Abstract
Polypyrimidine tract-binding protein 1 (PTBP1) is a multi-functional RNA-binding protein that is aberrantly overexpressed in glioma. PTBP1 and its brain-specific homologue polypyrimidine tract-binding protein 2 (PTBP2) regulate neural precursor cell differentiation. However, the overlapping and non-overlapping target transcripts involved in this process are still unclear. To determine why PTBP1 and not PTBP2 would promote glial cell-derived tumours, both PTBP1 and PTBP2 were knocked down in the human glioma cell lines U251 and LN229 to determine the role of these proteins in cell proliferation, migration, and adhesion. Surprisingly, removal of both PTBP1 and PTBP2 slowed cell proliferation, with the double knockdown having no additive effects. Decreased expression of both proteins individually and in combination inhibited cell migration and increased adhesion of cells to fibronectin and vitronectin. A global survey of differential exon expression was performed following PTBP1 knockdown in U251 cells using the Affymetrix Exon Array to identify PTBP1-specific splicing targets that enhance gliomagenesis. In the PTBP1 knockdown, previously determined targets were unaltered in their splicing patterns. A single gene, RTN4 (Nogo) had significantly enhanced inclusion of exon 3 when PTBP1 was removed. Overexpression of the splice isoform containing exon 3 decreased cell proliferation to a similar degree as the removal of PTBP1. These results provide the first evidence that RNA-binding proteins affect the invasive and rapid growth characteristics of glioma cell lines. Its actions on proliferation appear to be mediated, in part, through alternative splicing of RTN4.
Keywords: PTBP1, PTBP2, RTN4, glioma, RNA splicing
Introduction
Alternative RNA splicing can greatly affect protein levels and functions. In cancer, abnormal splicing often leads to tumour-promoting splice variants that are translated into activated oncogenes or inactivated tumour suppressors (Venables, 2004; Srebrow and Kornblihtt, 2006). Normal splicing patterns can be disrupted by either cis-acting mutations of splicing regulatory elements or changes in trans-acting splicing factors (Venables, 2004). An example of the latter is found in lung cancer where overexpression of the ASF/SF2 splicing factor generates splice isoforms in BIN1 and S6K1 that enhance tumour formation (Karni et al., 2007). Increases in the splicing factor Polypyrimidine Tract Binding protein (PTBP1, also known as PTB) that are associated with glioma malignancy could have similar oncogenic effects (Cheung et al., 2006).
The expression patterns and RNA-binding sites of both PTBP1 and its homolog, PTBP2 (also known as nPTB or brPTB) imply that they have similar, but distinct roles in brain development and function. In the mammalian foetal brain, PTBP1 and PTBP2 are expressed at high levels and then both transcripts decrease in the mature adult brain where staining patterns become mutually exclusive: PTBP1 in glial cells and PTBP2 mostly in neurons (Lillevali et al., 2001; Boutz et al., 2007; Makeyev et al., 2007). miRNA-124 may serve to reduce levels of PTBP1 to enable the PTBP2 expression that induces neuron precursor cell differentiation (Makeyev et al., 2007). The existence of similar regulatory pathways in glioma is unknown. Both homologs bind to RNA regulatory sequence motifs, CUCUCU and UUCUCU that occur at high frequencies within intronic sequences flanking brain-specific alternative exons (Brudno et al., 2001; Castle et al., 2008). Despite the similar binding sequences, these regulators of RNA expression have both overlapping and non-overlapping targets. The combination of differential expression and non-overlapping targets may affect glial and neuronal cell fate determination (Boutz et al., 2007; Grabowski, 2007). Therefore, we sought to determine the reasons behind the overexpression of PTBP1 and not PTBP2 in glioma in order to clarify their roles in neural progenitor cell growth and differentiation.
Several pieces of evidence support the notion that PTBP1 functions in cancer initiation and progression. In ovarian cancer, PTBP1 levels also correlate with the degree of malignancy (He et al., 2007). Higher amounts of PTBP1 occur in advanced, as compared to benign, ovarian tumours and PTBP1 increases when ovarian epithelial cells are immortalized (He et al., 2007). Removal of PTBP1 from ovarian tumour cells slows cell proliferation, anchorage-independent growth, and cell invasion. PTBP1 also enhances these aspects of tumour growth in PC3 and HeLa cells (Wang et al., 2008). Since PTBP1 has multiple functions in RNA metabolism, it could affect malignancy potential in different ways (Wagner and Garcia-Blanco, 2001). As a splicing factor, it induces exon skipping in genes that are involved in proliferation (FGFR1, FGFR2), invasion (CSRC), motility (ACTN, FBN), apoptosis (FAS, CASP2), and multi-drug resistance (ABCC1). In the cytoplasm, PTBP1 increases IRES-mediated translation of genes in proliferation (IGFR1), angiogenesis (VEGF), and apoptosis (APAF1). Furthermore, PTBP1 can form complexes with focal adhesion-encoding transcripts at the cell membrane, which may affect cell spreading (de Hoog et al., 2004). Taken together, mis-regulation of PTBP1 could cause multiple epigenetic changes in gene expression to promote tumourigenesis. We reduced levels of PTBP1 and PTBP2 in glioma cells and found decreased proliferation, migration, and increased adhesion of cells. A PTBP1-driven splicing change in RTN4 (also known as Nogo) appears to impact proliferation.
Materials and Methods
Cell culture and antisense morpholino experiments
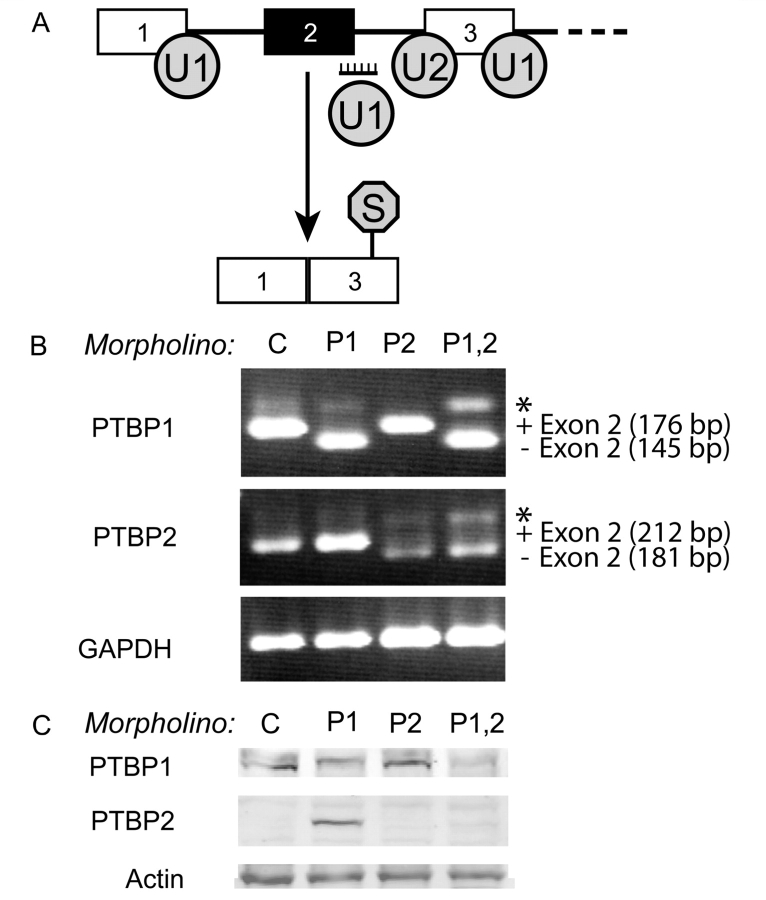
U251 and LN229 cells were grown in DMEM, high glucose with 5–10% FBS at 37°C, 5% CO2 using standard tissue culture methods. Knockdowns were achieved by using antisense morpholino oligonucleotides that target the 5′ splice sites in exon 2 within PTBP1 and/or PTBP2 primary transcripts. For both genes, induction of exon 2 skipping produces an RNA isoform containing a frameshift that introduces a premature termination codon (Fig. 2A). Morpholino oligonucleotides were introduced into cells by scrape delivery, as previously described (Bruno et al., 2004). Briefly, 106 cells plated in 100 mm culture dishes were scraped in the presence of 2.5–5 µM morpholino oligonucleotide in serum-free medium. After 72 h, the cells were scrape-loaded a second time in like manner and split into two 100 mm culture dishes. The following day, the cells were placed in serum-free media for 24 h before proliferation, wound-healing, and immunofluorescence assays were initiated, or left in serum-containing medium for the adhesion assay. The control morpholino oligo was 5′-CCTCTTACCTCAGTTACAATTTATA-3′, which can only target a mutant β-globulin mRNA with no predicted biological effect (Gene Tools, Philomath, OR). Morpholino oligos for PTBP1 and PTBP2 knockdown were 5′-AGTGAGAAGTGCCTACCTTTGTACC-3′ and 5′-CATAGTATTTTCCTACCTCACGCC-3′, respectively (Gene Tools, Philomath, OR).
Figure 2.
Targeted treatment with morpholino oligonucleotides downregulates PTBP1 and PTBP2. (A) Diagram illustrating the mechanism whereby targeting the exon 2 splice site induces skipping in either PTBP1 or PTBP2 transcripts through blocking of U1 snRNP binding to create mRNAs containing premature stop codons. (B) RT–PCR results for PTBP1, PTBP2 and GAPDH (loading control) after indicated morpholino treatments. Knockdown treatments included: control oligo (C), PTBP1-specific oligo (P1), PTBP2-specific oligo (P2), and a combination of both PTBP1- and PTBP2-specific oligos (P1,2). The asterisk denotes a heterodimer band between exon 2 +/− PCR products. (C) Western blot of PTBP1, PTBP2 and the actin loading control levels after the morpholino treatments described above. The observed PTBP1 doublet is derived from alternative mRNA isoforms.
Plasmid construction and U251 transfection
The MultiSite Gateway Technology (Invitrogen, Carlsbad, CA) was used to generate a plasmid that expresses Nogo-A cDNA under the expression of the CMV promoter. The entry vector pENTR-221 was purchased with the clone that includes exons 2 and 3 (pENTR™221 IOH53642, Accession Number: NM_020532.4). The clone was recombined into the destination vector pcDNA3.1/nV5-DEST to produce a v5-labelled Nogo-A designated as: pDEST-NogoA in a 1:1 ratio using the LR Clonase II enzyme mixture (Invitrogen, Carlsbad, CA). Clones were screened and sequenced for verification of insert integrity (Qiagen, Valencia, CA). Transient transfections were done in U251 cells. Total 250 000 cells were plated into each well of a 6-well dish. To prepare the transfection mixture, 2 μg of each plasmid was diluted into 100 μl of OPTIMEM medium (Invitrogen, Carlsbad, CA). Then, 5 μl of DreamFect transfection reagent (OZ Biosciences, Marseille Cedex 9, France) was added to 100 μl of OPTIMEM medium (Invitrogen, Carlsbad, CA). These tubes were mixed together and incubated at room temperature for 17 min. Then, the mixture was added to 2 ml of DMEM per well and placed at 37°C, 5% CO2. After 4 h, fresh media was added to the cells. The cells were incubated for another 2 days, after which the media was changed to G418 selection media (400 μg/ml) for 1 week. The Gateway plasmid was a gift from Dr Oliver Bogler (M.D. Anderson Cancer Centre, Houston, TX).
Standard and real-time RT–PCR
The RNA samples and standard RT–PCR used to monitor alternative splicing events were performed as previously described (Cheung et al., 2008). Briefly, 2 µg of RNA was used for reverse transcription: 1 µg primed with oligo-dT (50 ρmol for 1 µg of RNA) and the other 1 µg primed with random decamers (3 µg for 1 µg of RNA). The PCR reaction was carried out for 35–37 cycles, 55–62°C annealing depending on the primer set and transcript abundance. Taqman Assays Hs01106274_m1 and Hs00199671_m1 were used to detect levels of RTN4. Taqman Universal PCR mix 10 µl was used with 1 µl of Taqman probe, 1 µl of VIC-labelled PKC1 and 8 µl of 1:50 cDNA. The expression level of exon 3 was quantified by measuring the proportion of transcripts containing exon 3. The first Taqman assay (Hs01106274_m1) contains probes hybridizing to exons 3–4 and the other (Hs00199671_m1) measures the junction of exons 4–5. Comparative Ct was used. The ratio of exons 3–4/exons 4–5 was calculated, where exons 4–5 was used as the reference target. Taqman Universal Mix 10 μl was mixed with 1 μl of Taqman probe and 9 μl of 1:50 diluted cDNA. The assays were done in triplicate with a 95% confidence level.
Western blotting
Twenty-five micrograms of protein lysates in SDS buffer were separated on an 8% SDS–PAGE and transferred to PVDF membranes for 1 h. Primary antibodies were incubated at 4°C overnight with the following dilutions: 1:1000 for the mouse monoclonal DH7 PTBP1 antibody, 1:200–500 for the mouse monoclonal DH3 PTBP1 antibody, 1:1000 for the goat polyclonal PTBP1 antibody, 1:1000 for the rabbit polyclonal PTBP2 antibody, 1:2500 for the goat polyclonal actin antibody (Sigma Aldrich, St. Louis, MO). DH3 and DH7 antibodies were gifts from Dr. Sue Berget (Baylor College of Medicine, Houston, TX). The PTBP2 antibody was a gift from Dr. Doug Black (UCLA, Los Angeles, CA). Three washes in T-TBS were performed before addition of secondary antibodies at 1:5000 dilution in T-TBS for 1 h at room temperature. These were: IR680-labelled goat anti-mouse (Invitrogen, Carlsbad, CA), IR680-labelled donkey anti-goat (Li-Cor Biosciences, Lincoln, NE) and IR800-labelled goat anti-rabbit (Li-Cor Biosciences, Lincoln, NE). For Nogo-A, the isoform-specific rabbit polyclonal antibody (H-300, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was diluted (1:200) in T-TBS and incubated, shaking, overnight at 4°C. After three washes with T-TBS, IR680-labelled donkey anti-goat secondary antibody (Invitrogen, Carlsbad, CA) was diluted (1:5000) in T-TBS and added to the membrane for 1 h at room temperature. Three more washes in T-TBS were done before visualization. All results were scanned using the Odyssey imaging system (Li-Cor Biosciences, Lincoln, NE).
Cell growth curves
For each treatment, 10 000 cells were plated into wells of a 24-well dish following double morpholino treatment in serum-free medium (Day 0). Every day for a span of 5 days, quadruplicate wells were counted with a haemocytometer. The average and standard deviation of cells in the four wells were calculated and plotted.
Cell proliferation
Cell proliferation was monitored by 5′-bromo-2′-deoxyuridine (BrdU)-incorporation into cellular DNA using the Cell Proliferation ELISA BrdU colorimetric assay (Calbiochem, La Jolla, CA). Immediately following the second morpholino treatment, 5000 cells were seeded into a 96-well plate. After 24 h, the cells were washed with PBS, and fresh serum-free medium was added to the plate. On Day 3, cellular DNA was labelled for 4–10 h using BrdU labelling reagent. Average incorporation and standard deviation were calculated from quadruplicate samples. Significance was determined by one-way ANOVA and Fisher's least significant difference method.
Apoptosis assay
Caspase-3/7 activity was assessed using the SensoLyte homogenous AMC caspase-3/7 assay (AnaSpec, Inc., San Jose, CA). After the second morpholino treatment, 5000 cells were plated into a 96-well dish. The following day, cells were switched to fresh serum-free medium. Caspase activity was measured on Day 3 in quadruplicate samples and significance determined by one-way ANOVA and Fisher's least significant difference method.
Wound-healing assay
Control and targeted oligonucleotide-treated cells were plated to confluence in a 24-well dish in serum without medium. One day after plating, a scratch was made using a 200 µl pipette tip. The cells were incubated for 2 days in serum-free medium. Pictures were taken at 24 h (Day 1) and at 48 h (Day 2) after the scratch was made. To quantify glioblastoma cell migration we used Adobe Photoshop version 10 for Macintosh. The magic wand tool was utilized with a tolerance setting of three to determine the total pixel area that was unoccupied by cells. The average of three values for each treatment was compared to the unoccupied area at Day 0.
Cell adhesion assay
Following double-scraping, 50 000 cells of each morpholino treatment were plated in three to six wells of the matrices: fibronectin, vitronectin and collagen IV. Adhesion of cells to these matrices was measured according to the instructions given by the manufacturer (Chemicon International, Inc., Millipore, Billerica, MA). Cell number was indirectly quantified by measurement of absorbance levels after crystal violet staining (540 nm). Average absorbance readings from blank wells containing bovine serum albumin were subtracted from each sample well. The replicates were used to obtain an average reading with standard deviation. Significance was determined by one-way ANOVA and Fisher's least significant difference method.
Immunofluorescence
Approximately 10 000 double-scraped cells were plated onto round coverslips pre-coated with 1.25 mg/ml of poly-l-lysine in a 24-well dish for 48 h. Cells were then washed once with PBS and fixed with 4% paraformaldehyde for 15 min. Cells were rinsed with 0.1× PBS before permeabilization with 1% H2O2/PBS for 5 min followed by a 0.1× PBS wash for 5 min, a 5 min incubation in fresh 50 mM glycine, and a final 0.1× PBS wash for 5 min. Image-it Fx Enhancer (Invitrogen, Carlsbad, CA) was added at room temperature for 30 min, followed by two rinses in 0.2% gelatine–PBS, blocking in 5% goat serum in 0.2% gelatine–PBS for 30 min, and a 5 min rinse in 0.2% gelatine–PBS. Alexa Fluor 488-phalloidin was diluted 1:100 in methanol to 6.6 µM and added for 30 min at room temperature. DAPI-containing mounting media (Vector Laboratories, Burlingame, CA) was placed on a frost-free slide. The pictures were visualized at 40× magnification with a DeltaVision microscope (Applied Precision, Issaquah, WA) and pictures taken with a Photometrics Coolsnap HQ2 camera (Tucson, AZ). Deconvolution was done with the Photometrics softWoRx software.
Analysis of PTBP1 and PTBP2 expression datasets in glioma
To examine PTBP1 and PTBP2 expression we mined data publicly available at GEO (Barrett et al., 2007). Two datasets were chosen for the analysis. GSE4290 contained the larger dataset comprised of 180 patient samples that included 23 samples from epilepsy patients used as non-tumour samples, 26 astrocytomas (7 WHO grade II and 19 WHO grade III), 50 oligodendrogliomas (38 WHO grade II and 12 WHO grade III) and 81 glioblastomas (Sun et al., 2006). The GSE4412 dataset was comprised of 85 gliomas from 74 patients diagnosed as a WHO grade III (8 anaplastic astrocytomas, 7 anaplastic mixed oligo-astrocytomas and 11 anaplastic oligodendrogliomas) or WHO grade IV (59 glioblastomas) glioma (Freije et al., 2004). We used six probe-sets contained on the Affymetrix HG-U133A array mapped to PTBP1 and PTBP2 for our analysis (Affymetrix, Santa Clara, CA). Plots were generated using the arbitrary values provided for the probesets, averaging the five available PTBP1 probeset values. Statistical significance was determined using an equivalent rank-based test to generate a Pearson's product-moment correlation value.
Exon array analysis of morpholino-treated U251 cells
Isolation of RNA, preparation of target, hybridization, scanning and analysis of the Affymetrix Exon Human 1.0 ST arrays was performed as described previously (Cheung et al., 2008). RNA samples were prepared from U251 cells double-scraped treated with morpholinos as described above. Three independent plates were treated with control or PTBP1 Ex. 2 morpholinos for RNA isolation and array hybridization.
Results
PTBP1 and PTBP2 crosstalk is intact in glioma
A primary cellular function of both PTBP1 and PTBP2 is to auto- and co-regulate their levels in cell lines (Boutz et al., 2007; Spellman et al., 2007). PTBP1 mediates exon 10 removal from PTBP2 pre-mRNA introducing a premature stop codon (PTC), which causes nonsense mediated decay (NMD) of the mRNA (Spellman et al., 2007). This is an event that is accentuated by high levels of PTBP1 protein and results in low steady-state levels of PTBP2 protein despite its active transcription. PTBP1 also represses exon splicing of its own transcript, which also leads to degradation through the NMD pathway (Spellman et al., 2007). In glioma, there is significant overexpression of PTBP1 and it is unclear whether this regulatory pathway is functionally aberrant (McCutcheon et al., 2004). If intact, further reduction in PTBP2 levels from elevated PTBP1 could contribute to glioma formation. To explore PTBP1/PTBP2 regulatory interactions, microarray data from the largest available glioma dataset within the Gene Expression Omnibus (GEO) (see Materials and methods section) was used to compare probe hybridization intensities of both genes (Sun et al., 2006; Barrett et al., 2007). PTBP1 and PTBP2 probe-sets derived from the Affymetrix HG-U133A were used for analysis. Figure 1A compares the average PTBP1 probe hybridization intensities with those of PTBP2 probes for each sample. A slight trend of inverse expression is observed when samples are ordered by increasing PTBP1 expression. A rank-based Pearson's product-moment correlation using the average rank values for PTBP1 probes versus the PTBP2 probes demonstrated an inverse correlation (−0.64). Importantly, only 9 of 81 grade IV glioma samples were found to express PTBP1 levels lower than non-tumour controls and in each case PTBP2 expression was found to be elevated. These findings were reproducible in a second independent microarray dataset comprised of 85 samples from 74 patients with WHO grade III or IV glioma (also from GEO) (Fig. 1B) (Freije et al., 2004).
Figure 1.
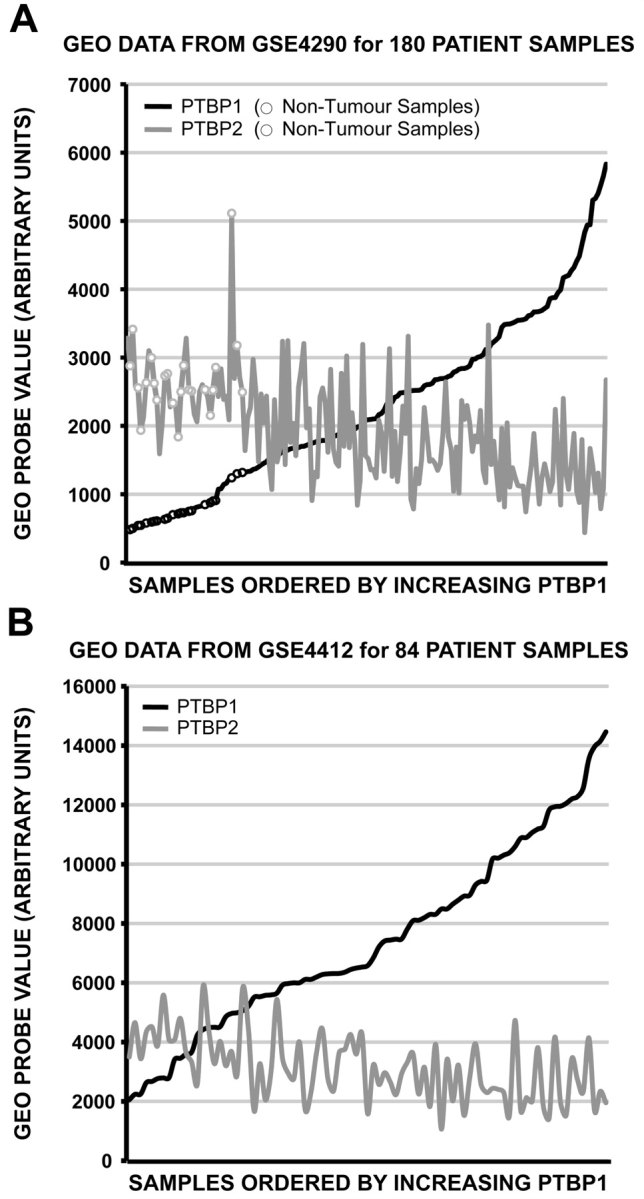
PTBP1 overexpression correlates with PTBP2 repression. Expression array values from two independent GEO datasets were plotted comparing PTBP1 and PTBP2 levels in the same sample. The patient samples are ordered by increasing PTBP1 expression. (A) Plot of GSE4290 dataset values. The expression levels for 23 non-tumour samples are indicated as open circles. (B) Plot of GSE4412 dataset values.
Overall, microarray results indicated that increased levels of PTBP1 would further decrease PTBP2 expression in glioma, although we cannot rule out biases that result from cell population heterogeneity. To directly test the interaction between PTBP1 and PTBP2 in glioma, an in vitro system was developed to specifically deplete PTBP1 and PTBP2 singly and in combination from glioma cells using antisense morpholino oligonucleotides (see Materials and methods section, Fig. 2A). PTBP1 protein is elevated in several glioma cell lines compared to normal human astrocytes and expression levels in these cells are similar those observed in WHO grade IV tumours (Cheung et al., 2006) (Supplementary Fig. 1A and data not shown). As shown in Fig. 2B, the control (C) had no effect on the endogenous levels of both PTBP1 and PTBP2 transcripts in U251 glioma cells, while the specific PTBP1 and PTBP2 morpholinos caused exon 2 skipping and subsequent generation of premature stop codon in their targeted transcripts. The premature stop codon in the PTBP1 mRNA did not affect overall transcript levels. In contrast, the premature stop codon-containing PTBP2 transcripts were markedly reduced, confirming an intact NMD pathway. Studies performed in other cell types demonstrate that high levels of PTBP2 transcripts do not result in correspondingly high levels of PTBP2 protein unless PTBP1 is low (Boutz et al., 2007; Makeyev et al., 2007; Spellman et al., 2007). This was also observed here as PTBP2 protein was only detected in cells with PTBP1 removal. In parallel, reducing PTBP2 levels had no effect on PTBP1 protein, as expected. In the last treatment, knockdown of PTBP1 and PTBP2 together resulted in a significant reduction of both PTBP1 and PTBP2 protein (Fig. 2C and Supplementary Fig. 1C). Similar effects on protein expression were seen in second glioma cell line, LN229 (Supplementary Fig. 1B and C). Therefore, both microarray (Fig. 1) and morpholino knockdown experiments confirmed that the co-regulation between PTBP1 and PTBP2 was intact in these tumour cells.
The differences in PTBP1 and PTBP2-mediated roles in glial cell transformation were further delineated using several assays to measure changes in proliferation, migration, and adhesion of morpholino-treated glioma cell lines. The double knockdowns appear to recapitulate the conditions found in normal adult brain where both of these proteins occur at low levels.
PTBP1 and PTBP2 play a minor role in promoting cellular proliferation
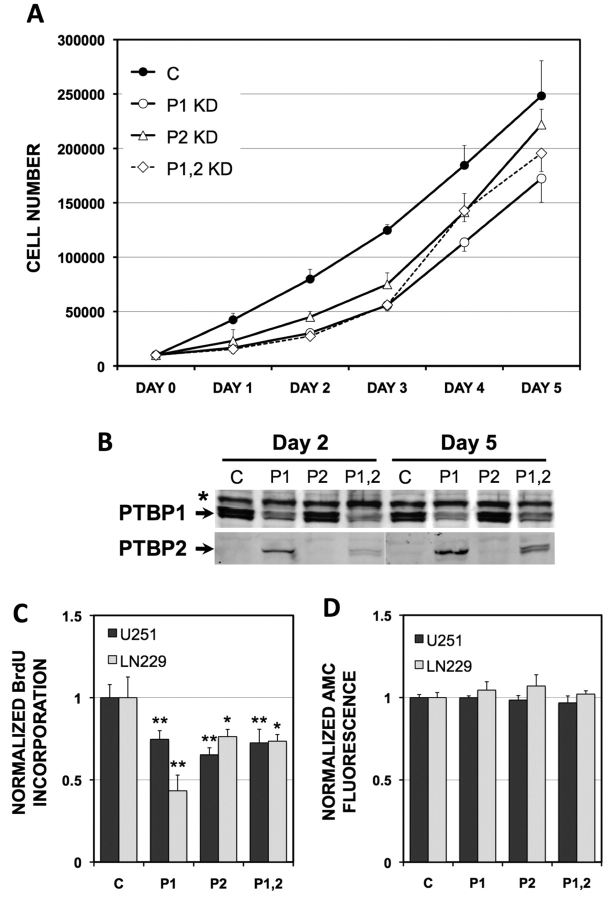
Reduction of PTBP1 inhibits tumour cell proliferation in culture, but has variable effects on other tumourigenic properties (He et al., 2007; Wang et al., 2008). It remains unclear whether PTBP2 affects these responses in cultured glioma cell lines. The morpholino knockdowns allowed us to test for overlapping and non-overlapping cellular responses to PTBP1 and PTBP2 in glioma cell lines. To measure changes in cell proliferation, we first examined linear growth curves of U251 cells plotted over 5 days after single and double PTBP1 and PTBP2 knockdowns (Fig. 3A and B). Both single and double knockdowns of PTBP1 and PTBP2 resulted in slower cellular proliferation as compared with the control-treated cells. The curves did not plateau, demonstrating that the cells did not senesce or undergo cell death when these proteins were absent. A direct effect of PTBP1 and PTBP2 on glioma cell growth was confirmed by examination of BrdU incorporation and caspase activation at Day 3. Significant reductions in BrdU incorporation (up to 56%) were seen in U251 cells and second human glioma cell line LN229, for both single and double knockdowns of PTBP1 and PTBP2 (Fig. 3C). These reductions occurred in the absence of any significant activation of apoptosis as assessed by measurement of caspase 3/7 activity (Fig. 3D). Therefore, these growth patterns suggested that both PTBP1 and PTBP2 positively affect glioma cell proliferation in culture.
Figure 3.
PTBP1 and PTBP2 slow glioma cell proliferation. (A) Cell growth curves were performed on U251 cells following morpholino treatment (see Materials and methods section). Treatments included: control oligo (C), PTBP1-specific oligo (P), PTBP2-specific oligo (P2) and a combination of both PTBP1- and PTBP2-specific oligos (P1,2). Cell numbers represent the average determination for 4 wells at each time-point ± standard deviations. (B) PTBP1 and PTBP2 expression levels were assessed at Days 2 and 5 by western blot. The asterisk indicates a non-specific band. Quantification of protein knockdown is provided in Supplementary Fig. 1. (C) BrdU proliferation assay was performed 3 days after the final morpholino treatment as in A. The relative incorporation of BrdU is indicated for U251 and LN229 glioma cells. Significant differences from control treated cells are indicated (*P < 0.05, **P < 0.001). (D) Caspase 3/7 activity was determined 3 days after the final morpholino treatment by the measurement of proteolytic cleavage of Ac-DEVD-AMC substrate (AMC) (see Materials and methods section). Enhanced fluorescence is direct indicator of AMC cleavage by caspase-3/7. No significant differences from control were observed.
PTBP1 and PTBP2 downregulation inhibited cell migration
The PTBP1 targets related to cell motility and focal adhesion complexes may be involved in the highly infiltrative nature of glioma cells (de Hoog et al., 2004). Therefore, we measured the effect of PTBP1 and PTBP2 depletion on glioma cell migration rates. Cell movement was assessed using the artificial wound-healing assay (Fig. 4 and Supplementary Fig. 2). Examination of the wound created in control-treated U251 and LN229 cells determined that 80%–100% of the gap was filled within 2 days (Fig. 4). In contrast, there was only partial filling of the gap for both single and double knockdown of PTBP1 and PTBP2. In U251 cells, movement of cells was affected more by PTBP2 knockdown than PTBP1 knockdown (23% versus 45% of the gap filled at Day 1). However, the most pronounced effect was observed for the double knockdown at Day 2. PTBP1 knockdown also reduced LN229 glioma cell migration. Unlike U251 cells, LN229 cell movement was inhibited to the same extent by either single knockdown or removal of both PTBP1 and PTBP2. Our results indicate removal of PTBP1 and PTBP2 impede glioma cell line motility.
Figure 4.
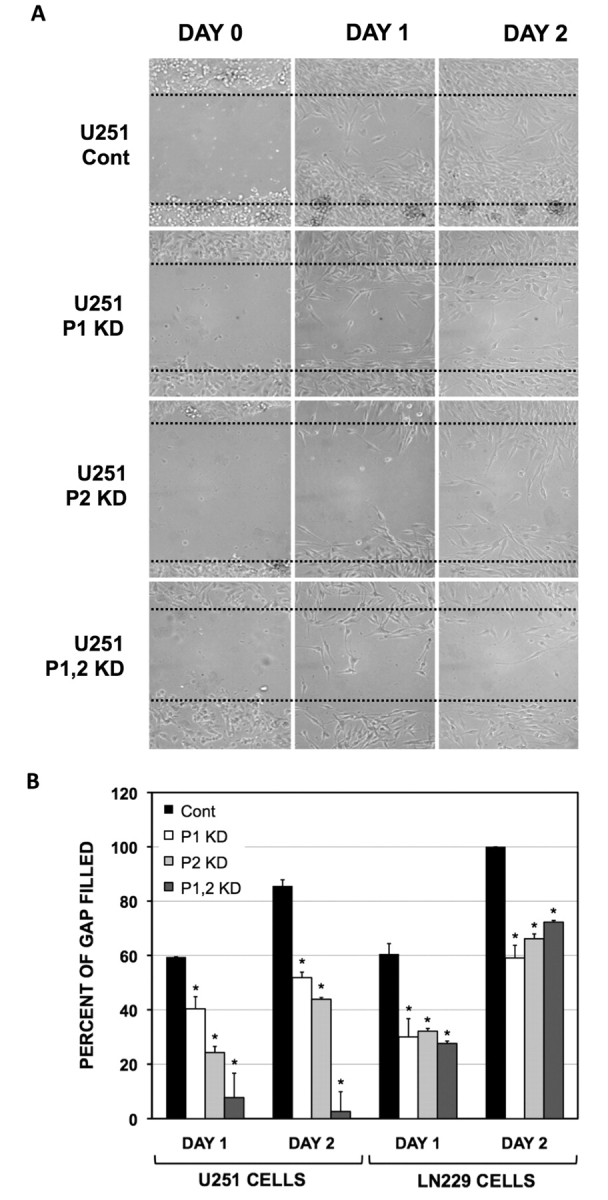
PTBP1 and PTBP2 impede glioma cell migration. Cell migration was assessed by the wound-healing assay following morpholino treatment. Treatments included: control oligo (Cont), PTBP1-specific oligo (P1), PTBP2-specific oligo (P2) and a combination of both PTBP1- and PTBP2-specific oligos (P1,2). U251 and LN229 glioma cells were plated to confluency with the scratch occurring on Day 0. The migration of cells was visually monitored with micrographs taken at Days 0, 1 and 2 as indicated. (A) Representative micrographs for morpholino-treated U251 glioma cells. (B) Quantification of cell migration as described in Materials and methods section. *P < 0.05 as determined for paired analysis with day-matched controls (Cont) by Fisher's least significant difference method. Representative micrographs for LN229 cells are shown in Supplementary Fig. 2..
PTBP1 and PTBP2 affect adhesion and distribution of actin filaments
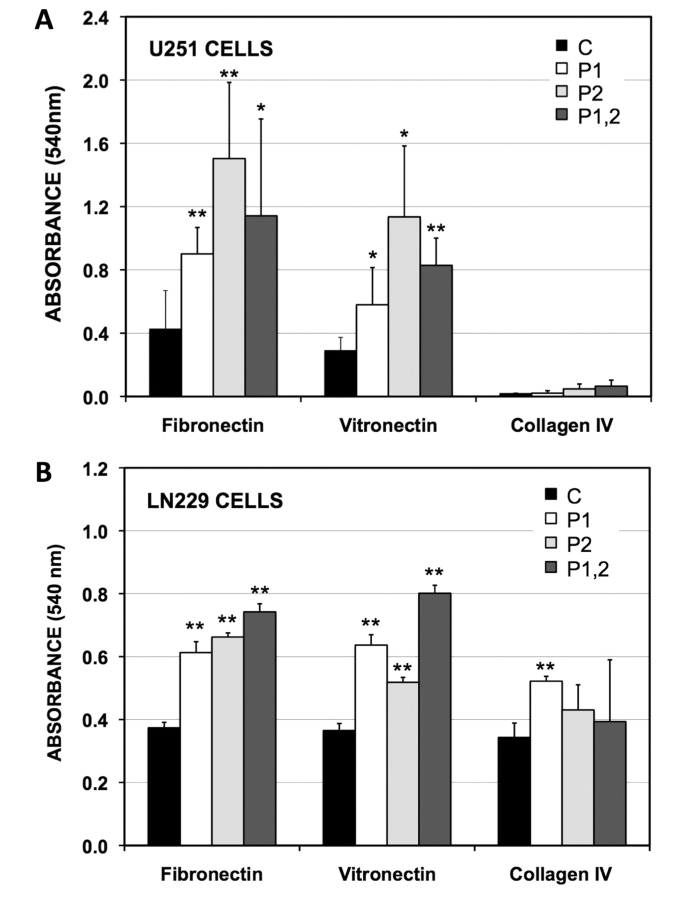
Since the absence of both PTBP1 and PTBP2 led to slower cellular mobility, we examined two components of cell movement: adherence to the extracellular matrix (ECM) and distribution of intracellular actin. Cells must be able to both attach and detach from the extracellular matrix in order to infiltrate (Friedl and Wolf, 2003). To test whether removal of PTBP1 and/or PTBP2 altered the attachment strengths glioma cells, we plated treated cells onto several extra-cellular matrices and quantified adherence using a colorimetric assay in which increased absorption equated with greater attachment (see Materials and methods section). The knockdown of PTBP1 and PTBP2, both singly and in combination resulted in a significant enhancement of attachment to fibronectin and vitronectin, but not collagen IV (Fig. 5). While this enhancement was observed for both U251 and LN229 cells, the effect was more pronounced in U251 cells, and consistent with the migration assay showed a greater dependence on PTBP2 knockdown. For U251 cells, the PTBP1 knockdown was also associated with 2-fold increases in both fibronectin and vitronectin receptor mRNA levels (Supplementary Table 1). These results suggest that loss of PTBP1 may slow cell mobility by increasing adherence to the extracellular matrix.
Figure 5.
PTBP1 and PTBP2 decrease adhesion to fibronectin and vitronectin. U251 (A) and LN220 (B) glioma cells were treated with morpholinos and plated in triplicate in wells coated with fibronectin, vitronectin and collagen IV. The cells were incubated for one hour, washed and stained with crystal violet (absorbance = 540 nm). The average absorbance of adhered cells was calculated ±SD. The morpholinos were: control oligo (C), PTBP1-specific oligo (P1), PTBP2-specific oligo (P2), and both PTBP1- and PTBP2-specific oligos combined (P1,2). Significant differences from control treated cells are indicated (*P < 0.05, **P < 0.001).
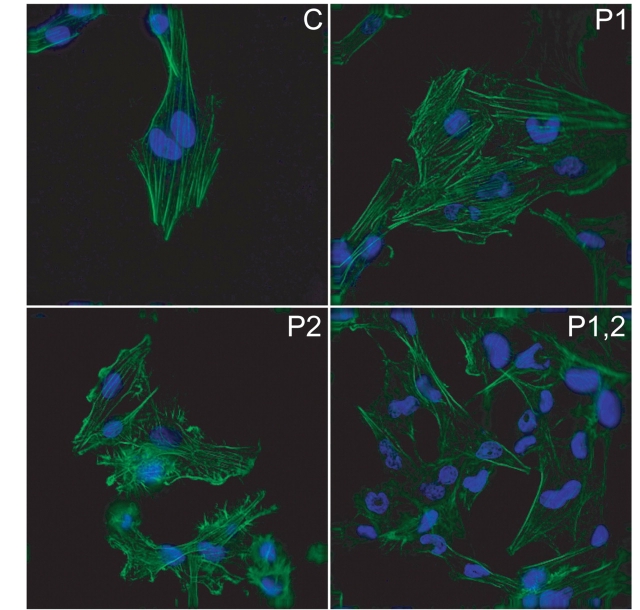
Cell movement also involves the formation of a leading edge at the cell membrane, made by protrusions of actin stress fibres along focal adhesion complexes (Machesky and Hall, 1997). Forward propelling motion is also facilitated by the tension between the leading and trailing edges of the cells that is created by actin filaments stretched across cells (Maidment, 1997). To monitor whether PTBP1 and PTBP2 affect these types of changes in actin distribution morpholino-treated cells were stained with phalloidin (Fig. 6). When cells were plated onto uncoated coverslips, no difference in stress fibre formations was seen in all treatment combinations (data not shown). However, poly-l-lysine-coating of coverslips affected actin distribution for both the single PTBP2 and double knockdowns. Removal of PTBP2 increased ruffles, decreased stress fibres, and increased filopodia. The double knockdowns had no ruffles, but both stress fibres and ruffles were absent from these cells. No difference was observed with the knockdown of PTBP1 alone. Thus, both PTBP1 and PTBP2 have individual responses in actin distribution. Some of the changes in morphology and mobility seen in the wound-healing assay may be attributed to these differences in actin regulation.
Figure 6.
Actin fibres show redistribution following PTBP2 reduction. Morpholino-treated U251 glioma cells were grown on poly-l-lysine and stained with phalloidin to highlight actin (green) and DAPI to identify nuclei (blue) nuclear staining is shown in blue. Representative deconvolution micrographs are provided for cell treated with: Control, nontargeting oligo (C), PTBP1-specific oligo (P1), PTBP2-specific oligo (P2), and both PTBP1- and PTBP2-specific oligos combined (P1,2).
Identification of PTBP1-mediated splicing targets
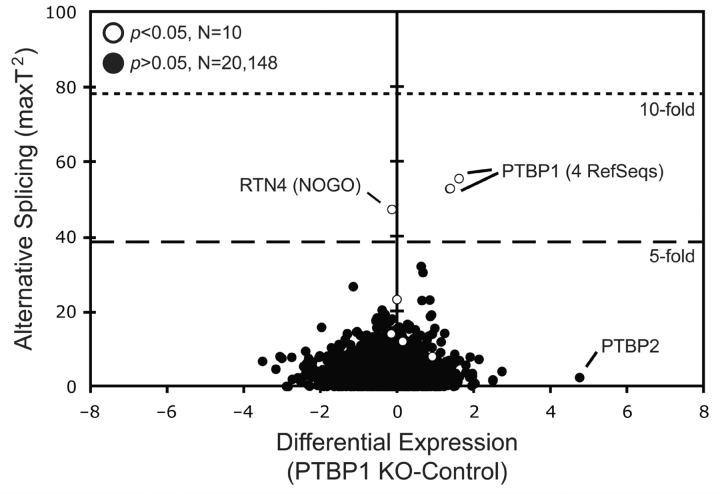
Given the observation that PTBP1 enhances U251 cell proliferation and motility, we performed global exon array studies to identify potential regulatory targets responsible for these changes. The effect of PTBP1 depletion on alternative RNA splicing and abundance was measured for 20 158 RefSeq transcripts in U251 glioma cells using the Affymetrix Human Exon array (Fig. 7). PTBP1 depletion identified six RefSeq transcripts with a >5-fold change in either alternative RNA splicing or expression (Fig. 7). Four of these transcripts were to PTBP1 RefSeq entries, which confirmed the morpholino-induced splicing changes used to knock down protein expression. The remaining transcripts belonged to PTBP2 and RTN4 (also known as Nogo), both recently reported targets of PTBP1 (Makeyev et al., 2007; Spellman et al., 2007). The primary effect of PTBP1 on PTBP2 was a >25-fold increase in steady-state levels resulting from enhanced inclusion of exon 10 to stabilize the mRNA, as expected (data not shown) (Boutz et al., 2007; Grabowski, 2007). The effect of PTBP1 on RTN4 was primarily on alternative RNA splicing (AS score = 47) with little change in transcript levels observed (DE score = −0.033). Significant RNA splicing changes were noted in other transcripts but the magnitude of their change did not approach that seen for RTN4. In addition, neither known PTBP1 targets nor previously defined glioma-specific splicing events showed significant deviations from zero. This was confirmed by standard RT-PCR and underscores the significance of RTN4 regulation (Cheung et al., 2008).
Figure 7.
Exon array analysis identifies RTN4/Nogo as a major target of PTBP1 splicing regulation. Comparative genome-wide exon expression analyses in U251 glioma cell. Differential Expression (DE) values are plotted against Alternative Splicing Scores (AS Score) for a defined set of 20 158 RefSeq entries. The plot shows the average of three independent experiments for U251 glioma cells in which PTBP1 exon 2 splicing was changed using targeted antisense oligonucleotide-treatment. The positions of 6 RefSeq entries representing PTBP1, PTBP2 and RTN4 are indicated. P-values are provided for transcripts with significant AS scores.
High levels of PTBP1 correlate with skipping of exon 3 in RTN4/Nogo
Transcript-specific hybridization maps were used to identify the specific RTN4 exon(s) responding to PTBP1 depletion in both morpholino-treated U251 cells and comparisons of non-tumour brain and glioblastoma patient samples (Supplementary Fig. 3). High levels of PTBP1 expression were associated with increased skipping of exon 3 in both GBM patients and in vitro systems. This splicing event illustrated in Fig. 8A creates the RTN4 gene products Nogo-A and Nogo-B‘. The correlation between PTBP1 levels and exon 3 skipping was also confirmed by both quantitative RT-PCR (Fig. 8B and data not shown) and isoform-specific Western analysis of Nogo-A (Fig. 8C). The regulation of exon 3 was specific to PTBP1 and not to PTBP2, as determined by comparing PTBP1 knockdown alone with the single PTBP2 and double PTBP1 and PTBP2 knockdowns. To confirm the in vivo relevance of this finding, we examined expression levels of exon 3 in non-tumour brain versus glioblastoma samples (Fig. 8D). The high levels of PTBP1 in the tumours correlated with lower levels of RTN4 exon 3 inclusion. Therefore, there is strong evidence in both of these datasets that more exon skipping occurred with increased PTBP1 levels.
Figure 8.
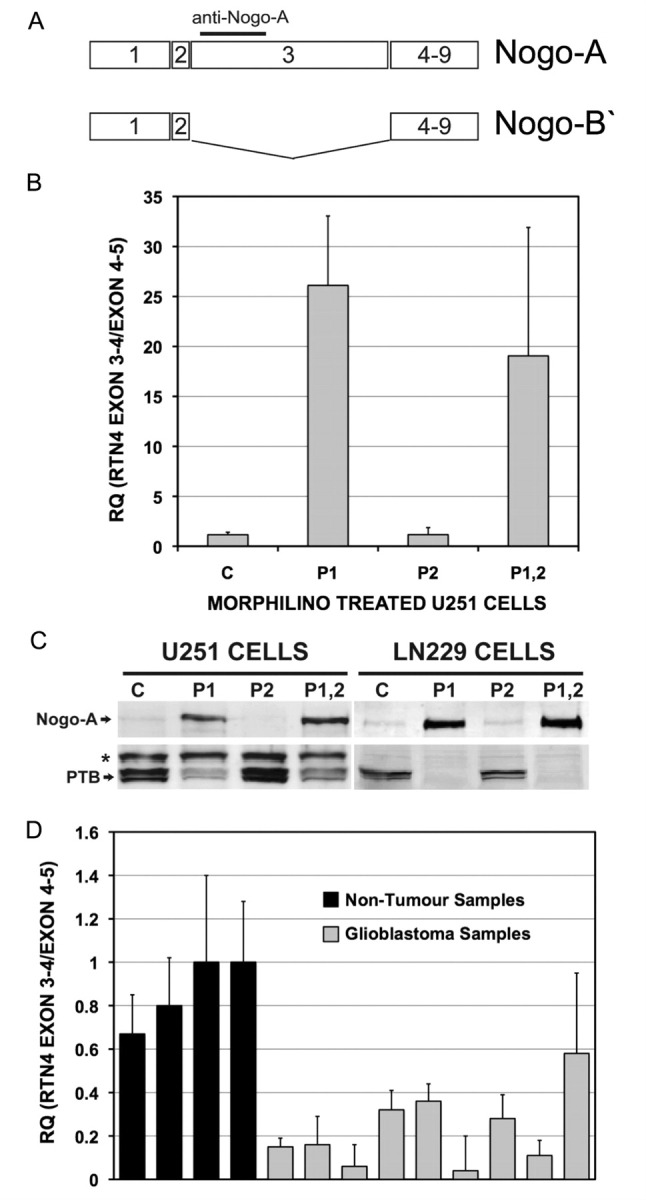
RTN4 exon 3 is specifically regulated by PTBP1 in glioma. (A) Schematic illustration of the RTN4 gene showing the exon 3-including Nogo-A and exon 3-skipped Nogo-B′ isoforms. The PTBP1-regulated exon 3 is 2400 bp long. Epitope-specific antibodies exist against exon 3 coding sequence detects the Nogo-A variant, but not Nogo-B′. (B) Quantitative RT-PCR was performed on U251 cells following morpholino treatment (see Materials and methods section). Treatments included: control oligo (C), PTBP1-specific oligo (P1), PTBP2-specific oligo (P2), and a combination of both PTBP1- and PTBP2-specific oligos (P1,2). Two RTN4 Taqman assays were selected to measure exon 3 splicing levels relative to the total number of RTN4 transcripts (exon 4–5). Values provided represent the RQ ratio ± RQ maximum. (C) Protein lysates prepared from U251 and LN229 glioma cell cultures were simultaneously probed with antibodies against PTBP1 and Nogo-A. The asterisk indicates a non-specific band. (D) Quantitative RT–PCR analysis of RTN4 exon 3 inclusion was performed as described in (B) on RNA isolated from four non-tumour brain samples and nine grade IV glioblastomas.
Overexpression of Nogo-A slowed proliferation of U251 glioma cells
The PTBP1 regulation of RTN4 exon 3 splicing suggests that either the aberrant expression of Nogo-B or the lack of Nogo-A expression in glioma cells could contribute to the previously observed growth and migration changes. To test one of these possibilities, we overexpressed Nogo-A in U251 cells. Empty and Nogo-A vectors were transfected into U251 cells and grown in G418-containing medium for one week to generate pooled stable cells. Overexpression of Nogo-A in the pDEST-NogoA-containing cells was confirmed using Western blotting (data not shown). Proliferation curves that were performed on pooled stable cells showed that transient transfection of Nogo-A slowed cell growth in a manner that was comparable to PTBP1 ablation (Fig. 9). Similar experiments performed in NIH 3T3 cells showed an even greater reduction in cell growth (Supplementary Fig. 4). Experiments examining U251 cell migration using the wound-healing assay found no effect of ectopic Nogo-A expression (data not shown). These data suggest that PTBP1 regulation of RTN4 splicing plays a role in proliferation. The data do not specifically address if Nogo-B’has direct effects on cell growth.
Figure 9.
Ectopic overexpression of Nogo-A slows U251 cell proliferation. Cell proliferation curves were performed on U251 cells following transfection and pooled selection with either empty vector or plasmid specifically expressing Nogo-A. Growth was monitored as described in Fig. 3. Significant differences are indicated (*P < 0.05, **P < 0.005).
Discussion
PTBP1 and its brain-specific homolog PTBP2 recognize similar binding motifs and have multiple overlapping target transcripts (Boutz et al., 2007). Their functional differences result from their effects on non-overlapping targets. Therefore, the upregulation of PTBP1 and not PTBP2 in glioma cell lines provides an opportunity to determine PTBP1-specific changes that enhance gliomagenesis. We measured PTBP1-induced effects on oncogenesis by removing PTBP1 and PTBP2 singly and in combination from glioma cell lines. Surprisingly, both single and double knockdowns resulted in slower proliferation and migration, and increased adhesion to the extracellular matrix. No changes were seen in either cell growth on soft agar or invasion (data not shown), which is consistent with the recent suggestion that these effects are cell-type-specific (Wang et al., 2008). The changes in proliferation, migration and adhesion for PTBP2 were unexpected as levels of PTBP2 protein were undetectable in these cells. Several possibilities may account for this. First, small amounts of PTBP2 may antagonize PTBP1 activity through competitive binding of overlapping targets (Boutz et al., 2007; Grabowski, 2007). Second, the knockdown of PTBP2, a major substrate of PTBP1, increases relative levels of PTBP1 that are available to act on other transcripts. However, experiments in other cell types show that ectopic overexpression of PTBP1 does not alter proliferation rates (Wang et al., 2008). Furthermore, mice overexpressing PTBP1 under the glial cell-specific promoter GFAP did not have any notable cellular phenotype (data not shown). Finally, the removal of trace PTBP2 protein could impede its effects on important, non-overlapping targets. This would support the proposed critical role for PTBP2 in neuroprogenitor cell differentiation (Makeyev et al., 2007). Clearly, glioma cell lines are sensitive to small changes in either PTBP1 or PTBP2.
Individual downregulation of PTBP1 and PTBP2 mildly impeded glioma cell line growth rate, which recapitulated observations in other cell types (Fig. 3) (He et al., 2007; Wang et al., 2008). Although PTBP1 may mediate cell type-specific effects, its positive effect on cell proliferation has been a consistent finding (Wang et al., 2008). Indeed, the complete absence of PTBP1 may result in cell lethality. Deletion of the Drosophila melangaster PTBP1 homolog, hephaestus, is embryonic lethal and that downregulation of PTBP1 debilitates the growth of normal human fibroblast cells to a greater degree than in tumour cells (Dansereau et al., 2002; Wang et al., 2008). However, it is important to consider that removal of both PTBP1 and PTBP2 resulted in a proliferative pattern that was similar to the single PTBP1 knockdown treatment and that the reduction in cell number was related to decreased cell division rather than an increase in apoptosis. Importantly, PTBP1 and PTBP2 do not have additive effects, but appear to act on an overlapping target(s) or proliferation pathway.
Cell motility, as measured by the wound-healing assay, was reduced substantially when either or both PTBP1 and PTBP2 were depleted from cells. These two proteins appeared to affect several fundamental aspects of cell motility. First, cellular processes resembling neuronal axons developed unexpectedly in single PTBP2 knockdown treatment and were more pronounced in the double knockdown U251 cells (Fig. 4 and Supplementary Fig. 5). These morphological changes were associated with increased expression of the neuronal marker MAP2 and decreases in the glial marker GFAP as detected by the exon array (Supplementary Fig. 5). This expression profile was most apparent in the double knockdown cells and implied that differentiation had occurred. Removing both PTBP1 and PTBP2 may enable neuron-like differentiation to be induced by serum withdrawal, which then renders the cells immobile. Importantly, these findings suggested that the differentiation pathways found in normal brain were maintained in glioma cells, where large decreases in PTBP1 and PTBP2 expression occur as neural stem cells commit to glial or neuronal lineages (Lillevali et al., 2001). Second, single and double knockdown cells had increased adherence to fibronectin and vitronectin, which occurred in parallel with upregulation of integrin receptor levels (Fig. 5 and Supplementary Table 1). These data appeared to correlate with the observed reduction in cell mobility. Third, the ability of U251 glioma cells to form fibres at focal adhesions for cell mobility was also decreased in single PTBP2 and double knockdowns. Cells with PTBP2 knockdown formed ruffles instead of pseudopodia on poly-l-lysine-coated coverslips and double knockdown had yet another phenotype of decreased stress fibre formation. Because lysine induces actin arrangements independently of integrins, specific PTBP1-mediated effects on cell adhesion clearly differ at a least one level compared with PTBP2 (Janmey et al., 1998). It is unclear whether the same changes occur in LN229 cells. The complex interactions between PTBP1 and PTBP2-mediated effects on cellular motility are to be uncovered.
Our data supported a role for PTBP1 in glioma cell proliferation, migration, adherence, and potentially dedifferentiation. The multiple effects of PTBP1 were consistent with its function in a broad and complex epigenetic control of gene expression. Surprisingly, our exon array experiments pointed to only two key regulatory targets of PTBP1: PTBP2 and RTN4/Nogo. While we cannot rule out that additional PTBP1 targets may be important, these two genes were altered several orders of magnitude greater than any other target found in this cellular context. Furthermore, the U251-derived results paralleled those observed in glioma patient samples for both genes. The targeting of RTN4 splicing differed from that of PTBP2 in that the final outcome produced a stable mRNA transcript and functionally distinct protein. As seen for PTBP2, RTN4 had a substantial increase in exon inclusion when PTBP1 was removed, which was consistent with PTBP1 being an inhibitor of exon splicing. Overall, PTBP2 and RTN4 may be the central mediators of PTBP1-dependent changes glioma cell in proliferation, cell motility and differentiation. Additional oncogenic targets that are absent from glioma cells could be responsible for the cell type-specific invasion and transformation observed with PTBP1 overexpression (He et al., 2007; Wang et al., 2008). Therefore, the concept of cell-specific RNA responses to PTBP1 knockdown is reiterated here, along with our own studies of removing PTBP1 from HeLa cells (Supplementary Fig. 6) (Boutz et al., 2007; Wang et al., 2008).
The targeting of PTBP2 and RTN4 by PTBP1 in glioma cell lines is not entirely unexpected. Both genes have expression patterns that are predominantly limited to the brain. RTN4 was first characterized for its inhibition on neurite outgrowth. It is a member of the reticulons, a family of transmembrane proteins that is enriched in the central nervous system and is linked to multiple neurodegenerative disorders (Yan et al., 2006). Every reticulon contains a functional domain that includes a ‘Nogo-66’ loop region. Through this loop, RTN4 triggers a rearrangement of actin filament extensions in neighbouring cells to inhibit neurite outgrowth (Schwab et al., 2006). RTN4 has multiple mRNA isoforms but only three characterized protein variants: Nogo-A, Nogo-B and Nogo-C whose unique cellular functions remain somewhat unclear (Oertle et al., 2003). All three isoforms contain the extracellular Nogo-66 loop. The longest isoform is Nogo-A includes the PTBP1-regulated exon 3, which contains a subdomain (delta 20) that further promotes neurite outgrowth inhibition. Our data suggest that Nogo-B’is the predominant mRNA isoform that is expressed in glioma cells where high levels of PTBP1 ensure that exon 3 is skipped. Forced expression of Nogo-A was associated with reduced U251 cell proliferation paralleling the effect observed for PTBP1 knockdown. This observation taken in the context of our exon array results implicates RTN4 as a primary mediator of PTBP1 action on cell proliferation. The specific mechanism through which RTN4 functions to regulate proliferation remains to be elucidated and may involve RTN receptors, contact inhibition, or actin fibres. We cannot completely rule out that PTBP-1-mediated induction of Nogo-A is playing a role in inhibition of cell migration. In fact, extracellular exposure to Nogo-A is associated with inhibition of cell spreading in numerous cell types including NIH3T3 and glioma cells (Oertle et al., 2003, Liao et al., 2004). Finally, it is important to note that the RTN4 gene was not a target of PTBP2, as judged by the fact that exon 3 splicing did not differ between PTBP1 single knockdown or the double knockdown of both PTBP1 and PTBP2. Therefore, PTBP2 has different downstream targets affecting cell proliferation and motility.
Overall, our data demonstrate apparent changes in proliferation, migration, and adhesion of glioma cell lines resulting from the single depletions of PTBP1 or PTBP2. Neither PTBP1 nor PTBP2 can compensate for the loss of the other, particularly in the migration and actin staining assays. This clearly points to non-overlapping target transcripts that mediate these effects. The more severe phenotypes of double knockdown cells as compared with the single knockdown cells highlight the importance of PTBP2 regulation by PTBP1.
Supplementary Material
Supplementary material is available at Brain online.
Funding
NCI #CA67946 (to G.J.C.); The John K. Funk Endowment (to G.J.C.), Rosalie B. Hite Foundation (to H.C.C.) and the Kleberg Foundation (to R.K.), with additional core grant support provided by NCI #CA16672 (DAF).
Supplementary Material
Acknowledgements
Henry Adams performed the deconvolution microscopy. Exon array samples were processed by Tamara J. Nixon and Valerie L. Neubauer.
Glossary
Abbreviations
- PTBP1
polypyrimidine tract-binding protein 1
- PTBP2
polypyrimidine tract-binding protein 2
References
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35:D760–5. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–52. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M, Gelfand MS, Spengler S, Zorn M, Dubchak I, Conboy JG. Computational analysis of candidate intron regulatory elements for tissue-specific alternative pre-mRNA splicing. Nucleic Acids Res. 2001;29:2338–48. doi: 10.1093/nar/29.11.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno IG, Jin W, Cote GJ. Correction of aberrant FGFR1 alternative RNA splicing through targeting of intronic regulatory elements. Hum Mol Genet. 2004;13:2409–20. doi: 10.1093/hmg/ddh272. [DOI] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, et al. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet. 2008;40:1416–25. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HC, Baggerly KA, Tsavachidis S, Bachinski LL, Neubauer VL, Nixon TJ, et al. Global analysis of aberrant pre-mRNA splicing in glioblastoma using exon expression arrays. BMC Genomics. 2008;9:216. doi: 10.1186/1471-2164-9-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HC, Corley LJ, Fuller GN, McCutcheon IE, Cote GJ. Polypyrimidine tract binding protein and Notch1 are independently re-expressed in glioma. Mod Pathol. 2006;19:1034–41. doi: 10.1038/modpathol.3800635. [DOI] [PubMed] [Google Scholar]
- Dansereau DA, Lunke MD, Finkielsztein A, Russell MA, Brook WJ. Hephaestus encodes a polypyrimidine tract binding protein that regulates Notch signalling during wing development in Drosophila melanogaster. Development. 2002;129:5553–66. doi: 10.1242/dev.00153. [DOI] [PubMed] [Google Scholar]
- de Hoog CL, Foster LJ, Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117:649–62. doi: 10.1016/s0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;10:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–10. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- Grabowski PJ. RNA-binding proteins switch gears to drive alternative splicing in neurons. Nat Struct Mol Biol. 2007;14:577–9. doi: 10.1038/nsmb0707-577. [DOI] [PubMed] [Google Scholar]
- He X, Pool M, Darcy KM, Lim SB, Auersperg N, Coon JS, et al. Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene. 2007;26:4961–8. doi: 10.1038/sj.onc.1210307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA, Kas J, Shah JV, Allen PG, Tang JX. Cytoskeletal networks and filament bundles: regulation by proteins and polycations. Biol Bull. 1998;194:334–5. doi: 10.2307/1543105. (Discussion 335–6) [DOI] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–93. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Duka T, Teng FYH, Sun L, Bu W, Sohail A, et al. Nogo-66 and myelin-associated glycoprotein (MAG) inhibit the adhesion and migration of Nogo-66 receptor expressing human glioma cells. J Neurochem. 2004;90:1156–1162. doi: 10.1111/j.1471-4159.2004.02573.x. [DOI] [PubMed] [Google Scholar]
- Lillevali K, Kulla A, Ord T. Comparative expression analysis of the genes encoding polypyrimidine tract binding protein (PTB) and its neural homologue (brPTB) in prenatal and postnatal mouse brain. Mech Dev. 2001;101:217–20. doi: 10.1016/s0925-4773(00)00566-9. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Hall A. Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization. J Cell Biol. 1997;138:913–26. doi: 10.1083/jcb.138.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidment SL. The cytoskeleton and brain tumour cell migration. Anticancer Res. 1997;17:4145–9. [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–48. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon IE, Hentschel SJ, Fuller GN, Jin W, Cote GJ. Expression of the splicing regulator polypyrimidine tract-binding protein in normal and neoplastic brain. Neuro Oncol. 2004;6:9–14. doi: 10.1215/S1152851703000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertle T, van der Haar ME, Bandtlow CE, Robeva A, Burfeind P, Buss A, et al. Nogo-A inhibits neurite outgrowth and cell spreading with three discrete regions. J Neurosci. 2003;23:5393–406. doi: 10.1523/JNEUROSCI.23-13-05393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab JM, Tuli SK, Failli V. The Nogo receptor complex: confining molecules to molecular mechanisms. Trends Mol Med. 2006;12:293–7. doi: 10.1016/j.molmed.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Spellman R, Llorian M, Smith CW. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell. 2007;27:420–34. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–41. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–54. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol. 2001;21:3281–8. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Norton JT, Ghosh S, Kim J, Fushimi K, Wu JY, et al. Polypyrimidine tract-binding protein (PTB) differentially affects malignancy in a cell line-dependent Manner. J Biol Chem. 2008;283:20277–87. doi: 10.1074/jbc.M803682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Shi Q, Hu X, Zhou X. Reticulon proteins: emerging players in neurodegenerative diseases. Cell Mol Life Sci. 2006;63:877–89. doi: 10.1007/s00018-005-5338-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.