Introduction
The virtual physiological human (VPH) initiative is intended to support the development of patient-specific computer models and their application in personalised and predictive healthcare. The VPH, a core target of the European Commission's 7th Framework Programme, will serve as a ‘methodological and technological framework that, once established, will enable collaborative investigation of the human body as a single complex system' (http://www.europhysiome.org/roadmap/). As such, the VPH initiative constitutes an integral part of the international Physiome Project (http://www.physiome.org.nz/), a worldwide public domain effort to develop a computational framework for the quantitative description of biological processes in living systems across all relevant levels of structural and functional integration, from molecule to organism, including the human (Kohl et al, 2000; Bassingthwaighte et al, 2009).
So, what is the connection between this grand challenge and systems biology? To explore this, we must first agree on what we take systems biology to mean.
Systems biology
Description versus definition
Descriptions of systems biology range from the view that it is merely ‘new wording, more fashionable, for physiology' (http://is.gd/tQJL), to the all-inclusive ‘systems biology involves the application of experimental, theoretical, and computational techniques to the study of biological organisms at all levels, from the molecular, through the cellular, to the organ, organism, and populations. Its aim is to understand biological processes as integrated systems instead of as isolated parts' (http://is.gd/tQK0).
At the same time, attempts to concisely define systems biology have not yielded definitive form of words that is acceptable to the majority of researchers engaged in what they consider to be systems biology.
One of the reasons for this situation may be that many different scientific streams have come together in the systems biology pool (see also Bassingthwaighte et al, 2009), each with its own conceptual and terminological legacy.
But, another possible explanation for this apparent shortcoming is that systems biology may constitute an approach (as detailed below), rather than a discipline (such as biology), or a destination (such as the VPH). Such a scientific approach can be explained descriptively, but cannot necessarily be defined prescriptively.
In either case, the lack of a generally acceptable definition of systems biology need not be regarded as a surprise, or even as a disadvantage, as the artificial uniformity that could be associated with a definition might exclude important current or future work.
Terminological origins
It may be helpful, at this stage, to step back and consider the etymology of terms, before discussing their possible interrelation.
Biology is contracted from bios (Greek for ‘life') and logos (Greek for ‘reasoned account'). It is the science, or the logic, of life (Boyd and Noble, 1993).
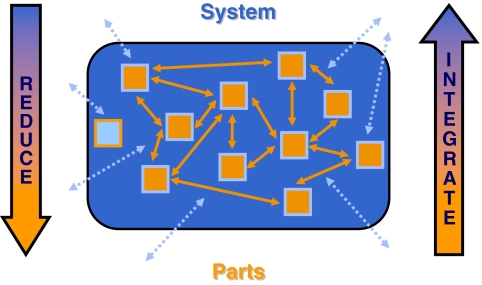
A system is ‘the object' of the activity synthithemi (Greek for ‘I put together') and has been defined as follows: ‘A system is an entity that maintains its existence through the mutual interaction of its parts' (von Bertalanffy, 1968). In keeping with this concept (Figure 1), research into systems therefore must combine:
the identification and
detailed characterisation of the parts, with the
investigation of their interaction with each other and
with their wider environment, to
elucidate the maintenance of the entity.
Figure 1.
A system as an ‘entity that maintains its existence through the mutual interaction of its parts' (von Bertalanffy, 1968). Systems research must combine the (i) identification and (ii) detailed characterisation of parts (orange boxes, as opposed to ‘look-alikes', pale blue box, which need to be identified and excluded), with the exploration of their interactions (iii) with each other (orange arrows), and (iv) with the environment (pale blue dashed arrows affecting parts either directly, or indirectly through modulation of internal interactions), to develop a (v) systemic understanding (an important, but often overlooked, aspect is that the system itself not only enables, but also restricts, the type and extent of functions and interactions that may occur; dark-blue box). Systems research therefore requires a combination of reductionist and integrative tools and techniques.
Subject matter
On the basis of the definition of a system, systems biology can be seen as a conceptual approach to biological research that consciously combines ‘reductionist' (parts; points i and ii) and ‘integrationist' (interactions; points iii and iv) research, to understand the nature and maintenance of entities (point v). In biological systems, preservation of entity includes a broad range of behaviours, including growth and development, adaptation and maladaptation, and progeny, which explains why streams from so many different research directions must be pooled.
In addition, the ‘parts' of a biological system (e.g. organs of a body, or tissues within an organ, etc.) can usually be broken down into smaller biologically relevant entities (such as cells, proteins, amino acids), which—when focussing at a lower level of structural integration—form ‘systems' in their own right. This illustrates two further points: first, systems biology as an approach can be applied to research targets independent of their ‘scale', that is, their level of structural and functional complexity and second, no particular scale has privileged relevance for systems biology (Noble 2008a, 2008c). From the multi-scale nature of biological systems, it follows further that systems biology inherently involves a multi-scale approach (see below).
So, does this mean that there is nothing special about systems biology? Is it really just another, more fashionable label for good old physiology?
Probably not. Systems biology forms a logical juxtaposition to the recently prevailing ‘reductionist' drive, serving as the ‘post-genomic' manifestation of the need to balance dissection and synthesis. Certain aspects of systems biology do indeed mirror the ‘pre-genomic' approach of subjects such as physiology, but at a higher level. Thus, Claude Bernard showed the way as early as the 19th century and specifically called for the mathematical analysis of biological phenomena (see Noble, 2008a). However, with a few notable exceptions, such as the Hodgkin–Huxley equations for the nerve impulse (Hodgkin and Huxley 1952), their application to the heart (Noble, 1962), or the early ideas of Guyton for a quantitative model of the circulation (Guyton et al, 1972), classic physiology largely lacked the ability to pursue the quantitative integration of observed behaviour. This may be one reason why it failed to compete with the rise of molecular biology, which was perceived to be more solidly quantitative. In fact, many academic departments of physiology became molecular or cellular, in focus and in name.
Having turned full circle on what the dialectic method depicts as a three-dimensional spiral of development, we have come ‘back to the future', now that bio-science can harness the power of mathematics and computation and apply it to a re-integration of the pieces of the jigsaw—which have been amply provided by reductionist research approaches. Systems biology therefore thrives on the revolutionary improvement of experimental techniques to investigate system components and their interactions, and on significant advances in computational power, tools, and techniques, which allow quantitative modelling and reintegration at hitherto unimaginable detail and breadth. Modern computational models thus address points (i) to (v) above, and project between them, while observing elementary rules such as conservation of mass, energy, and matter and taking into account natural restrictions imposed on parts and interactions by the system's own properties (e.g. a water-based solute system will impose different constraints compared to a hydro-carbon based one; dark-blue background in Figure 1).
So, perhaps this is where the essence of systems biology lies: by providing a framework for the re-unification of biological studies with ‘the other' sciences, and their joint application to iterative reduction and synthesis, it forms the approach on which quantitative descriptions of parts (i and ii) and their interactions (iii and iv) give rise to an understanding of the maintenance of biological entities (v) across all relevant levels of structural and functional integration (Figure 2).
Figure 2.
Our understanding of ‘real world systems' (top left) usually forms a simplified representation (top right) of that reality, and therefore represents a model in its own right. The progressive development of this understanding is based on the application and analysis of experimental and theoretical models. For biological systems research, these models allow the exploration of partial systems behaviour at all relevant structural levels between body and molecule. ‘Wet' experimental models are developed through a broad range of research directions and provide increasingly detailed data on structure–function relations and their change over time. This can be re-integrated using ‘dry' conceptual (thought) and formal (computation) models. Many of these developments occur in parallel. Systems biology provides the framework for the targeted interrelation of these different facets of model application to bio-medical research and development. Note that, for simplicity, this diagram depicts models by horizontal arrows, although models can involve multiple scales.
An important aspect of this summary is the plural of ‘quantitative description'. Like their experimental counterparts, computational models are—by the very definition of the term ‘model'—simplified representations of reality. Like tools in a toolbox, models for biomedical research, whether ‘wet' or ‘dry', have a range of applications for which they are suitable. This suitability is affected by the extent to which models are representative of the aspect of reality that they mimic; relevant for the question under investigation; reasonable in terms of their cost (including not merely financial considerations, but also resources such as time, training requirements, or ethical dimensions); and reproducible (a challenge also for computational models, not only when they include descriptions of stochasticity, but also when they exhibit language-, compiler-, or hardware-dependence) (Kohl et al, 2006). Thus, the multi-level nature of biological systems must find suitable reflection in an integrated set of multiple models, both experimental and computational. This will be discussed next in the context of the VPH initiative.
Systems biology and the VPH
The VPH initiative
As its name suggests, the VPH initiative targets the whole human body as the system of interest. But, it does not herald a return to classical top-down physiology from entity to parts. The aim is to understand human physiology quantitatively, as a dynamic system, and at all relevant levels between genes and the organism.
Equally, it is not a bottom-up analysis from parts to entities. This would be impossible, both conceptually (as the ‘parts' of the whole organism form systemic ‘entities' of their own), and practically (as the number of possible combinations of interactions between the products of 25 000 genes is simply too vast (Feytmans et al, 2005)).
The approach is better characterised by a term introduced by Sydney Brenner, ‘middle-out' (Brenner et al, 2001), which is based on conceptualising insight at whichever level there is a good understanding of data and processes, and on then connecting this to higher and lower levels of structural and functional integration. In a system of multi-level interactions that involves both regulatory feedforward and feedback pathways, as well as environmentally prescribed parameter constraints, there is really no alternative to breaking in at one level (the ‘middle' part of the metaphor) and then reaching ‘out' to neighbouring levels using appropriate, experimentally founded and validated mathematical methods (Bassingthwaighte et al, 2009).
Of course, one has to be aware of the possible (and in the present case counterproductive) association of the expressions ‘higher' or ‘lower' level with ‘superior' or ‘inferior' in terms of relevance for systems function. Regulatory interactions are, by definition, two-way (‘regulatory loop'), and the metaphoric use of high and low is associated here simply with the notion of spatial scale, not relevance. Furthermore, it is important to realize that influences from ‘outer' levels to the ‘middle' are equally relevant. One might call this an outside-in approach, illustrating the utility and limitations of metaphors, simplified representations of a concept or idea (models!), which are not necessarily of much help when used outside the applicable contextualisation for which they were developed.
A lead example: systems biology of the virtual heart
We will illustrate the ideas discussed above by considering the modelling of cardiac structure and function, partly because that is the area of our own research, but also because, by common consent, it is the most highly developed example of a virtual organ, with applications already within the pharmaceutical industry and in the development of medical devices (Hunter et al, 2001; Noble 2008b). There are three reasons for this situation.
First, cardiac cell models have now benefited from a track record of nearly 50 years of iterative interaction between modelling and experimentation, with an accumulating body of insights derived as much from the ‘failures' as from the ‘successes' of theoretical prediction and experimental validation (Noble 2002). In fact, the contradiction of predictions—whether based on hypotheses formed in thought experiments (conceptual models) or quantitative simulation (computer models)—is usually more instructive than their confirmation. Although confirmation increases the confidence associated with a particular concept or model, contradiction highlights shortcomings in the quality and/or quantity of data input, processing, or interpretation. This will prompt additional observation, consideration, and conceptualisation, with the potential of advancing models and insight (Kohl et al, 2000).
Second, despite its complexity, the heart shows pronounced spatial regularity in structural properties (from the tissue level right through to the arrangement of subcellular protein- and membrane-structures), and it is governed by a very high degree of spatio-temporal coordination of key functional behaviour (such as the spreading wave of electrical excitation that invokes every single cardiomyocyte during each heartbeat, or the highly orchestrated sequence of ionic fluxes and protein interactions that give rise to remarkably optimised pressure generation some 2.5 billion times in the healthy human heart during a life time).
Third, systems of interaction in the heart show a considerable degree of modularity. Basic models of cardiac electrophysiology, for example, do not need to take into account interactions with cardiac mechanics, circulation, metabolism, and so on, to predict important aspects of the interplay between ion distributions, currents, and voltage changes. As they become increasingly detailed, however, wider interactions become more and more relevant, as entities that were classically believed to be linked in a one-directional manner are subject to cross-talk and interaction. Examples include the interdependence of cardiac structure and function (Allessie et al, 2002), of ion channels and cell or tissue behaviour (Hodgson et al, 2003), or of electrophysiology and mechanics (Kohl et al, 2006).
Work on the virtual heart has advanced with progressively increasing complexity. The earliest cell models had just three differential equations that represented the summary kinetics of multiple ‘lumped' electrical mechanisms which, by and large, had not yet been identified and were not, therefore, strictly related to individual protein channel subtypes as we know them now. Cell models today may contain 50 or more equations (Ten Tusscher et al, 2004), depending on the extent to which individual ion handling mechanisms are represented (e.g. through Markov models of ion channels (Clancy and Rudy, 1999)) and the complexity with which intracellular structural features are simulated (Pásek et al, 2008). The insertion of such models into tissue and organ models has also occurred at different levels of tissue size and complexity. Although the goal of reconstructing the whole organ with representative histo-anatomical detail is important for some applications (Burton et al, 2006; Plank et al, 2009), much insight can be gleaned from multi-cellular simulations using one-dimensional strands of cells, two-dimensional sheets, and three-dimensional simplified tissue geometries (Garny et al, 2005). The overall lesson from these simulations has been that theoretical models of biological behaviour are most efficient when they are as complex as necessary, yet as simple as possible.
Extension of principles from heart to other systems: opportunities and challenges
We do not have the space here to review the modelling of other organs and systems. Readers can find out more by accessing the websites of the Physiome Project (http://www.physiome.org.nz/) and the VPH (http://www.vph-noe.eu/). However, some of the approaches and principles developed for, and applied to, cardiac modelling may be transferrable to other aspects of the VPH initiative. Among the features that are already being tackled with some success by the Physiome community are several general issues related to the various types of modelling approaches and their role in the discovery process (Box 1). These principles have emerged largely from grass-roots development of model systems in the cardiac field. Although instructive, there is of course no reason to regard them as prescriptive indicators of how other VPH-related projects should be pursued.
Box 1 General principles learned from the cardiac modelling field.
Conceptual Duality: the combined application of reductionist and integrationist tools and concepts lies at the very heart of successful development of a quantitative understanding of systems behaviour. The analysis of heart rhythm resulting from individual protein interactions (reductionist aspect) and their integration through feedback from the overall cell electrical activity (integration) is a good example (Noble, 2006, chapter 5).
Iteration of Theory and Practice: ‘wet' experimental and ‘dry' theoretical models need to be developed in continuous iteration, where new experimental (or clinical) data feed model development and/or refinement, while computational predictions are used to guide hypothesis formation and experimental design, the outcome of which is the used to validate model predictions. A good example of this approach can be found in the papers of Lei and Kohl (1998) and Cooper et al (2000), which used modelling to interpret experiments showing an unexpected effect of cell swelling on pacemaker frequency, leading to work using axial stretch to yield the expected result, also explained by the modelling.
Structure–Function Relationship: biological function cannot be dissociated from underlying structure. This finds a reflection in modelling, whether using ‘lumped parameters' to describe general compartmentalisation (Orchard et al, 2009) or detailed representations of three-dimensional morphology of proteins (Young et al, 2001), cells (Iribe et al, 2009), or organs (Zhao et al, 2009). Increasingly, this effort benefits from standards, tools, and markup languages, such as SBML (http://sbml.org/Main_Page), CellML (http://www.cellml.org/) and FieldML (http://www.fieldml.org/).
Multi-Scale Modelling: models at different scales of structural integration are required to explore behaviour from molecule to organ or organism. This applies equally to ‘wet' and ‘dry' research, and involves bridging spatial scales of (at least) nine orders of magnitude (from nm to m) and temporal scales spanning 17 orders of magnitude or more (from nanoseconds for description of molecular motion, to years or decades, for longitudinal assessment of human development in norm and disease (Hunter and Borg, 2003). This requires application of ‘new maths' to systems modelling, for example, scale relativity theory (Auffray and Nottale, 2008; Nottale and Auffray, 2008).
Multiplicity of Models (at each individual level): the availability of models of differing levels of complexity, even at the same level of structural integration, allows the treatment of the same biological aspect in different ways, depending on the nature of the question being addressed (for examples see Noble and Rudy, 2001). Although this is common practice in ‘wet' studies, it is often questioned in ‘dry' research.
Multi-dimensional Modelling: models from 0D to 3D+Time are needed to analyse parts of the system that may, in some situations, be regarded as point-sources (e.g. cell electrophysiology when looking at gross electrical behaviour such as reflected in the electrocardiogram), and in others as complex spatio-temporally structured reaction environments (such as the same cell when considering signal transduction cascades). For an Open Source environment designed to address this see Bernabeu et al (2009).
Multi-physics Modelling: addressing questions of varying character, from the stochastic behaviour of ion-channel-interactions to deterministic links between events, or from multiple ODE systems to soft tissue mechanics and fluid dynamics, require different implementations (e.g. finite differences, finite elements, or boundary element methods, Hodgkin–Huxley versus Markov formalisms (see e.g. Fink and Noble, 2009), diffusion reaction versus Monte Carlo approaches, etc).
Modularity of Models: a desirable but thus far ill-implemented need is the definition of model interfaces. These may range from true modularity of components, to translation tools or black-box style parameter inheritance. Likewise, model mapping is an area where much more research into theoretical understanding and practical tools is called for (Terkildsen et al, 2008).
High-Speed Simulation: application to real-world scenarios, in particular for time-critical emergency settings, calls for faster-than-real-time simulation. The new generation of supercomputers (e.g. the 10 petaflop machine being constructed for RIKEN in Kobe, Japan) combined with improved algorithms is expected to make this possible (Bordas et al, 2009).
Interactivity: interactive assessment of model behaviour is relevant for efficient implementation of ‘dry' experiments, as well as for training, education, and interaction between experts from different professional backgrounds (Garny et al, 2009).
The reason for this is straightforward and bears relevance for systems biology in general: we simply do not know what approach will eventually succeed. Researchers pursuing a systems approach can be likened more to people finding their way through unchartered territory, than to those walking a path that has already been mapped. Contrary to the Genome Project, we do neither know the smallest part that we need to identify (there is no elementary set of generic building blocks from which we can assemble the jigsaw), nor the extent of the overall entity (in terms of the types and number of interactions that need to be quantified). We have to determine the best approach as we try out various ideas on how to modularise, simplify, connect multiple levels, relate different aspects at the same level, and incorporate increasingly fine-grained structural and functional data. At the same time, we are also seeking mathematical approaches and computational resources that will enable models to be run in a reasonable period of time (Fink and Noble, 2009), while using user interfaces that allow utilisation by non-experts in computational modelling (Garny et al, 2003). These considerations are associated with a number of additional challenges that have also been experienced in the cardiac modelling field, but are far from being resolved (some examples are listed in Box 2).
Box 2 Issues and Challenges.
Model Curation and Preservation: the long-term preservation of data and models and the maintained ability to access digital data formats are recognised challenges of modern IT infrastructures. They also present key concerns for the VPH initiative.
Tools, Standards, Ontologies and Access: concerted efforts have been launched to facilitate the identification of suitable tools, standards, and ontologies to support model development, interaction, and access (Hucka et al, 2003). This is one of the declared aims of the VPH initiative and requires a willingness to
contribute to the development of standards;
adhere to ‘good practice', once standards are agreed; and
share and publish data, metadata, and models in a suitably annotated, re-usable format.
Patient-specific Analysis and Treatment: as non-invasive data-rich imaging methods are becoming increasingly productive in the clinical setting, the goal of incorporating patient-specific data into models for use in diagnosis, treatment planning, and prevention is beginning to become a reality. This goal is desirable for a variety of reasons, ranging from economic (it makes sense to choose treatments that are tailor-made for the patient, rather than block-buster medicines that often miss the target) to ethical (we should look forward to the day when we no longer tolerate disastrous side-effects that could be eliminated by stratifying the patient population) and scientific considerations (prevent, and if that is not possible, treat the patient—not the disease).
Of particular relevance, in our view, is the need to establish public access to data and models derived from publicly funded work. This could be regarded as a make-or-break issue, as crucial for systems biology as was the decision by a majority of Genome Project investigators to publish and share information on annotated gene sequences, obtained through publicly funded research (rather than patenting them, which would have invoked a whole host of ethical, scientific, and socioeconomic dilemmas).
In this context, a range of ethical issues arise. We will refer briefly to just three of them here. The first is one of scientific integrity and social responsibility (and inherently underlies the drive towards public access to data and models): to serve the usual criteria of scientific scrutiny and public accountability, and to avoid ‘re-inventing wheels', it is required to enable others to review, (re-)use, develop, and efficiently apply prior work. From this, a second issue arises, related to professional development and career progression: as long as the prevailing approach to assessing ‘academic merit' disproportionately rewards ‘peer-reviewed' publications as the output of academic endeavour, compared with the (often very time consuming) development of ‘peer-used' tools, sharing data and models may end up disadvantaging those professionals who generate them (by relieving them of control over and, conceivably, co-authorship in their follow-on use). A third ethical aspect is the obvious need to protect the privacy of individuals' data (a common challenge to using, re-using, and sharing human data). An international solution to these challenges may be regarded as a second make-or-break issue for systems biology and the VPH.
Conclusions
Systems biology may be interpreted as a scientific approach (rather than a subject or destination) that consciously combines ‘reductionist' (identification and description of parts) and ‘integrationist' (internal and external interactions) research, to foster our understanding of the nature and maintenance of biological entities. During the decade or so in which systems biology has become popular, it has often been interpreted as an extension of molecular biology, here to foster the understanding of subcellular regulation networks and interaction pathways, essentially equating ‘system' with ‘cell'. While representing an important aspect of the systems approach, there is no a priori reason why one level of structural or functional complexity should be more important than any other (Noble, 2008a). Work involving more complex levels of structural and functional integration is essential if systems biology is to deliver in relation to human physiology and health care. In addition to this vertical integration across multiple scales, we also need horizontal integration across boundaries such as between organ systems, and between ‘wet' and ‘dry' modelling. Often, the best results are obtained when theoretical work is pursued in close and continuous iteration with experimental and/or clinical investigations. An essential task for systems biology is therefore the quantitative integration of in-silico, in-vitro, and in-vivo research. Key make-or-break issues are the extent to which we can harvest the synergies between the multiple international efforts in the field by sharing data and models, and the question of how to address the ethical dimensions of relevant research and development in this area.
Editorial Note
This Guest Editorial was commissioned on the occasion of the EMBL/EMBO Science & Society Conference on ‘Systems and Synthetic Biology: Scientific and Social Implications', Heidelberg, November 7–8, 2008. Additional contributions from several speakers are available on the EMBO Reports website (http://www.nature.com/embor).
Acknowledgments
Work in the authors' laboratory is supported by the European FP6 BioSim network and the normaCOR grant; by the European FP7 VPH NoE, preDiCT, and EU-Heart projects, as well as by the UK's Biotechnology and Biological Sciences Research Council, the Engineering and Physical Sciences Research Council, the Medical Research Council and The Wellcome Trust. PK is a Senior Fellow of the British Heart Foundation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allessie M, Ausma J US (2002) Electrical, contractile and structural remodeling during atrial fibrillation. Cardiovasc Res 54: 230–246 [DOI] [PubMed] [Google Scholar]
- Auffray C, Nottale L (2008) Scale relativity theory and integrative systems biology 1. Founding principles and scale laws. Progress Biophys Mol Biol 97: 79–114 [DOI] [PubMed] [Google Scholar]
- Bassingthwaighte JB, Hunter PJ, Noble D (2009) The Cardiac Physiome: perspectives for the future. Exp Physiol 94: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabeu MO, Bordas R, Pathmanathan P, Pitt-Francis J, Cooper J, Garny A, Gavaghan DJ, Rodriguez B, Southern JA, Whiteley JP (2009) Chaste: incorporating a novel multiscale spatial and temporal algorithm into a large scale open source library. Philos Trans R Soc A 367: 1907–1930 [DOI] [PubMed] [Google Scholar]
- Bordas R, Carpentieri B, Fotia G, Maggio F, Nobes R, Pitt-Francis J, Southern JA (2009) Simulation of cardiac electrophysiology on next-generation high-performance computers. Philos Trans R Soc A 367: 1951–1970 [DOI] [PubMed] [Google Scholar]
- Boyd CAR, Noble D (ed) (1993) The Logic of Life. Oxford: OUP [Google Scholar]
- Brenner S, Noble D, Sejnowski T, Fields RD, Laughlin S, Berridge M, Segel L, Prank K, Dolmetsch RE (2001) Understanding complex systems: top-down, bottom-up or middle-out?. In Novartis Foundation Symposium: Complexity in Biological Information Processing, Bock G, Goode J (eds), Vol. 239, pp 150–159Chichester: John Wiley [Google Scholar]
- Burton RAB, Plank G, Schneider JE, Grau V, Ahammer H, Keeling SJ, Lee J, Smith NP, Gavaghan D, Trayanova N, Kohl P (2006) Three-dimensional models of individual cardiac histo-anatomy: tools and challenges. Ann NY Acad Sci 1080: 301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy CE, Rudy Y (1999) Linking a genetic defect to its cellular phenotype in a cardiac arrhythmia. Nature 400: 566–569 [DOI] [PubMed] [Google Scholar]
- Cooper PJ, Lei M, Cheng LX, Kohl P (2000) Axial stretch increases spontaneous pacemaker activity in rabbit isolated sino-atrial node cells. J Appl Physiol 89: 2099–2104 [DOI] [PubMed] [Google Scholar]
- Feytmans E, Noble D, Peitsch M (2005) Genome size and numbers of biological functions. Trans Comput Syst Biol 1: 44–49 [Google Scholar]
- Fink M, Noble D (2009) Markov models for ion channels - versatility vs. identifiability and speed. Philos Transact A Math Phys Eng Sci 367: 2161–2179 [DOI] [PubMed] [Google Scholar]
- Garny A, Kohl P, Noble D (2003) Cellular open resource (COR): a public CellML based environment for modelling biological function. Int J Bifurcat Chaos 13: 3579–3590 [Google Scholar]
- Garny A, Noble D, Hunter PJ, Kohl P (2009) Cellular open resource (COR): current status and future directions. Philos Trans R Soc A 367: 1885–1905 [DOI] [PubMed] [Google Scholar]
- Garny A, Noble D, Kohl P (2005) Dimensionality in cardiac modelling. Prog Biophys Mol Biol 87: 47–66 [DOI] [PubMed] [Google Scholar]
- Guyton AC, Coleman TG, Granger HJ (1972) Circulation: overall regulation. Ann Rev Physiol 34: 13–46 [DOI] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF (1952) A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol 117: 500–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson DM, Zingman LV, Kane GC, Perez-Terzic C, Bienengraeber M, Ozcan C, Gumina RJ, Pucar D, O'Coclain F, Mann DL, Alekseev AE, Terzic A (2003) Cellular remodeling in heart failure disrupts KATP channel-dependent stress tolerance. EMBO J 22: 1732–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ et al. (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models Bioinformatics 4: 524–531 [DOI] [PubMed] [Google Scholar]
- Hunter PJ, Borg TK (2003) Integration from proteins to organs: the physiome project. Nat Rev Mol Cell Biol 4: 237–243 [DOI] [PubMed] [Google Scholar]
- Hunter PJ, Kohl P, Noble D (2001) Integrative models of the heart: achievements and limitations. Philos Trans R Soc A 359: 1049054-1 [Google Scholar]
- Iribe G, Ward CW, Camelliti P, Bollensdorff C, Mason F, Burton RA, Garny A, Morphew MK, Hoenger A, Lederer WJ, Kohl P (2009) Axial stretch of rat single ventricular cardiomyocytes causes an acute and transient increase in Ca2+ spark rate. Circ Res 104: 787–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl P, Bollensdorf C, Garny A (2006) Effects of mechanosensitive ion channels on ventricular electrophysiology: experimental and theoretical models. Exp Physiol 91: 307–321 [DOI] [PubMed] [Google Scholar]
- Kohl P, Noble D, Winslow R, Hunter PJ (2000) Computational modelling of biological systems: tools and visions. Philos Trans R Soc A 358: 579–610 [Google Scholar]
- Lei M, Kohl P (1998) Swelling-induced decrease in spontaneous pacemaker activity of rabbit isolated sino-atrial node cells. Acta Physiol Scand 164: 1–12 [DOI] [PubMed] [Google Scholar]
- Noble D (1962) A modification of the Hodgkin-Huxley equations applicable to Purkinje fibre action and pacemaker potentials. J Physiol 160: 317–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble D (2002) Modelling the heart: insights, failures and progress. BioEssays 24: 1155–1163 [DOI] [PubMed] [Google Scholar]
- Noble D (2006) The Music of Life Oxford: OUP [Google Scholar]
- Noble D (2008a) Claude Bernard, the first Systems Biologist, and the future of Physiology. Exp Physiol 93: 16–26 [DOI] [PubMed] [Google Scholar]
- Noble D (2008b) Computational models of the heart and their use in assessing the actions of drugs. J Pharmacol Sci 107: 107–117 [DOI] [PubMed] [Google Scholar]
- Noble D (2008c) Genes and causation. Philos Trans R Soc A 366: 3001–3015 [DOI] [PubMed] [Google Scholar]
- Noble D, Rudy Y (2001) Models of cardiac ventricular action potentials: iterative interaction between experiment and simulation. Philos Trans R Soc A 359: 1127–1142 [Google Scholar]
- Nottale L, Auffray C (2008) Scale relativity and integrative systems biology 2. Macroscopic quantum-type mechanics. Prog Biophys Mol Biol 97: 115–157 [DOI] [PubMed] [Google Scholar]
- Orchard CH, Pásek M, Brette F (2009) The role of mammalian cardiac t-tubules in excitation-contraction coupling: experimental and computational approaches. Exp Physiol 94: 509–519 [DOI] [PubMed] [Google Scholar]
- Pásek M, Brette F, Nelson A, Pearce C, Qaiser A, Christe G Orchard CH (2008) Quantification of t-tubule area and protein distribution in rat cardiac ventricular myocytes. Prog Biophys Mol Biol 96: 244–257 [DOI] [PubMed] [Google Scholar]
- Plank G, Burton RAB, Hales P, Bishop M, Mansoori T, Bernabeu M, Garny A, Prassl AJ, Bollensdorf C, Mason F, Mahmood F, Rodriguez B, Grau V, Schneider JE, Gavaghan D, Kohl P (2009) Generation of histo-anatomically representative models of the individual heart: tools and application. Philoso Trans R Soc A 367: 2257–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Tusscher KHWJ, Noble D, Noble PJ, Panfilov AV (2004) A model of the human ventricular myocyte. Am J Physiol 286: H1573–H1589 [DOI] [PubMed] [Google Scholar]
- Terkildsen JR, Niederer S, Crampin E, Hunter PJ, Smith NP (2008) Using physiome standards to couple cellular functions for rat cardiac excitation-contraction. Exp Physiol 93: 919–929 [DOI] [PubMed] [Google Scholar]
- von Bertalanffy L (1968) General System Theory New York: George Braziller, Inc [Google Scholar]
- Young HS, Jones LR, Stokes D (2001) Locating phospholamban in co-crystals with Ca(2+)-ATPase by cryoelectron microscopy. Biophys J 81: 884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Trew ML, Legrice IJ, Smaill BH, Pullan AJ (2009) A tissue-specific model of reentry in the right atrial appendage. J Cardiovasc Electrophysiol 20: 675–684 [DOI] [PubMed] [Google Scholar]