Summary
Recurrent, prognostically significant chromosomal abnormalities occur in approximately 75% of pediatric acute lymphoblastic leukemia (ALL), but only infrequently in children with Down syndrome (DS) and ALL. Recently, novel somatic activating mutations in Janus kinase 2 (JAK2) were reported in 18% of DS ALL. Here we report identification and clinical correlates of JAK2 mutations in an independent cohort. JAK2 activating mutations occurred in 10 of 53 DS ALL cases (18.9%). Mutations were overrepresented in males (p<0.03), occurred once in association with high hyperdiploidy, and were not significantly correlated with age, initial white blood count, or event-free survival. Our results confirm significance of JAK-STAT pathway activation in DS ALL.
Keywords: Down syndrome, pediatric, acute lymphoblastic leukemia, JAK2
Introduction
Children with Down syndrome (DS) have a 10–20 fold increased risk of developing acute leukemia, either acute lymphoblastic leukemia (ALL) or acute myeloid leukemia (AML) (Hasle et al, 2000). A unique genetic lesion, mutations in the transcription factor GATA1, has been implicated in pathogenesis of AML in patients with DS (Wechsler et al, 2002). However, no such characteristic abnormality unique to DS ALL had been identified until the recent discovery of novel mutations in the Janus kinase 2 (JAK2) gene in 18% of DS ALL cases, nearly all occurring at a specific site, arginine 683 (R683) (Bercovich et al, 2008). Another JAK2 mutation, V617F (valine to phenylalanine), occurs in over 90% of polycythemia vera and approximately half of other myeloproliferative disorders. However, mutations involving R683 have been identified only in cases of DS ALL (and one ALL case with possible Down syndrome mosaicism (Bercovich et al, 2008)). Reported cases include two DS ALL cases with complex in-frame insertion and deletion events proximal to R683 (Bercovich et al, 2008); one case with a 5-amino acid deletion including R683 (Malinge et al, 2007); and the majority with R683 point mutations (Bercovich et al, 2008;Kearney et al, 2008). Three of these point mutations have been shown to cause JAK2 constitutive activation and cytokine-independent growth in vitro (Bercovich et al, 2008;Kearney et al, 2008). Since only 29 cases have been reported in the literature to date (Malinge et al, 2007;Bercovich et al, 2008;Kearney et al, 2008), we sought to confirm the incidence and associated features of JAK2 R683 mutations in an independent cohort of DS ALL cases.
Materials and methods
Patient samples
The study population included diagnostic and remission bone marrow aspirates obtained from 40 DS and 6 non-DS patients with B-precursor ALL diagnosed between 1998–2007 and treated on or according to Pediatric Oncology Group protocols, and 13 Italian DS patients with B-precursor ALL diagnosed between 1989–2000 and treated on protocols of the Associazione Italiana Ematologia ed Oncologia Pediatrica (AIEOP). All patient samples were obtained with informed consent under protocols approved by institutional review board (U.S.) or ethics committee (Italy), and the study was approved by the Baylor College of Medicine institutional review board. DNA was extracted from the bone marrow specimens using standard methods (Qiagen Allprep DNA/RNA Mini Kit and QIAamp DNA Blood Mini Kit, Valencia, CA). Samples were linked to demographic and outcome data obtained from the Children’s Oncology Group (COG) statistical office or abstracted from the medical record (for non-COG samples).
JAK2 sequence analysis
Samples were analyzed by Sanger di-deoxy sequencing, pyrosequencing, or both. For Sanger sequencing, we utilized previously published primer sequences (Bercovich et al, 2008), with addition of the universal M13 sequence, to amplify JAK2 exon 16 which contains R683. Approximately 25 ng total genomic DNA was used for polymerase chain reaction (PCR) amplification using 2x polymerase master mix (Promega, Madison, WI). PCR products were sequenced in both forward and reverse directions and sequence analysis was performed using Sequencher 4.8 software (Gene Codes Corporation, Ann Arbor, MI). For pyrosequencing, a 200 base-pair region containing the R683 site was amplified by PCR using the following primers (5′ to 3′): forward ATGTGCGTTTAACTCTAATAG; biotin-labeled reverse GCAAAACTGTAATACTAATGC; pyrosequencing GCCAAAAATATTCTGCT.
Statistical analysis
Patient demographic characteristics were compared using Fisher’s exact test for categorical variables and two-sample t test for continuous variables. Event-free survival estimates were obtained using the Kaplan-Meier method, and standard errors of the estimates were calculated by the method of Peto and Peto (Peto et al, 1977). Time to event was calculated as the time from study entry to first event (relapse, secondary malignancy, or death) or date of last contact. The log-rank test was used for comparison of survival curves by JAK2 mutation status.
Results and Discussion
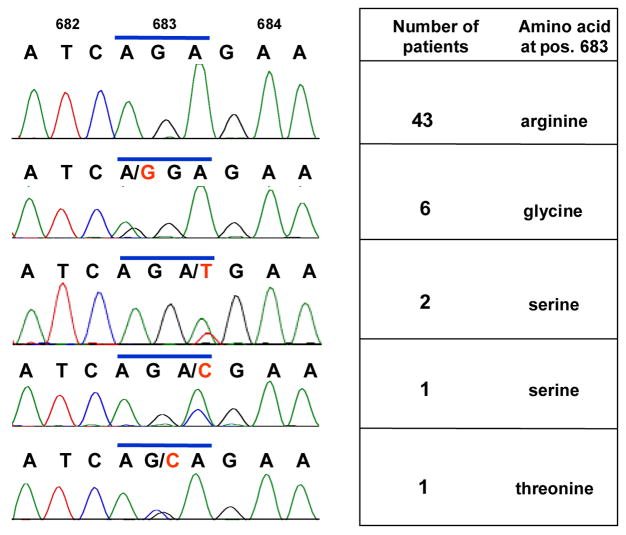
We sequenced JAK2 exon 16, which contains R683, in an independent, previously unstudied cohort of 53 pediatric DS ALL patients, as well as six control ALL patients without DS. For 25 of the 53 DS patients, paired nonleukemic, germline DNA was also available for sequence analysis. Ten of 53 (18.9%) DS ALL cases carried a point mutation at R683, virtually identical to the incidence reported by Bercovich et al (2008) and slightly lower than the incidence reported by Kearney et al (2008) (Fig 1). No mutations other than point mutations were observed. Six mutations resulted in substitution of glycine; three serine; and one threonine, the latter being an amino acid substitution not previously reported. All cases for which Sanger sequence analysis demonstrated point mutations were confirmed by pyrosequencing (data not shown). Five of the DS ALL cases bearing JAK2 R683 mutations had paired germline DNA samples available, and none of these demonstrated mutations, suggesting that the mutations arose as somatic events. All mutations appeared heterozygous, as did all but one in the Bercovich et al study (2008).
Fig 1.
JAK2 mutations in pediatric DS ALL patients. Chromatograms correspond to amino acids 682-684, with the top row depicting a wild-type case (R683), and subsequent rows depicting representative heterozygous point mutations substituting glycine (R683G), serine (R683S) and threonine (R683T).
Comparison of JAK2 mutated and wild-type cases in our cohort showed no significant differences in patient age or initial WBC, but JAK2 mutations occurred significantly more often in males (Table I). This is in contrast to the report by Bercovich et al. (2008) that JAK2 mutations occurred more frequently in younger patients, with a trend toward a higher initial white blood count (WBC), and with no significant gender predisposition. Our reports agreed in finding no significant correlation between JAK2 mutation status and 5-year event-free survival, with median follow-up of 30 months. One JAK2 mutated case in the present study had cytogenetic evidence of high hyperdiploidy (51 chromosomes), whereas prior studies reported no common chromosomal abnormalities except a single case of 9p21 deletion among their JAK2 mutated cases (Bercovich et al, 2008; Kearney et al, 2008).
Table I.
Clinical and demographic features of JAK2 mutated versus JAK2 wild-type DS ALL cases.
| Parameters | JAK2 wild type | JAK2 mutated | p value |
|---|---|---|---|
| Age (years, mean ± std. dev.) | 6.6 ± 4.2 | 5.8 ± 3.3 | 0.28 |
| Initial white blood count (×103 cells/μl, mean ± std. dev.) | 40.0 ± 63.2 | 35.2 ± 67.5 | 0.42 |
| Male gender (n, %) | 16 (37%) | 8 (80%) | 0.03 |
| Recurrent cytogenetic abnormality* (n, %) | 3 (10.3%) | 1 (20%) | 0.49 |
| 5-year event-free survival (percent ± standard error) | 76.3 ± 11.2% | 87.5 ± 17.9% | 0.97 |
Recurrent cytogenetic abnormalities are defined as TEL-AML1, E2A-PBX1, BCR-ABL, MLL rearrangement, or high hyperdiploidy (>50 chromosomes).
Our results confirm that nearly one-fifth of DS ALL cases bear mutations of JAK2 at a unique site, R683. It is noteworthy that among the 36 R683 point mutations collectively reported to date, all but a single lysine substitution (Bercovich et al, 2008) result in replacement of the strongly basic amino acid arginine with a neutral amino acid, perhaps reflecting selective pressure for mutations which all effect a similar and marked alteration in protein binding characteristics. Novel features of the present study include the association of JAK2 mutations with male gender; and lack of association with age or initial WBC. Identification of a JAK2 mutation in a case with concomitant high hyperdiploidy suggests that at least on occasion, JAK2 mutations may arise in cooperation with abnormalities found in non-DS ALL. Further studies, with greater patient numbers, will be necessary to conclusively resolve these questions, and to further elucidate the role of the JAK-STAT pathway in the pathogenesis of DS ALL.
Acknowledgments
We thank Renee Webb and Carlos Vega for assistance with pyrosequencing, and the patients and families whose participation made this work possible. This work was supported in part by an American Society of Hematology Trainee Award (C.R.); Bear Necessities Pediatric Research Foundation Grant (K.R.); Children’s Cancer Research Foundation Grant (K.R.); and National Institutes of Health Pediatric Oncology Clinical Research Training Grant CA90433-06 (K.R.).
Footnotes
Institution where work was performed: Texas Children’s Cancer Center, Baylor College of Medicine, Houston, Texas
References
- Bercovich D, Ganmore I, Scott LM, wainreb G, Birger Y, Elimelech A, Shochat C, Cazzaniga G, Biondi A, Basso G, Cario G, Schrappe M, Stanulla M, Strehl S, Haas OA, Mann G, Binder V, Borkhardt A, Kempski H, Trka J, Bielorei B, Avigad S, Stark B, Smith O, Dastugue N, Bourquin JP, Tal NB, Green AR, Izraeli S. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down’s syndrome. Lancet. 2008 doi: 10.1016/S0140-6736(08)61341-0. [DOI] [PubMed] [Google Scholar]
- Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukaemia and solid tumours in individuals with Down’s syndrome. Lancet. 2000;355:165–169. doi: 10.1016/S0140-6736(99)05264-2. [DOI] [PubMed] [Google Scholar]
- Kearney L, Gonzalez De CD, Yeung J, Procter J, Horsley SW, Eguchi-Ishimae M, Bateman CM, Anderson K, Chaplin T, Young BD, Harrison CJ, Kempski H, So CW, Ford AM, Greaves M. A specific JAK2 mutation (JAK2R683) and multiple gene deletions in Down syndrome acute lymphoblastic leukaemia. Blood. 2008 doi: 10.1182/blood-2008-08-170928. [DOI] [PubMed] [Google Scholar]
- Malinge S, Ben-Abdelali R, Settegrana C, Radford-Weiss I, Debre M, Beldjord K, Macintyre EA, Villeval JL, Vainchenker W, Berger R, Bernard OA, Delabesse E, Penard-Lacronique V. Novel activating JAK2 mutation in a patient with Down syndrome and B-cell precursor acute lymphoblastic leukemia. Blood. 2007;109:2202–2204. doi: 10.1182/blood-2006-09-045963. [DOI] [PubMed] [Google Scholar]
- Peto R, Pike MC, Armitage P, Breslow NE, Cox DR, Howard SV, Mantel N, McPherson K, Peto J, Smith PG. Design and analysis of randomized clinical trials requiring prolonged observation of each patient. II analysis and examples. Br J Cancer. 1977;35:1–39. doi: 10.1038/bjc.1977.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler J, Greene M, McDevitt MA, Anastasi J, Karp JE, Le Beau MM, Crispino JD. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]