive carcinoma, grade 3; 4, infiltrating ductal carcinoma and DCIS, grade 2; and 5, extensive DCIS, undefined grade. C, gene array datasets were screened for differential TTP mRNA levels using Oncomine v3. Median TTP mRNA levels are shown by solid lines within each box on distribution plots. Box upper and lower limits represent the 75th and 25th percentiles, respectively, while the extended lines indicate 10th and 90th percentiles. Analysis methods, statistical comparisons, and dataset sources are included in Supplementary Table S2.
The Cancer Profiling Array experiments showed that TTP mRNA was suppressed in many tumors and cancer cell lines. Since this method was limited to ≤10 patients for each tissue type, we used two further approaches to validate these findings across larger sample populations. First, gene array surveys using the Oncomine utility (18) identified numerous patient populations exhibiting significant differences in TTP mRNA levels (P < 0.001). For example, TTP expression was repressed in lung carcinomas relative to healthy lung tissues in three independent patient pools (Fig. 2C and Supplementary Table S2). TTP mRNA levels were similarly decreased in ovarian, cervical, and liver tumors. Contrasting this trend was smoldering multiple myeloma, where TTP mRNA was induced relative to healthy bone marrow. Prostate cancer represented yet another case, where three separate studies showed significantly less TTP mRNA in metastases versus primary tumors, suggesting that suppression of TTP might not be linked solely to oncogenesis, but also to the stage of tumor development. Our second approach to survey tumor-dependent changes in TTP mRNA levels used the Cancer Genome Anatomy Project database, based on the frequency of Expressed Sequence Tag (EST) and Serial Analysis of Gene Expression (SAGE) hits across large sample pools (19). In libraries derived from cancerous tissues, TTP tags were recovered 40–50% less frequently than from normal tissues by both EST and SAGE approaches (Supplementary Table S3), supporting our observation that TTP expression is frequently repressed in tumors.
Restoring TTP alters cell morphology and suppresses tumorigenic phenotypes in HeLa cells
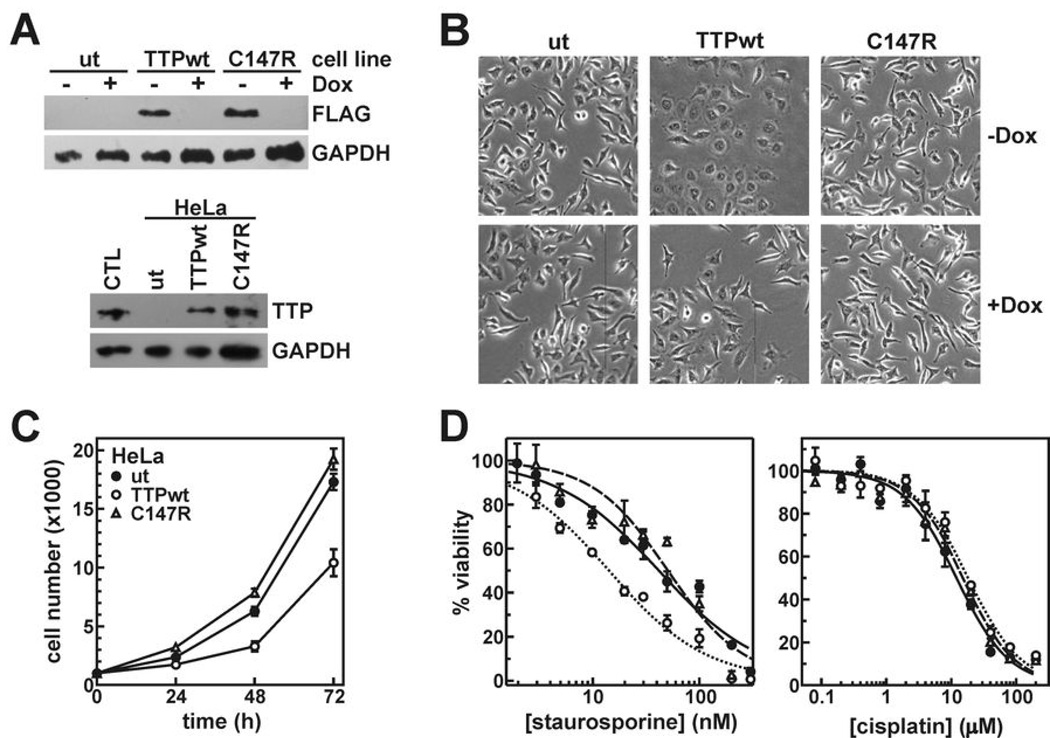
Conceivably, decreasing TTP expression could influence many cellular functions, depending on the population of TTP substrate mRNAs present. To identify TTP-responsive tumorigenic phenotypes, we constructed stably transfected HeLa/Tet-Off cell clones expressing FLAG-tagged TTP from a tetracycline-responsive cassette. Additional clones expressed the TTP mutant C147R; disrupting this Zn2+-coordinating residue within the C-terminal zinc finger abrogates RNA-binding activity (20). Three principal observations prompted use of the HeLa/Tet-Off model. First, HeLa cells are derived from a cervical adenocarcinoma, where TTP expression is frequently repressed (Fig. 1 and Fig. 2). Second, endogenous TTP mRNA is virtually undetectable in HeLa cells (Fig. 2A). Third, potential TTP-dependent influences on cell proliferation or survival necessitated tight regulation of transgene expression to prevent negative selection during clonal expansion.
Dox potently inhibited expression of FLAG-TTPwt or -C147R proteins in HeLa/Tet-Off clones (Fig. 3A). However, removing Dox induced FLAG-TTP to levels consistent with normal cervical tissue. Restoring wild type TTP in HeLa cells induced morphological changes; cells became flattened, increased surface contact area with culture plates, and developed irregular membrane edges (Fig. 3B). These phenomena specifically accompanied expression of FLAG-TTPwt, since morphology did not noticeably change upon induction of the C147R mutant, or in TTPwt clones when the transgene was repressed (+Dox).
Figure 3.
Influence of TTP on tumor cell phenotypes. A, Western blots showing expression of FLAG-TTPwt and -C147R in HeLa/Tet-Off clones 24 hours after removal of Dox (top) and compared to endogenous TTP protein in a cervical tissue lysate (CTL) using anti-TTP antibodies (bottom). B, phase contrast photomicrographs of HeLa clones before (+Dox) and after (-Dox) induction of FLAG-TTPwt and -C147R. C, proliferation of untransfected HeLa cells (ut, closed circles) or cells expressing FLAG-TTPwt (open circles) or FLAG-C147R (triangles). Each point represents the mean ± SD of at least five cell populations. Triplicate independent experiments yielded similar results. D, untransfected or TTPwt/C147R-expressing HeLa cells were counted 24 hours after treatment with various concentrations of staurosporine and cisplatin. Symbol assignments are identical to C and represent the mean ± SD of four cell populations. IC50 values from multiple independent experiments are summarized in Supplementary Table S4.
Since accelerated cell growth is a fundamental characteristic of tumors (1), we measured the effects of TTP on cell proliferation. Following Dox removal, HeLa cells expressing FLAG-TTPwt proliferated approximately 40% slower than untransfected cells (Fig. 3C; P = 0.001). An independent HeLa/FLAG-TTPwt cell clone yielded identical results (data not shown). This effect required the RNA-binding activity of TTP, since FLAG-C147R did not influence cell proliferation. Another property of tumor cells is the ability to evade apoptosis (1). While transient TTP overexpression induces apoptosis in some cultured cell models (21), restoring TTP to physiological levels in HeLa/FLAG-TTPwt clones did not induce significant cell death (data not shown). However, to determine whether TTP could influence apoptotic pathways, we measured the sensitivity of these cultures to two different pro-apoptotic stimuli. Staurosporine is a general protein kinase inhibitor that activates mitochondrial-directed apoptosis (22). Comparing IC50 values between HeLa clonal lines indicates that cells expressing FLAG-TTPwt are 2 to 2.5-fold more sensitive to staurosporine then either untransfected cells or those expressing the C147R mutant (Fig. 3D and Supplementary Table S4; P = 0.002). PARP cleavage analysis (23) verified that cell death following staurosporine treatment was apoptotic (Supplementary Fig. S2). However, TTP did not enhance sensitivity to all pro-apoptotic stimuli. Cisplatin, which induces apoptosis by covalently modifying and distorting DNA (24), killed all HeLa clones with equal efficacy, whether functional TTP was expressed or not (Fig. 3D).
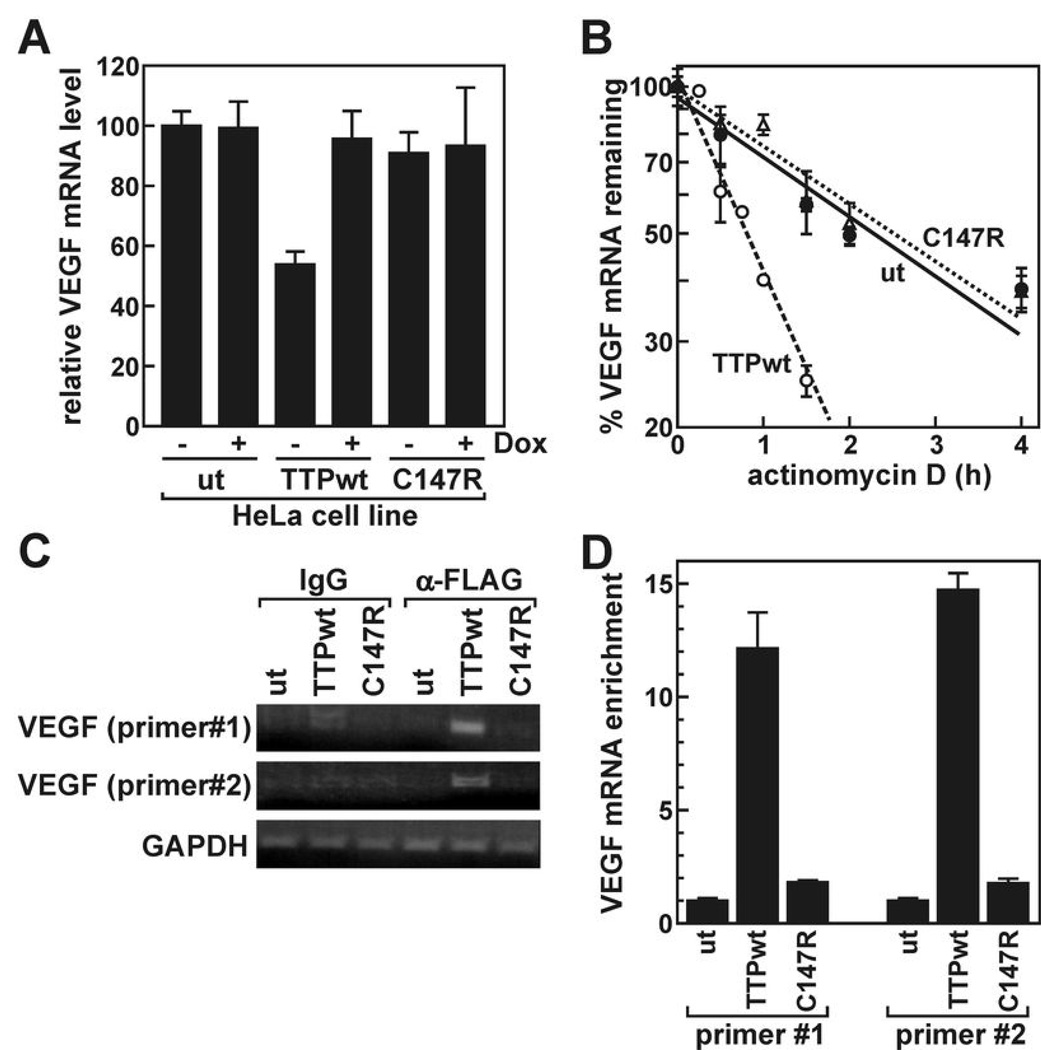
TTP limits VEGF expression in HeLa cells by accelerating VEGF mRNA decay
Aggressive tumors frequently enhance production of pro-angiogenic factors like VEGF (1). Several sequences resembling the UUAUUUAUU high affinity TTP-binding motif (13) are located within the 3’UTR of VEGF mRNA (data not shown), and this transcript is targeted by TTP in Rastransformed fibroblasts and glioma cells (10, 25). In our HeLa clones, expression of FLAG-TTPwt suppressed VEGF mRNA by 50% (Fig. 4A; P = 0.0004) relative to untransfected cells or those expressing C147R. To determine whether FLAG-TTPwt limits VEGF mRNA levels by accelerating its decay, we measured VEGF mRNA stability using actD time course assays (Fig. 4B). These experiments revealed that VEGF mRNA decays more rapidly in HeLa cells expressing FLAG-TTPwt (t1/2 = 0.83 ± 0.11 h, n = 3) than in untransfected cells (t1/2 = 2.3 ± 0.1 h, n = 2; P = 0.0008) or cells expressing the C147R mutant (t1/2 = 2.6 ± 0.1 h, n = 2). Next, we used RNP-IP assays to determine whether TTP binding accompanied accelerated VEGF mRNA turnover. Using both conventional RT-PCR (Fig. 4C) and qRT-PCR (Fig. 4D), VEGF mRNA was easily detected in RNP-IPs from cells expressing FLAG-TTPwt, but not from untransfected HeLa cells or those expressing the C147R mutant, indicating selective association of this transcript with FLAG-TTPwt.
Figure 4.
Recognition and regulation of VEGF mRNA by TTP. A, VEGF mRNA levels were measured by qRT-PCR in untransfected HeLa cells (ut) or cells stably transfected with FLAG-TTPwt or -C147R prior to (+Dox) and 24 hours following (-Dox) transgene induction. Bars represent the mean ± SD of three independent samples normalized to GAPDH mRNA. B, actD time courses measuring VEGF mRNA decay kinetics in each HeLa line 24 hours following transgene induction. mRNA half-lives resolved from multiple independent experiments are given in the text. C, RNP-IP experiments were performed using control IgG or anti-FLAG antibodies and lysates from indicated HeLa lines, then screened for VEGF and GAPDH mRNAs by qualitative RT-PCR. D, VEGF mRNA levels were measured from anti-FLAG RNP-IPs by qRT-PCR and normalized to GAPDH mRNA (mean ± SD of three reactions). An independent replicate experiment yielded similar results.
Cellular consequences of TTP depletion are cell type-specific
To this point, our survey of TTP-responsive tumorigenic phenotypes was limited to highly transformed cells. To evaluate TTP involvement in these pathways using a non-transformed background, MEFs were isolated from TTP knockout mice (TTP−/−; Ref. 26) and wild type littermates (TTP+/+). A Western blot verified the absence of TTP in TTP−/− MEFs (Supplementary Fig. S3A). Unlike our observations in HeLa cells, TTP expression enhanced proliferation of MEFs 3-fold (Supplementary Fig. S3B; P < 0.0001). Similar results were observed using MEFs derived from independent TTP+/+ and TTP−/− littermates (data not shown). Furthermore, TTP expression dramatically decreased the sensitivity of MEFs to staurosporine-induced apoptosis (Supplementary Fig. S3C and Supplementary Table S4; P < 0.0001), unlike the drug-sensitizing effect observed in HeLa cells. Cisplatin sensitivity experiments revealed that both TTP+/+ and TTP−/− MEFs were highly resistant (IC50 > 50 µM) to this compound.
In contrast to the divergent effects of TTP on the proliferation and staurosporine sensitivity of MEFs versus HeLa cells, TTP suppressed VEGF expression similarly in both cell backgrounds (Supplementary Fig. S3). VEGF mRNA levels were 45% lower in TTP+/+ versus TTP−/− MEFs (n = 3; P = 0.027). Also, actD time course assays revealed that VEGF mRNA was degraded more quickly in TTP+/+ MEFs (t1/2 = 1.03 ± 0.08 h, n = 2) than TTP−/− MEFs (t1/2 = 2.6 ± 0.1 h, n = 2; P = 0.003 versus TTP+/+), analogous to observations in HeLa cells.
Suppressed TTP expression is a negative prognostic indicator in breast cancer
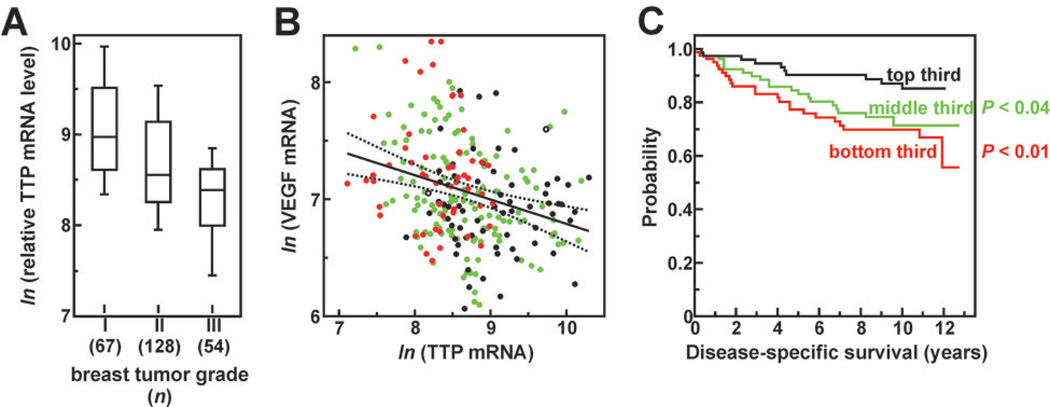
If decreased TTP expression is associated with tumor progression or aggressiveness, we predicted that tumors with varying TTP expression levels would behave differently in the patient population. To test this, we surveyed TTP expression across a gene array dataset derived from the excised breast tumors of 251 patients (27). Considering TTP mRNA levels for each patient as a function of tumor grade revealed a negative correlation between these parameters, with more advanced tumors typically showing the weakest TTP expression (Fig. 5A). A second prediction is that, if TTP destabilizes VEGF mRNA, then VEGF expression should be enhanced in tumors that express TTP poorly. Plotting relative TTP and VEGF mRNA levels for all breast tumors indicated a modest but statistically significant negative correlation between these transcripts (Fig. 5B). However, similar analysis using a gene array dataset from 89 prostate tumors (28) revealed no correlative relationship between these mRNAs (Supplementary Fig. S4), even though TTP expression was potently suppressed in prostate metastases (Supplementary Table S2). This observation was another example where the consequences of TTP suppression in tumors varied with tissue type. Finally, we ranked all 251 breast tumors by TTP mRNA level, and then divided them into 3 equally sized cohorts. Kaplan-Meier analyses of patient outcomes indicated that patients with intermediate or low tumor TTP mRNA levels were 2- to 3-fold more likely to die from recurrent breast cancer following tumorectomy than patients whose tumors strongly expressed TTP at excision (Fig. 5C). Coincident correlation of low TTP mRNA levels with higher tumor grade, elevated VEGF expression, and poor patient survival indicates that suppressed TTP expression in tumors may represent a negative prognostic indicator in breast cancer.
Figure 5.
Correlation analyses of TTP expression versus pathological features and clinical outcomes in breast cancer. TTP mRNA levels were extracted from an array dataset containing expression profiles for 251 human breast tumors. This dataset (GEO Acc# GSE3494) is described in Ref. 27 and includes the Elston-Ellis pathological grade of each tumor (40) and patient mortality from recurrent breast cancer over the subsequent 13 years. A, TTP expression correlates negatively with breast tumor grade (r = −0.431, P = 1.1×10−12). Distribution plots are as described in Fig. 2 except that TTP mRNA is plotted as the ln of normalized array signal intensity. B, VEGF mRNA levels correlate negatively with TTP mRNA in breast tumors (r = − 0.281, P = 5.9×10−6). Black circles indicate grade I tumors, green circles grade II, red circles grade III, and open circles are undefined grade. Dotted lines indicate 95% confidence intervals of the regression solution. C, Kaplan-Meier analyses of patient cohorts ranked by tumor TTP mRNA expression. P values indicate cohort comparisons to patients expressing the highest TTP mRNA levels (black line).
Discussion
The mRNAs encoding many proteins contributing to oncogenesis and tumor progression contain ARE-like sequences within their 3’UTRs, and as such, may be regulated post-transcriptionally by one or more ARE-BPs. While individual ARE-BPs have been linked to specific tumorigenic phenotypes in selected experimental systems (described above), it was unclear whether these mechanisms contribute significantly to clinical neoplastic syndromes. In this study, comparing mRNA levels for four ARE-BPs in tumors versus patient-matched non-transformed tissues presented examples where expression of each RNA-binding factor was dramatically induced or inhibited in cancerous cells. However, several lines of evidence indicated that the mRNA-destabilizing factor TTP is frequently and dramatically suppressed in solid tumors. First, TTP mRNA levels were substantially decreased (≥ 50%) in the majority of tumors versus patient-matched peripheral tissues across 154 patients (Fig. 1), a trend also observed at the protein level among a small cohort of breast tumors (Fig. 2B). Second, constitutive expression of TTP mRNA is very weak in many cancer cell lines relative to untransformed human tissues (Fig. 2A). Third, meta-analysis of gene array datasets revealed decreased TTP mRNA levels in a variety of tumors relative to healthy tissue (Supplementary Table S2). Finally, EST and SAGE tags encoding TTP mRNA fragments were recovered at a significantly lower frequency from cancerous versus normal tissues (Supplementary Table S3).
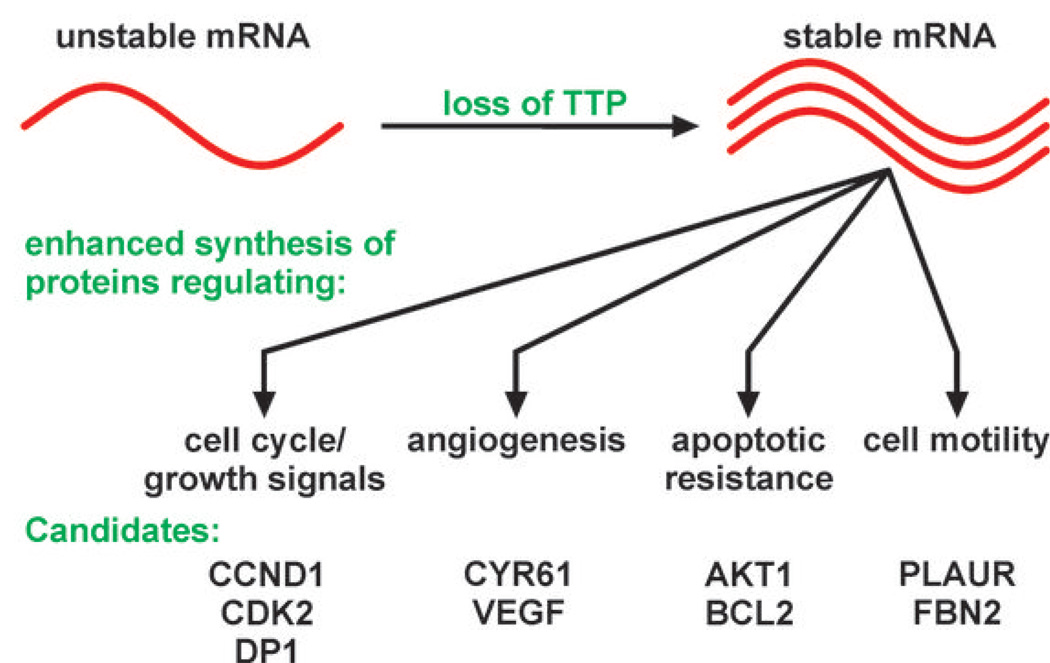
In cells, TTP recognizes selected ARE-containing mRNAs and targets them for decay, which limits the expression of their encoded gene products (12). However, in tumor cells that suppress TTP, we predict that this ARE-containing mRNA population will be stabilized, leading to increases in their cytoplasmic concentrations (Fig. 6). It follows, therefore, that TTP suppression may enhance cellular tumorigenic phenotypes by overexpression of proteins encoded by this mRNA population. To identify TTP substrate mRNAs that might encode protumorigenic factors, we surveyed the ARED database (brp.kfshrc.edu.sa/ARED/; Ref. 29) for mRNAs containing high affinity TTP binding sites in their 3’UTRs. Conceivably, overexpression of even a subset of these proteins following TTP suppression could enhance tumorigenic phenotypes in a variety of ways (Fig. 6 and Supplementary Table S5). For example, mRNAs encoding the cell cycle/growth regulatory proteins cyclin D1 (CCND1) and CDK2 both contain high affinity TTP-binding motifs in their 3’UTRs, as do mRNAs encoding the anti-apoptotic regulators AKT1 and BCL2. Consistent with these possibilities, we observed that HeLa cells lacking TTP exhibited both increased proliferation rates and partial resistance to staurosporine-induced apoptosis (Fig. 3). Our HeLa cell model also revealed that TTP limits the expression of VEGF, which can limit the aggressiveness of many cancers by abrogating the potential of VEGF to promote tumor vascularization (30). Finally, high affinity TTP binding sites were also observed in mRNAs encoding factors that regulate cell motility or invasiveness, including urokinase plasminogen activator receptor (PLAUR) and fibrillin-2 (FBN2), suggesting that loss of TTP may increase the production of these proteins.
Figure 6.
A model for control of pro-oncogenic post-transcriptional gene regulatory networks by TTP. Features of the model are described in the Discussion. Potential TTP substrate mRNAs encoding factors promoting tumor progression were identified using the ARED database as described in Supplementary Table S5.
While the influence of TTP on HeLa cells is consistent with a classical tumor suppressor, the tumorigenic consequences of TTP suppression can vary widely in different cell/tissue types. For example, in MEFs TTP enhanced proliferation and decreased sensitivity to staurosporine-induced apoptosis (Supplementary Fig. S3), directly opposing our observations in HeLa cells. Experiments in mast tumor cells presented a third outcome, whereby TTP did not alter cell proliferation in culture, but precluded tumor growth in murine xenografts (9). Conversely, inhibiting VEGF expression represents a more consistent tumor suppressing function for TTP. Here, VEGF mRNA was destabilized by TTP in both HeLa (Fig. 4B) and MEF (Supplementary Fig. S3) cells, and VEGF mRNA levels correlated inversely with TTP mRNA across 251 breast tumors (Fig. 5B). TTP also destabilizes VEGF mRNA in glioma cells (25) and Ras-transformed fibroblasts (10), inhibiting vascularization of tumor xenografts from the latter. However, the absence of any correlation between VEGF and TTP mRNAs in prostate tumors (Supplementary Fig. S4) suggests that even induction of VEGF expression is not a uniform consequence of TTP suppression. As an example of how TTP may modulate tumorigenic phenotypes in a cell type-specific manner, let us consider the differential effects of TTP on the staurosporine sensitivity of HeLa versus MEF cells. Staurosporine promotes mitochondria-mediated cell death signaling by several distinct routes, including mitochondrial recruitment of p53 and inhibition of protein kinase B/AKT signaling pathways (31, 32). While the molecular basis for modulation of apoptosis by TTP is unknown, two mRNAs recently shown to bind TTP in macrophages (33) provide some intriguing mechanistic possibilities. One TTP-binding mRNA encodes RBBP6 (also called PACT or P2P-R), a protein that binds p53 and inhibits its proapoptotic activity (34). By binding and destabilizing RBBP6 mRNA, TTP should decrease expression of this factor, thus diminishing its ability to suppress p53-dependent apoptosis. In this case, TTP could enhance sensitivity to some pro-apoptotic stimuli, as was observed in HeLa cells. Another mRNA targeted by TTP in macrophages encodes the transcription factor ST18 (33), which induces expression of many pro-apoptotic and pro-inflammatory genes in fibroblasts (35). By destabilizing ST18 mRNA, TTP should suppress expression of St18-inducible genes, thus decreasing apoptotic sensitivity analogous to our observations in MEFs. Given the variety of complex and often interconnected pathways directing apoptosis, we propose three mechanisms that may account for differential apoptotic sensitivity of HeLa versus MEF cells in response to TTP. First, TTP can only regulate substrate mRNAs with which it is presented; hence, the unique transcriptional profile of each cell type will dictate the mRNA substrate pool subject to TTP activity. Considering the above examples, it is possible that the genes encoding RBBP6 and/or ST18 may be transcribed in only selected cell models. Second, TTP is but one of a diverse collection of 20 or more ARE-BPs identified to date (4, 12, 36). While many ARE-BPs present distinct mRNA substrate preferences (13, 16, 37, 38), significant overlap between binding determinants can permit competitive, additive, or redundant functional relationships between these proteins. Accordingly, while suppression of TTP may enhance RBBP6 and/or ST18 expression by stabilizing their mRNAs, it is also possible that loss of TTP may permit redundant or even more potent mRNA-destabilizing activities access to one or both of these mRNAs in a cell-specific manner. Finally, the relative influences of various pro- and anti-apoptotic pathways on staurosporine-induced cell death are likely to be unique in each cell model. As such, suppression of RBBP6 or ST18 (or other TTP substrate mRNAs) may influence the apoptotic sensitivity of HeLa cells differently than MEFs. Considering these possibilities, we believe that rigorous identification of TTP substrate mRNA populations across different cell types will be required to delineate the specific post-transcriptional regulatory networks controlled by this factor.
The central findings of this work are that TTP expression is frequently repressed in human cancers and that diminution of functional TTP can modulate diverse tumorigenic phenotypes. In breast cancer, decreased TTP expression may be a negative prognostic indicator, since patients with low tumor TTP mRNA levels disproportionately presented more advanced tumors and increased risk of mortality from recurrent cancer. However, frequent suppression of TTP in tumors from many tissue types suggests that its utility as a prognostic indicator may be much more inclusive. Other data suggest that measuring tumor TTP expression may be diagnostically useful. For example, TTP-dependent effects on the sensitivity of HeLa and MEF cells to staurosporine indicates that some apoptotic signaling pathways may involve factors encoded by TTP-regulated mRNAs. Because radiation and many chemotherapeutic strategies function by promoting tumor cell apoptosis, it follows that altered target cell sensitivity to such stimuli resulting from TTP suppression may limit (or enhance) treatment efficacy. The TTP-related protein BRF1 has been linked to apoptosis in cultured head and neck squamous cell carcinoma lines, where cisplatin sensitivity varies inversely with BRF1 levels (39). Curiously, TTP did not influence the sensitivity of either HeLa or MEF cells to cisplatin, possibly reflecting yet another example of the cell type- and/or protein-specific influence of ARE-BPs on tumorigenic phenotypes.
Supplementary Material
Acknowledgments
Grant Support: American Cancer Society grant RSG-07-293-01-GMC and NIH grant R01 CA102428 (to G.M.W.), and the Intramural Research Program of the NIA-NIH (M.G. and Y.K.).
Footnotes
Potential conflicts of interest: The authors have filed a patent application covering the measurement of TTP expression as an oncological diagnostic and prognostic tool.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Chen CYA, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 3.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 4.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang T, Kruys V, Huez G, Gueydan C. AU-rich element-mediated translational control: complexity and multiple activities of trans-acting factors. Biochem Soc Trans. 2002;30:952–958. doi: 10.1042/bst0300952. [DOI] [PubMed] [Google Scholar]
- 6.Gouble A, Grazide S, Meggetto F, Mercier P, Delsol G, Morello D. A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res. 2002;62:1489–1495. [PubMed] [Google Scholar]
- 7.Lin S, Wang W, Wilson GM, et al. Down-regulation of cyclin D1 expression by prostaglandin A2 is mediated by enhanced cyclin D1 mRNA turnover. Mol Cell Biol. 2000;20:7903–7913. doi: 10.1128/mcb.20.21.7903-7913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denkert C, Weichert W, Winzer KJ, et al. Expression of the ELAV-like protein HuR is associated with higher tumor grade and increased cyclooxygenase-2 expression in human breast carcinoma. Clin Cancer Res. 2004;10:5580–5586. doi: 10.1158/1078-0432.CCR-04-0070. [DOI] [PubMed] [Google Scholar]
- 9.Stoecklin G, Gross B, Ming XF, Moroni C. A novel mechanism of tumor suppression by destabilizing AU-rich growth factor mRNA. Oncogene. 2003;22:3554–3561. doi: 10.1038/sj.onc.1206418. [DOI] [PubMed] [Google Scholar]
- 10.Essafi-Benkhadir K, Onesto C, Stebe E, Moroni C, Pages G. Tristetraprolin inhibits Ras-dependent tumor vascularization by inducing vascular endothelial growth factor mRNA degradation. Mol Biol Cell. 2007;18:4648–4658. doi: 10.1091/mbc.E07-06-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian Q, Streuli M, Saito H, Schlossman SF, Anderson P. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell. 1991;67:629–639. doi: 10.1016/0092-8674(91)90536-8. [DOI] [PubMed] [Google Scholar]
- 12.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 13.Brewer BY, Malicka J, Blackshear PJ, Wilson GM. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: Conformational changes coupled to the bipartite nature of AU-rich mRNA-destabilizing motifs. J Biol Chem. 2004;279:27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- 14.Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol Cell Biol. 2006;26:9196–9208. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki A, Tsutomi Y, Akahane K, Araki T, Miura M. Resistance to Fas-mediated apoptosis: activation of caspase 3 is regulated by cell cycle regulator p21WAF1 and IAP gene family ILP. Oncogene. 1998;17:931–939. doi: 10.1038/sj.onc.1202021. [DOI] [PubMed] [Google Scholar]
- 16.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrick DM, Blackshear PJ. Comparative expression of tristetraprolin (TTP) family member transcripts in normal human tissues and cancer cell lines. Arch Biochem Biophys. 2008;462:278–285. doi: 10.1016/j.abb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lal A, Lash AE, Altschul SF, et al. A public database for gene expression in human cancers. Cancer Res. 1999;59:5403–5407. [PubMed] [Google Scholar]
- 20.Lai WS, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: Non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J Biol Chem. 2002;277:9606–9613. doi: 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- 21.Johnson BA, Geha M, Blackwell TK. Similar but distinct effects of the tristetraprolin/TIS11 immediate-early proteins on cell survival. Oncogene. 2000;19:1657–1664. doi: 10.1038/sj.onc.1203474. [DOI] [PubMed] [Google Scholar]
- 22.Jarvis WD, Turner AJ, Povirk LF, Traylor RS, Grant S. Induction of apoptotic DNA fragmentation and cell death in HL-60 promyelocytic leukemia cells by pharmacological inhibitors of protein kinase C. Cancer Res. 1994;54:1707–1714. [PubMed] [Google Scholar]
- 23.Boulares AH, Yakovlev AG, Ivanova V, et al. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J Biol Chem. 1999;274:22932–22940. doi: 10.1074/jbc.274.33.22932. [DOI] [PubMed] [Google Scholar]
- 24.Jordan P, Carmo-Fonseca M. Molecular mechanisms involved in cisplatin cytotoxicity. Cell Mol Life Sci. 2000;57:1229–1235. doi: 10.1007/PL00000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suswam E, Li Y, Zhang X, et al. Tristetraprolin down-regulates interleukin-8 and vascular endothelial growth factor in malignant glioma cells. Cancer Res. 2008;68:674–682. doi: 10.1158/0008-5472.CAN-07-2751. [DOI] [PubMed] [Google Scholar]
- 26.Taylor GA, Carballo E, Lee DM, et al. A pathogenic role for TNFα in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 27.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu YP, Landsittel D, Jing L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 29.Bakheet T, Williams BRG, Khabar KSA. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carmeliet P. VEGF as a key mediator of angiogenesis in cancer. Oncology. 2005;69 Suppl. 3:4–10. doi: 10.1159/000088478. [DOI] [PubMed] [Google Scholar]
- 31.Charlot JF, Prétet JL, Mougin C. Mitochondrial translocation of p53 and mitochondrial membrane potential (ΔΨm) dissipation are early events in staurosporine-induced apoptosis of wild type and mutated p53 epithelial cells. Apoptosis. 2004;9:333–343. doi: 10.1023/b:appt.0000025810.58981.4c. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Zhang B, Peng X, Perpetua M, Harbrecht BG. Bcl-XL prevents staurosporine-induced hepatocyte apoptosis by restoring protein kinase B/mitogen-activated protein kinase activity and mitochondrial integrity. J Cell Physiol. 2007;215:676–683. doi: 10.1002/jcp.21350. [DOI] [PubMed] [Google Scholar]
- 33.Stoecklin G, Tenebaum SA, Mayo T, et al. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem. 2008;283:11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L, Deng B, Xing G, et al. PACT is a negative regulator of p53 and essential for cell growth and embryonic development. Proc Natl Acad Sci USA. 2007;104:7951–7956. doi: 10.1073/pnas.0701916104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Siqueira MF, Behl Y, Alikhani M, Graves DT. The transcription factor ST18 regulates proapoptotic and proinflammatory gene expression in fibroblasts. FASEB J. 2008;22:3956–3967. doi: 10.1096/fj.08-111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson GM, Brewer G. The search for trans-acting factors controlling messenger RNA decay. Prog Nucleic Acids Res Mol Biol. 1999;62:257–291. doi: 10.1016/s0079-6603(08)60510-3. [DOI] [PubMed] [Google Scholar]
- 37.Fialcowitz EJ, Brewer BY, Keenan BP, Wilson GM. A hairpin-like structure within an AU-rich mRNA-destabilizing element regulates trans-factor binding selectivity and mRNA decay kinetics. J Biol Chem. 2005;280:22406–22417. doi: 10.1074/jbc.M500618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez de Silanes I, Galban S, Martindale JL, et al. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SK, Kim SB, Kim JS, et al. Butyrate response factor 1 enhances cisplatin sensitivity in human head and neck squamous cell carcinoma cell lines. Int J Cancer. 2005;117:32–40. doi: 10.1002/ijc.21133. [DOI] [PubMed] [Google Scholar]
- 40.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.