Abstract
Objectives
The high prevalence of isoniazid-resistant Mycobacterium tuberculosis is often explained by a high mutation rate for this trait, although detailed information to support this theory is absent. We studied the development of isoniazid resistance in vitro, making use of a laboratory strain of M. tuberculosis.
Methods
Spontaneous isoniazid-resistant mutants were characterized by molecular methods allowing identification of the most commonly encountered resistance-conferring mutations. Additionally, we determined the in vitro mutation rates for isoniazid and rifampicin resistance, and characterized the genome of a triple-resistant strain.
Results
Results confirm that the in vitro mutation rate for isoniazid resistance (3.2 × 10−7 mutations/cell division) is much higher than the rate for rifampicin resistance (9.8 × 10−9 mutations/cell division). However, in the majority of the in vitro mutants katG was partially or completely deleted and neither of the two most common in vivo mutations, katG-S315T or inhA-C(-)15T, were found in 120 isogenic mutants. This implies that clinically prevalent resistance mutations were present in <0.8% of isoniazid-resistant strains selected in vitro (95% CI 0%–2.5%). The triple-resistant strain had acquired isoniazid resistance via a 49 kbp deletion, which included katG. Apart from previously identified resistance-conferring mutations, three additional point mutations were acquired during sequential selection steps.
Conclusions
These outcomes demonstrate that the in vivo mechanism of isoniazid resistance is not reflected by in vitro experiments. We therefore conclude that the high in vitro mutation rate for isoniazid resistance is not a satisfactory explanation for the fact that isoniazid monoresistance is significantly more widespread than monoresistance to rifampicin.
Keywords: tuberculosis, drug resistance, mutation rate
Introduction
Although the global incidence of tuberculosis (TB) is declining, drug resistance is rapidly emerging and spreading.1 Insufficient control measures have led to a drastic increase in the prevalence of drug-resistant strains and, more importantly, the degree of drug resistance; in certain high-burden countries the nature of multidrug-resistant TB (MDR-TB) has rapidly evolved from incidental to epidemic.1
The production of new and more effective antibiotics partially addresses this problem, but without a more detailed comprehension of the molecular mechanisms that lead to successful (M)DR strains, these new compounds may also be quickly rendered ineffective by the generation of extensively drug-resistant TB (XDR-TB).
Isoniazid and rifampicin are the two most commonly used anti-TB drugs and the backbone of any successful TB treatment programme. MDR-TB is consequently defined as TB that is resistant to at least isoniazid and rifampicin. Monoresistance to isoniazid is significantly more prevalent than monoresistance to rifampicin and MDR-TB; based on the most recent data it is estimated that 13.3% of all TB cases globally involve isoniazid monoresistance, versus 0.6% rifampicin monoresistance and 5.3% MDR-TB cases.1
Shortly after the introduction of isoniazid in the 1950s, the first resistant Mycobacterium tuberculosis mutants were isolated and it was observed that many of these strains had reduced catalase activity.2 It was first assumed that deletion of katG, coding for the only identified catalase–peroxidase in M. tuberculosis, was the main mechanism responsible for resistance to isoniazid.3 However, subsequent studies demonstrated that, although conferring high levels of resistance, this is not the primary mechanism.4–7 Although any mutation reducing KatG activity results in resistance, relatively few mutations in katG account for the majority of isoniazid resistance in clinical isolates; resistance to isoniazid is primarily conferred by the mutations katG-S315T (45%–75%) and inhA-C(-)15T (15%–25%).8–13
Biochemical and in vitro studies have shown that the most prevalent mutant, katG-S315T, still retains a significant part of its catalase activity,7,14–17 which appears to be sufficient for the preservation of virulence.18,19 Strains carrying this mutation typically have an intermediate (1–5 mg/L) level of resistance to isoniazid,11,15,20,21 since KatG has reduced abilities to activate isoniazid, but has not completely lost this function. Mutations in the regulatory region of inhA usually lead to a low-level resistance (0.2–1.0 mg/L).15,21
Mutations that lead to complete loss of function of KatG, such as deletions, insertions or other frameshifts, are found in a small proportion of the isoniazid-resistant strains.22 On average, partial or complete katG deletion mutants are reported in only ∼5% of resistant isolates,13,15,20,23,24 but higher proportions have been reported on occasion.5,6 Additionally, strains with isoniazid resistance due to deletion of the katG gene are less likely to be clustered than strains with the katG-S315T mutation, a mutation significantly associated with successful dissemination.12,13,25,26
Isolates resistant due to inactivation of KatG often have additional mutations in the regulatory region or coding sequence of ahpC, coding for an alkylhydroperoxidase.27,28 Originally, researchers assumed these mutations were alternative mechanisms of isoniazid resistance,29,30 but subsequent studies showed that the majority of the mutations in ahpC or the intergenic region oxyR-ahpC were compensating for the loss of katG function.31,32
It is frequently assumed that the mutation rate for isoniazid resistance is ∼100 times higher than for rifampicin and, consequently, that isoniazid resistance precedes resistance to rifampicin in the development of MDR-TB. However, there are no empirical grounds for these assumptions. We therefore studied the emergence of isoniazid resistance in vitro, making use of well-characterized M. tuberculosis laboratory strains. We determined the in vitro mutation rate to isoniazid and characterized spontaneous isoniazid-resistant mutants by sequencing, multiplex ligation-dependent probe amplification (MLPA)33 and comparative genome hybridization (CGH).
Materials and methods
Growth of bacteria
The M. tuberculosis parent strains that all experiments were conducted with, MTB72 (ATCC 35801) and H37Rv (ATCC 27294), were initially susceptible to all antimicrobials used in this study. Bacteria were cultured in Middlebrook medium 7H9 (Difco, BD, Sparks, MD, USA), supplemented with oleic acid/albumin/dextrose/catalase (OADC Enrichment, BD, Sparks, MD, USA), in a shaking incubator at 37°C. A liquid starting culture was made by inoculating pure colonies from Löwenstein–Jensen or Coletsos slopes into 10 mL of culture medium. When these cultures reached the logarithmic growth phase, 0.5 mL was transferred to 9.5 mL of fresh, non-selective medium and bacteria were cultured until mid-logarithmic growth. Oxidative stress was applied by addition of H2O2 (31% solution Ultrapur, Merck KGaA, Darmstadt, Germany) at a subinhibitory concentration of 50 mM, 24 h before the bacteria were harvested. All procedures with live mycobacteria were performed in a Biosafety Level III laboratory.
Growth in macrophages
THP-1 cells
Human macrophage-like THP-1 cells (ATCC TIB-202) were grown in 8 mL of RPMI 1640 medium supplemented with 5% fetal bovine serum, 1% HEPES and 1% l-glutamine (all from Gibco, Invitrogen, Breda, The Netherlands). The cells were grown in 75 cm2 flasks at 37°C in a humidified chamber with a 5% CO2 atmosphere. When a density of ∼1 × 106 cells/mL was reached, cells were treated with 16 nM phorbol 12-myristate 13-acetate (PMA; Sigma–Aldrich Chemie, Zwijndrecht, The Netherlands). After 1 h, M. tuberculosis bacteria were added and after 5 h the PMA and bacteria that were not phagocytosed were washed away with medium twice.
M. tuberculosis
Log-phase cultures of M. tuberculosis strain MTB72 were allowed to stand for 5 min and then the upper 500 µL was filtered with a 5 µm filter (Millipore). After filtration, the concentration of the dispersed bacterial suspensions was determined by measuring the absorbance at 420 nm, an A420 of 0.15 corresponding to 1 × 108 bacteria/mL. Bacteria were added to the macrophages in such a way that a multiplicity of infection of ∼1/10 was maintained. An acid-fast stain was made and analysed by light microscopy (magnification: ×100) in order to determine if bacteria were properly associated with the macrophages.
Selection of isoniazid-resistant mutants
In three experiments, intracellular isoniazid-resistant bacteria were selected by addition of 1 mg/L isoniazid to the flasks after 3 days growth of macrophages + bacteria. After 4 h of incubation with isoniazid, the supernatant was discarded and cells were washed with fresh medium. Macrophages were scraped off the bottom of the flask with a cell scraper and taken up in 2 mL sterile water in a new tube. The macrophages were disrupted by application of osmotic stress and sonication in a water bath, and bacteria were subsequently plated on solid Middlebrook medium (MB7H11) containing 1 mg/L isoniazid.
In one experiment, the pre-selection of resistant mutants in the cells was omitted and after disruption of the macrophages an aliquot of the bacteria was used for determination of cfu. From this experiment, the mutation frequency in macrophages was calculated.
Proportion of mutants
To determine the mutation frequency (i.e. the proportion of resistant mutants), four 0.5 mL aliquots of log-phase cultures were plated onto solid MB7H11 medium (Difco, BD, Sparks, MD, USA) containing 0.4, 1.0 or 20 mg/L isoniazid (Sigma–Aldrich Chemie). Serial dilutions of the cultures were plated on non-selective medium to determine the cfu present. The mutation frequency was calculated by dividing the number of resistant colonies by the cfu plated. From each experiment, a number of resistant mutants were picked from the antibiotic-containing plates and DNA was isolated for further molecular characterization.
Fluctuation assay
The mutation rates (i.e. the chance of a mutation occurring per generation) for isoniazid and rifampicin were determined by performing a fluctuation assay, making use of the p0-method.34
A 10 mL starting culture was prepared by inoculating strain MTB72 from a slope, to minimize the presence of spontaneous mutants. When bacterial growth reached mid-logarithmic phase, the culture was vortexed vigorously to homogenize the bacterial suspension. Clumped cells were allowed to settle for 3 min and 55 new 1 mL cultures were made by transferring 1 µL of this cell suspension (∼1000 bacteria) to a 2 mL screwcap tube containing 1 mL of MB7H9 medium + OADC. Two to three sterile glass beads were added to each culture, to ensure a continuous homogeneous suspension. The 1 mL cultures were incubated in a shaking incubator at 37°C for ∼10 generations (∼8 days), ensuring a ≥1000-fold increase in cell number. The cultures were then centrifuged at 5000 g for 8 min and 850 µL of supernatant was discarded. For each culture the remaining 150 µL was plated in one well of a square 25-well replica plate (Greiner, Germany) containing 3 mL of MB7H11 supplemented with either 8 mg/L rifampicin (Sigma–Aldrich Chemie) or 1.0 mg/L isoniazid. The number of cfu plated in each well was estimated by making serial dilutions of the five remaining 1 mL cultures and plating onto non-selective MB7H11. The two 25-well replica plates were allowed to dry in a biosafety laminar flow cabinet, until all liquid was absorbed into the solid medium. The plates were then sealed and incubated at 37°C. After 3–4 weeks incubation, the proportion of squares without growth was counted and the mutation rate was calculated.34
DNA isolation
Separate mutant colonies, selected on isoniazid-containing solid medium, were picked and suspended in 150 µL of Tris/EDTA buffer containing 1% Triton X-100 (BDH Laboratory Supplies, Poole, England) and then heated at 95°C for 30 min. After lysis, cells were centrifuged at 5000 g for 3 min and 130 µL of the supernatant was collected as a DNA sample.
For the experiment using pooled isolates, four separate colonies were suspended in a single tube and DNA was extracted simultaneously.
PCR and sequencing
A 233 bp fragment, surrounding codon 315 of katG, was amplified from selected isolates by PCR using forward primer katG-315_FW 5′-CATGAACGACGTCGAAACAG-3′ and reverse primer katG-315_RV 5′-CGAGGAAACTGTTGTCCCAT-3′. In addition, a 300 bp fragment of katG surrounding codon 463 was amplified using forward primer katG-463_FW 5′-TCCCGTTGCGAGATACCTT-3′ and reverse primer katG-463_RV 5′-AGGGTGCGAATGACCTTG-3′. Isolates were scored as ΔkatG if one or both of the PCR products were absent. From a proportion of the strains that were positive for the katG-315 PCR, the PCR fragment was sequenced in both directions according to protocols previously published.35
From another subselection of our isoniazid-resistant isolates, a 251 bp fragment spanning the intergenic region of oxyR-ahpC was amplified and sequenced, using forward primer oxyR-ahpC_FW 5′-TGCTGAACCACTGCTTTGC-3′ and reverse primer oxyR-ahpC_RV 5′-GGTCACCGCCGATGAGAGC-3′.
Results of all PCRs were analysed by agarose gel electrophoresis using a 2% agarose gel (Multi-Purpose agarose, Roche Diagnostics GmbH, Mannheim, Germany).
MLPA
M. tuberculosis-specific MLPA was performed as previously published,33 except in the present study only four probes were included (Table 1), allowing results to be easily interpreted on an agarose gel. The 16S rRNA probe (202 bp) targets a region specific for members of the MTB complex and was used as internal control. Probes katG-315 (160 bp) and inhA-15 (178 bp) identify the two most prevalent isoniazid resistance-conferring mutations. Probe katG-463 targets a point mutation in codon 463 of katG specific for principal genotypic groups (PGGs) 2 and 3 that was present in all strains used in this study. The absence of the katG-463 or katG-315 products identified a deletion of these regions.
Table 1.
MLPA probes used in this study to screen isoniazid-resistant mutants
| MLPA probe | Target | Length of ligated probes | Phenotype |
|---|---|---|---|
| 16S rRNA | 16S rRNA gene | 202 bp | member of MTB complex |
| katG-315 | katG codon 315 ACC | 160 bp | isoniazid-resistant via katG-S315T |
| inhA-15 | inhA (-)15 ‘T’ allele | 178 bp | isoniazid-resistant via inhA-C(-)15T |
| katG-463 | katG codon 463 CGG | 319 bp | PGG 2 or 3 |
Indicated are the name of the probe, the region or point mutation it targets, the length of the ligated probes and the phenotype that is associated with the point mutation.
CGH
Strain RB14, a rifampicin- and rifabutin-resistant isolate derived from MTB72,35,36 was plated on solid medium containing 20 mg/L isoniazid. Upon growth of isoniazid-resistant mutants, 13 colonies were picked, purified and analysed by PCR and MLPA, as described in this study. Seven out of 13 (54%) were negative for both the katG-315 and the katG-463 PCR, indicating a substantial deletion of the katG region. We selected one of these deletion mutants of RB14, RB14H5, for further analysis. In order to determine the exact size of the deletion, the genomes of strain RB14H5 and MTB72, its wild-type parent, were compared by CGH (NimbleGen CGH Whole-Genome Tiling Arrays, Roche NimbleGen, Madison, WI, USA). DNA was extracted from liquid cultures and processed, purified and shipped according to the company's requirements. Data obtained from the CGH experiment was interpreted and analysed with SignalMap v1.9 and Microsoft Excel. Primers were designed to amplify likely regions of difference identified and any mutations present were confirmed by cycle sequencing [Table S1, available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/)].
Results
Deletions in katG are the most common mechanism of resistance to isoniazid in vitro
Two initial independent experiments with MTB72 were performed, using 20 mg/L isoniazid to select resistant mutants. Mutation frequencies of 4.40 × 10−6 and 7.56 × 10−6 were measured, and PCR assays targeting the regions flanking codons 315 and 463 of katG identified deletions in 4/20 and 18/20 mutants, respectively (Table 2). None of the mutants with an intact katG gene carried a mutation, as determined by sequencing, except for one strain that had acquired a TGG(W)→CGG(F) mutation in codon 321. Deletions in katG were confirmed by MLPA, as well as the absence of typical mutations katG-S315T or inhA-C(-)15T in all the characterized mutants.
Table 2.
Characterization of the spontaneous isoniazid-resistant mutants derived from strain MTB72
| Antibiotic concentration | Selection condition | Mutation frequency | Mutation rate | No. of ΔkatG (%) | No. of targeted mutations |
|---|---|---|---|---|---|
| 20 mg/L INH | — | 7.56 × 10−6 | — | 18/20 (90%) | 0/20 |
| — | 4.40 × 10−6 | — | 4/20 (20%) | 0/20 | |
| +H2O2 | 3.60 × 10−6 | — | 17/20 (85%) | 0/20 | |
| 1 mg/L INH | — | 4.17 × 10−6 | — | 15/20 (75%) | 0/20 |
| — | 1.78 × 10−6 | — | 13/37 (35%) | 0/37 | |
| +H2O2 | 1.60 × 10−5 | — | 14/20 (70%) | 0/20 | |
| +THP1 | 1.84 × 10−6 | — | 5/24 (21%) | 0/24 | |
| +THP1 + INH | — | — | 1/10 (10%) | 0/10 | |
| +THP1 + INH | — | — | 0/5 (0%) | 0/5 | |
| +THP1 + INH | — | — | 6/20 (30%) | 0/20 | |
| 1 mg/L INH | — | 2.98 × 10−6a | 3.20 × 10−7 | — | — |
| 8 mg/L RIF | — | 6.86 × 10−8b | 9.81 × 10−9 | — | — |
INH, isoniazid; RIF, rifampicin; THP1, macrophage; MTB, M. tuberculosis.
The concentration of isoniazid was the concentration in the solid medium on which the bacteria were selected. The selection conditions are additional criteria to select against ΔkatG mutants, in an attempt to favour the selection of clinically relevant mutants. Mutation frequency is the proportion of mutants. Mutation rate is the estimated number of mutants per generation. No. of ΔkatG is the number of katG deletion mutants (of the characterized mutants). No. of targeted mutations is the number of katG-S315T or inhA-C(-)15T found in the characterized mutants.
aAverage of data depicted in this table.
bAverage of five independent experiments.
We hypothesized that the high mutation frequencies and domination of katG deletions over isoniazid resistance-conferring mutations were either due to the absence of significant oxidative stress in vitro or the concentration of isoniazid used. To test the first hypothesis, the experiment was repeated after exposure to 50 mM H2O2, in an attempt to select against the occurrence of katG deletions in favour of clinically relevant mutations. The mutation frequency was comparable to that obtained in previous results (3.6 × 10−6, see Table 2) and 17/20 (85%) selected mutants had a deletion in katG. None of the analysed strains had acquired resistance mutations in katG-315 or inhA-(-)15.
We then lowered the concentration of isoniazid to 1 mg/L. Again, two independent experiments were performed, both resulting in average to high proportions of katG deletion mutants (13/37 and 15/20) and none of the targeted clinically prevalent mutations in 57 colonies analysed. The mutation frequencies were 1.78 × 10−6 and 4.17 × 10−6, respectively. Addition of H2O2 to the medium slightly increased the mutation frequency to 1.60 × 10−5, but the majority (14/20) of the selected mutants had still acquired a deletion in katG (Table 2).
Five independent experiments were also conducted to determine the mutation frequency for rifampicin, resulting in an average mutation frequency of 6.86 × 10−8 using 8 mg/L rifampicin (Table 2). This was 43.4 times lower than the average mutation frequency observed with 1 mg/L isoniazid. In addition, fluctuation assays were performed to determine the mutation rates for isoniazid and rifampicin. The estimated mutation rate for 1 mg/L isoniazid was 3.2 × 10−7 mutations/cell division, 32.3 times higher than the estimated rate for 8 mg/L rifampicin (9.81 × 10−9 mutations/cell division, Table 2).
Less than 1% of in vitro selected isoniazid-resistant mutants carry a clinically prevalent mutation
Being unsuccessful in our attempt to select for isoniazid-resistant strains with either of the two most clinically prevalent mutations, we decided to screen a larger pool of isoniazid-resistant mutants derived from MTB72, in order to estimate the prevalence of the most common clinical mutations in vitro. We used the methods described above to select isoniazid-resistant mutants with 0.4 mg/L isoniazid.
The mutation frequency was 8.02 × 10−6 (Table 3) and an initial characterization of 29 mutants showed that 28% (8/29, Table 3) had acquired a deletion in katG. To increase the chance of finding clinically prevalent mutations, additional mutants were picked from the original plates (0.4 mg/L isoniazid) and plated in duplicate on plates containing 0.4 mg/L isoniazid and 2.0 mg/L isoniazid. Thirty colonies were picked that grew on 0.4 mg/L isoniazid but showed no growth on 2.0 mg/L isoniazid and, thus, had an MIC between 0.4 and 2.0 mg/L isoniazid. Of these 30 selected mutants, three (10%) had acquired deletions in katG (Table 3). No mutations were found in katG-315 or inhA-(-)15 by MLPA, but sequencing of katG from 10 of these low-MIC mutants with intact katG revealed an ACT(T)→ATT(I) mutation in codon 271 of katG in one of the strains.
Table 3.
Characterization of MTB72-derived isoniazid-resistant mutants initially selected with 0.4 mg/L isoniazid
| Acquired MIC | Mutation frequency | No. of ΔkatG (%) | No. of targeted mutations (%) |
|---|---|---|---|
| Undefined | 8.02 × 10−6 | 8/29 (28%) | 0/29 (<3.5%) |
| 0.4 < MIC < 2.0 | — | 3/30 (10%) | 0/30 (<3.3%) |
| MIC > 2.0a | — | — | 0/120 (<0.8%) |
A subdivision of two groups of mutants was made on the basis of their acquired isoniazid MIC, indicated in the column ‘acquired MIC’. MIC is expressed in terms of mg/L isoniazid. Mutation frequency is the proportion of mutants. No. of ΔkatG is the number of katG deletion mutants. No. of targeted mutations is the number of katG-S315T or inhA-C(-)15T found in the characterized mutants.
aDNA samples were derived from mutants pooled in sets of four.
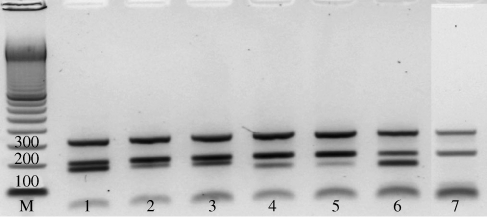
From the panel of mutants selected with 0.4 mg/L isoniazid, an additional subpopulation with an MIC > 2.0 mg/L was analysed by MLPA, targeting the inhA-15 and katG-315 mutant alleles. In order to facilitate analysis, mutant colonies were pooled in sets of four before DNA was extracted and a total of 120 mutants were screened in this fashion. The suitability of the method for detecting a single mutant in a mixed sample using MLPA was confirmed by analysis of artificial mixtures made with wild-type M. tuberculosis DNA and DNA from clinical strains carrying either the katG-S315T or inhA-C(-)15T mutation (Figure 1). We were unable to measure the number and percentage of ΔkatG mutants with this method, since samples were mixtures of four colonies and only samples in which all four colonies had a deletion in katG would be detected. No mutations were found in these 120 isoniazid-resistant in vitro mutants, indicating that the prevalence of clinically relevant mutations [katG-S315T or inhA-C(-)15T] in vitro is <0.8% (95% CI 0%–2.5%), under the conditions described (Table 3).
Figure 1.
Agarose gel showing separation of MLPA products. Samples shown are artificial mixtures made with DNA from isoniazid-resistant clinical isolates and DNA from wild-type strain MTB72. From top to bottom, bands represent amplified probes targeting katG-L463R (319 bp), 16S rRNA (202 bp) and inhA-C(-)15T (178 bp, lanes 1–3) or katG-S315T (160 bp, lanes 4–6). Lane 1, 100% inhA-C(-)15T mutant; lane 2, 10% inhA-C(-)15T mutant + 90% wild-type; lane 3, 20% inhA-C(-)15T mutant + 80% wild-type; lane 4, 20% katG-S315T mutant + 80% wild-type; lane 5, 10% katG-S315T mutant + 90% wild-type; lane 6, 100% katG-S315T mutant; lane 7, 100% wild-type. M denotes a 100 bp marker, numbers on the first three bands correspond to the respective sizes (in bp).
Representative experiments were also performed with model strain H37Rv. Selection of spontaneous resistant mutants with 0.4 mg/L isoniazid resulted in a mutation frequency of 2.3 × 10−6. From the resistant mutants, a subselection was made of strains with an MIC between 0.4 and 2.0 mg/L isoniazid. MLPA analysis of 30 of these selected strains showed that 3.3% (1/30) had acquired a deletion in katG, but no mutations were detected in katG-315 or inhA-(-)15. This indicates that also for strain H37Rv <3.3% of the isoniazid-resistant H37Rv mutants carried a clinical mutation.
No adaptive mutations were found in the oxyR-ahpC intergenic region of a random selection of ΔkatG mutants
In order to exclude the possibility that the high prevalence of ΔkatG mutants in our experiments was due to the acquisition of mutations in the oxyR-ahpC intergenic region, we sequenced a 251 bp fragment spanning the oxyR-ahpC intergenic region of a selection of our MTB72-derived isoniazid-resistant mutants. This panel of strains represented the different isoniazid concentrations and additional selection conditions used in this study, and all strains had acquired deletions in various regions of katG. In total, we screened 21 ΔkatG mutants for adaptive mutations in oxyR-ahpC. The sequence of all analysed mutants was identical to that of the wild-type parent MTB72, indicating that none of the mutants had acquired mutations in this region.
In vitro-selected ΔkatG mutant has a 49 kbp deletion, but is viable in vitro
The genomes of MTB72 and RB14H5 were screened by CGH, in order to obtain information about the mutations that RB14H5 acquired during the sequential selection steps. Purified DNA from both strains was compared with the sequence of strain CDC1551 (GenBank AE000516). The CGH data indicated a large deletion in the RB14H5 genome, compared with that of MTB72 [Figure S1, available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/)]. A PCR with primers targeting the flanking regions confirmed that a 49 kbp fragment spanning 47 genes (MT1928–MT1976), including the whole of katG, was deleted from the RB14H5 genome [Table S2, available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/), and Figure S1]. The exact location of the deletion, 2127129–2176164, was determined by sequencing of this PCR product.
Additional SNPs were accumulated during in vitro selection
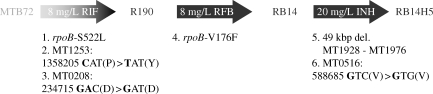
With the data obtained by CGH, not only the extent of the katG deletion could be determined, but also other mutations throughout the genome of strain RB14H5 could be identified. The two previously identified mutations in rpoB conferring resistance to rifampicin and rifabutin, S522L and V176F, respectively,35,36 were confirmed by CGH. In addition, three single nucleotide polymorphisms (SNPs) were identified in genes MT1253, MT0208 and MT0516, corresponding to genes Rv1215c, Rv198c and Rv0498 in H37Rv (GenBank AL123456), respectively. PCR and sequencing of these regions in RB14H5 confirmed a C→T mutation in MT1253, a G→A mutation in MT0208 and a C→G mutation in MT0516, of which only the mutation in MT1253 was non-synonymous (Figure 2).
Figure 2.
Schematic representation of the three rounds of antibiotic selection that strain MTB72 was subjected to. The selection steps finally resulted in the triple-resistant strain RB14H5. The position of the drug resistance-conferring mutations (1, 4 and 5) and additional mutations (2, 3 and 6) indicates in which selection step they were acquired. Gene ID and coordinates of mutations 2, 3, 5 and 6 are based on the annotated genome sequence of strain CDC1551 (GenBank AE000516). R190 and RB14 are the names of the intermediate strains. RIF, rifampicin; RFB, rifabutin; INH, isoniazid; kbp, kilobase pairs; del., deletion.
The regions where mutations were identified in RB14H5 were screened in the intermediate strains isolated after each antibiotic selection step, to determine when these additional mutations occurred. After selection with rifampicin, two SNPs, in MT1253 and MT0208, were identified in addition to the mutation in rpoB. Selection with rifabutin resulted only in a resistance-conferring mutation, rpoB-V176F, but the 49 kbp deletion was accompanied by the mutation in MT0516 after selection with isoniazid (Figure 2).
Discussion
Despite the increasing prevalence of resistant M. tuberculosis, isoniazid remains an essential component of current TB treatment programmes. In an attempt to shed more light on the molecular mechanisms leading to the development of MDR-TB, we initially screened in vitro-selected isoniazid-resistant mutants of M. tuberculosis strain MTB72, using different concentrations of isoniazid. None of the characterized strains carried the mutations that are predominantly responsible for isoniazid resistance in clinical isolates [katG-S315T or inhA-C(-)15T]. Moreover, a large proportion of the resistant clones acquired resistance to isoniazid by deletion of katG, a genotype that is rarely observed in clinical isolates. In agreement with a previous report,37 we measured a higher mutation rate for isoniazid resistance than for rifampicin resistance in vitro. However, the dramatic difference between the in vivo and in vitro spectrum of isoniazid resistance mutations suggests that an in vivo mutation rate for isoniazid resistance equal to or potentially lower than the rifampicin resistance rate (∼10−8 mutations/cell division) is much more feasible. These findings indicate the importance of genetic characterization of mutants before results obtained in the laboratory are extrapolated to the clinical setting.
KatG protects bacteria from the oxidative burst encountered when M. tuberculosis cells reside in macrophages or granuloma.18,38 In vitro conditions are significantly less challenging, not in the least because of the extracellular catalases in the medium, protecting the bacteria from harmful reactive oxygen species. We speculated that these protective culture conditions allowed for the observed high in vitro prevalence of ΔkatG mutants, which are highly resistant to isoniazid, but susceptible to oxidative insult.28,31,38 However, addition of 50 mM H2O2 did not alter the spectrum of mutations (Table 2). Attempts to select isoniazid-resistant mutants from mycobacteria grown in medium without added catalase have been unsuccessful, as bacterial growth of the wild-type strain MTB72 was inadequate in medium without catalase.
Conditions that seemed to have an effect on the proportion of katG deletion mutants, but not on the presence of katG-S315T or inhA-C(-)15T mutants, were selection of mutants with low levels of isoniazid (0.4–2.0 mg/L, Tables 2 and 3) and growth/selection in human macrophages. The average proportion of ΔkatG mutants selected with 20 mg/L was 65% (39/60), with 1 mg/L was 55% (42/77) and with 0.4 mg/L was 28% (8/29, Tables 2 and 3), suggesting that ΔkatG mutants are more likely to be highly resistant.
Moreover, from the mutants selected with 0.4 mg/L isoniazid that had an MIC between 0.4 and 2.0 mg/L, only 10% (3/30, Table 3) had a deletion in the katG region. Growth in THP1 cells reduced the proportion of deletion mutants, selected with 1 mg/L isoniazid, to 20% (12/59, Table 2). These results show that it is possible to reduce the proportion of deletion mutants under specific conditions, but even in strains selected under these conditions neither of the two most prevalent clinical mutations were identified.
The mutants selected with 0.4 mg/L isoniazid (Table 3) that were stratified on the basis of their MICs were all originally selected on the same plate. In total, 179 isolates selected with 0.4 mg/L isoniazid (29 + 30 + 120, Table 3) were analysed by MLPA from this plate. None of these 179 isolates acquired either of the targeted mutations; thus, the prevalence of these mutations in vitro was <0.6% (<1/179, 95% CI 0%–1.6%) under the conditions described. Similar results were also obtained with strain H37Rv (<3.3%), suggesting that this is a general phenomenon, rather than a strain-specific effect. This observation is also supported by the absence of reports of spontaneous in vitro isoniazid-resistant mutants that carry clinical mutations. In fact, we are aware of only one study that reports the selection of an isoniazid-resistant katG-S315T mutant,21 but this mutant was acquired after a mouse was infected and treated with isoniazid. It is possible that none of the selection conditions we used were suitable for the selection of the targeted mutations, perhaps because further adaptation is required to obtain an MIC > 0.4 mg/L. However, this would be true for all previous reports of in vitro studies.
The general assumption is that isoniazid resistance in vivo is much more widespread due to a higher mutation rate, particularly in katG.39 Indeed, the M. tuberculosis katG gene is located within a genetically unstable region,40 suggesting an increased propensity for mutation. Based on this instability, one would expect to observe a whole spectrum of katG mutations in resistant clinical isolates, but in practice the majority of clinically resistant strains contain the same mutations. However, the genetic instability of the katG region is a likely explanation for the high prevalence of ΔkatG mutants in our and other laboratory studies.
The empirical evidence for the high mutation rate hypothesis is extracted from a few studies in which fluctuation assays with isoniazid were performed.37 These studies report a significantly higher mutation rate for isoniazid compared with that for rifampicin, although mutants were not genetically characterized. We obtained similar results for the in vitro mutation rates when selecting with isoniazid and rifampicin (Table 2). However, we suspect that this does not explain the high prevalence of isoniazid resistance in vivo, since none of the isoniazid-resistant mutants we screened carried the mutations that are most frequently observed in clinical isolates. In fact, merely two or three mutations account for isoniazid resistance in the majority of resistant clinical isolates.8–13 For rifampicin resistance there are three rpoB mutations that are seen in 80%–95% of clinical isolates.9,41−43 The in vivo mutation rate for isoniazid resistance is therefore more likely to be similar to or lower than the rate for rifampicin resistance, as was also suggested by others.44 This would still lead to a higher prevalence of isoniazid-monoresistant isolates if one takes into account that isoniazid has been used to treat TB much longer than rifampicin. In addition, isoniazid has been regularly administered as monotherapy, whereas rifampicin is almost exclusively part of a combination therapy, thereby reducing the chances of drug resistance.
Previously, researchers have shown that a whole spectrum of different ΔkatG mutants can be found both in vitro and in vivo.13,15,40,45 In our study, partial deletions of the katG gene close to the 5′ terminus would not have been detected with the methods used, possibly resulting in an underestimation of the deletion mutants present. The few in vitro studies on isoniazid resistance that have been performed have indeed determined that the majority of the mutants were catalase-negative.45 Furthermore, one group of researchers have implicitly stated that almost all in vitro isoniazid mutants identified in their laboratory were ΔkatG strains, regardless of the concentrations of isoniazid used.44
Throughout the course of this work, only two isoniazid-resistant mutants were identified that contained point mutations in the katG gene: one mutant acquired a TGG(W)→CGG(F) mutation in codon 321; and the other an ACT(T)→ATT(I) mutation in codon 271. Neither of these mutations has been observed in clinical isolates, although other mutations in codon 321 have been reported and associated with very low quantities of KatG, and, therefore, significantly reduced catalase/peroxidase activity.7,16,22
None of our selected isoniazid-resistant strains carried a mutation in the oxyR-ahpC intergenic region, even after exposure to H2O2. Sherman et al.28 suggested that either a second selection event or passaging was probably necessary for in vitro-generated mutants to acquire the additional mutation in oxyR-ahpC. This may, in part, explain why this mutation was not identified in any of our strains, although there is at least one report of an isoniazid-resistant mutant that had acquired a mutation in oxyR-ahpC in vitro.46
The failure to detect any of the clinically prevalent mutations in katG, inhA or ahpC in our study, in combination with results of others, suggests that the current in vitro model is not representative of the in vivo situation. These observations also argue against the view that the primary route to MDR-TB is invariably via isoniazid monoresistance. In fact, there is evidence that isoniazid resistance does not always precede rifampicin resistance in the development of MDR in clinical settings.
In a recent report, a rifampicin-monoresistant M. tuberculosis strain was disseminated and subsequently acquired isoniazid and other drug resistance via the common resistance mechanisms, including katG-S315T.47 Moreover, a recent outbreak in London started with an isoniazid-resistant strain, but upon dissemination the strain acquired rifampicin resistance via unusual mutations in rpoB.48 This is interesting as the genetic background of bacteria probably plays a role in the evolutionary routes they can and will follow. The presence of pre-existing drug resistance mutations can be of influence on the acquisition of additional drug resistance36 and several studies have shown that different genotypes of M. tuberculosis appear to have different preferred resistance mechanisms.12,13,49–51 Similar observations were made in our laboratory and by others.35,52 The occurrence of atypical drug resistance mutations may therefore be an indication of suboptimal genetic routes or unusual conditions.
We used CGH to compare the genomes of triple-resistant strain RB14H5 and that of its wild-type parent strain, MTB72. Earlier, we had determined that RB14H5 has a non-functioning catalase–peroxidase in addition to a double mutation in rpoB, which moderately increases the stress levels.36 Remarkably, strain RB14H5 had acquired a 49 kbp deletion, spanning 47 genes including the complete katG gene, but nonetheless it remained viable in vitro. We are aware of only one other report of a strain with an equally large deletion in the katG region.40
Strain RB14H5 acquired three other SNPs in addition to the major deletion and the drug resistance mutations (Figure 2). It is unknown if these SNPs are adaptive mutations or more opportunistic ‘hitchhike’ mutations and whether results obtained are representative of the in vivo situation.
The genetic analysis of bacteria is generally focused on previously identified targets, such as genes involved with antibiotic resistance or repetitive sequences for fingerprinting purposes. One consequence of this bias is that additional mutations elsewhere in the genome are missed. Isogenic strains generated in the laboratory may not be as isogenic as assumed and new phenotypes may consequently be attributed to the wrong mutation.53 CGH allows the whole genome to be screened for additional mutations that may have occurred in a strain of interest compared with the wild-type template. It would be interesting to compare clinical strains in this fashion, for instance sequential isolates obtained from a single patient.
Additionally, CGH can be valuable in detecting drug resistance-conferring mutations;54 in some geographical locations up to 25% of clinical strains have acquired isoniazid resistance via unidentified mechanisms.10,13,39,43
The present study shows that characterization of in vitro mutants is essential in order to draw accurate conclusions and that results obtained in the laboratory are not automatically applicable to the in vivo situation. Combined MLPA and CGH analysis of representative clinical isolates should be used to elucidate preferred routes to MDR. Based on our results it can be concluded that the high in vitro mutation rate for isoniazid resistance is not a satisfactory explanation for the widespread isoniazid resistance found in clinical isolates, as there is no substantial evidence that the in vivo mutation rate for isoniazid resistance is significantly higher than that for rifampicin resistance. Moreover, the outcomes of our study in combination with reports of others lead us to question the current view that development of a successful MDR strain predominantly begins with isoniazid monoresistance. A more detailed study of the genetic mechanisms by which M. tuberculosis strains adapt to their environment is warranted, hopefully leading to identification of genotypes or drug resistance profiles that are most likely to result in virulent strains.
Funding
This work was partly funded by the EU 6th Framework Programme TBAdapt (project no. 037919).
Transparency declarations
None to declare.
Supplementary data
Supplementary Material
Acknowledgements
We would like to thank MRC-Holland (Amsterdam, The Netherlands) for providing all MLPA reagents and the RIVM (Bilthoven, The Netherlands) for providing us with strain H37Rv. We also thank Dr Arend Kolk and Sjoukje Kuijper for help with the macrophage growth experiments.
References
- 1.World Health Organization. Anti-tuberculosis Drug Resistance in the World. Report No. 4. Geneva: WHO; 2008. WHO/HTM/TB/2008.394. [Google Scholar]
- 2.Middlebrook G. Isoniazid-resistance and catalase activity of tubercle bacilli. Am Rev Tuberc. 1954;69:471–2. doi: 10.1164/art.1954.69.3.471. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Heym B, Allen B, et al. The catalase–peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–3. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 4.Altamirano M, Marostenmaki J, Wong A, et al. Mutations in the catalase–peroxidase gene from isoniazid-resistant Mycobacterium tuberculosis isolates. J Infect Dis. 1994;169:1162–5. doi: 10.1093/infdis/169.5.1162. [DOI] [PubMed] [Google Scholar]
- 5.Stoeckle MY, Guan L, Riegler N, et al. Catalase–peroxidase gene sequences in isoniazid-sensitive and -resistant strains of Mycobacterium tuberculosis from New York City. J Infect Dis. 1993;168:1063–5. doi: 10.1093/infdis/168.4.1063. [DOI] [PubMed] [Google Scholar]
- 6.Goto M, Oka S, Tachikawa N, et al. KatG sequence deletion is not the major cause of isoniazid resistance in Japanese and Yemeni Mycobacterium tuberculosis isolates. Mol Cell Probes. 1995;9:433–9. doi: 10.1006/mcpr.1995.0066. [DOI] [PubMed] [Google Scholar]
- 7.Rouse DA, DeVito JA, Li Z, et al. Site-directed mutagenesis of the katG gene of Mycobacterium tuberculosis: effects on catalase–peroxidase activities and isoniazid resistance. Mol Microbiol. 1996;22:583–92. doi: 10.1046/j.1365-2958.1996.00133.x. [DOI] [PubMed] [Google Scholar]
- 8.Herrera-León L, Molina T, Saíz P, et al. New multiplex PCR for rapid detection of isoniazid-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother. 2005;49:144–7. doi: 10.1128/AAC.49.1.144-147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Rie A, Warren R, Mshanga I, et al. Analysis for a limited number of gene codons can predict drug resistance of Mycobacterium tuberculosis in a high-incidence community. J Clin Microbiol. 2000;39:636–41. doi: 10.1128/JCM.39.2.636-641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M, Yue J, Yang YP, et al. Detection of mutations associated with isoniazid resistance in Mycobacterium tuberculosis isolates from China. J Clin Microbiol. 2005;43:5477–82. doi: 10.1128/JCM.43.11.5477-5482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalla Costa ER, Ribeiro MO, Silva MS, et al. Correlations of mutations in katG, oxyR-ahpC and inhA genes and in vitro susceptibility in Mycobacterium tuberculosis clinical strains segregated by spoligotype families from tuberculosis prevalent countries in South America. BMC Microbiol. 2009;9:39. doi: 10.1186/1471-2180-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gagneux S, Burgos MV, DeRiemer K, et al. Impact of bacterial genetics on the transmission of isoniazid-resistant Mycobacterium tuberculosis. PLoS Pathog. 2006;2:e61. doi: 10.1371/journal.ppat.0020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hazbón MH, Brimacombe M, Bobadilla del Valle M, et al. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2006;50:2640–9. doi: 10.1128/AAC.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wengenack NL, Uhl JR, St Amand AL, et al. Recombinant Mycobacterium tuberculosis KatG(S315T) is a competent catalase–peroxidase with reduced activity toward isoniazid. J Infect Dis. 1997;176:722–7. doi: 10.1086/514096. [DOI] [PubMed] [Google Scholar]
- 15.Coll P, Aragón LM, Alcaide F, et al. Molecular analysis of isoniazid and rifampin resistance in Mycobacterium tuberculosis isolates recovered from Barcelona. Microb Drug Resist. 2005;11:107–14. doi: 10.1089/mdr.2005.11.107. [DOI] [PubMed] [Google Scholar]
- 16.Mo L, Zhang W, Wang J, et al. Three-dimensional model and molecular mechanism of Mycobacterium tuberculosis catalase–peroxidase (KatG) and isoniazid-resistant KatG mutants. Microb Drug Resist. 2004;10:269–79. doi: 10.1089/mdr.2004.10.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saint-Joanis B, Souchon H, Wilming M, et al. Use of site-directed mutagenesis to probe the structure, function and isoniazid activation of the catalase/peroxidase, KatG, from Mycobacterium tuberculosis. Biochem J. 1999;338:753–60. [PMC free article] [PubMed] [Google Scholar]
- 18.Pym AS, Saint-Joanis B, Cole ST. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect Immun. 2002;70:4955–60. doi: 10.1128/IAI.70.9.4955-4960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Kelley C, Collins F, et al. Expression of katG in Mycobacterium tuberculosis is associated with its growth and persistence in mice and guinea pigs. J Infect Dis. 1998;177:1030–5. doi: 10.1086/515254. [DOI] [PubMed] [Google Scholar]
- 20.Abe C, Kobayashi I, Mitarai S, et al. Biological and molecular characteristics of Mycobacterium tuberculosis clinical isolates with low-level resistance to isoniazid in Japan. J Clin Microbiol. 2008;46:2263–8. doi: 10.1128/JCM.00561-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo H, Seet Q, Denkin S, et al. Molecular characterization of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis from the USA. J Med Microbiol. 2006;55:1527–31. doi: 10.1099/jmm.0.46718-0. [DOI] [PubMed] [Google Scholar]
- 22.Heym B, Alzari PM, Honoré N, et al. Missense mutations in the catalase–peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol Microbiol. 1994;15:235–45. doi: 10.1111/j.1365-2958.1995.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 23.Ramaswamy SV, Reich R, Dou SJ, et al. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2003;47:1241–50. doi: 10.1128/AAC.47.4.1241-1250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marttila HJ, Soini H, Huovinen P, et al. katG mutations in isoniazid-resistant Mycobacterium tuberculosis isolates recovered from Finnish patients. Antimicrob Agents Chemother. 1996;40:2187–9. doi: 10.1128/aac.40.9.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Soolingen D, de Haas PE, van Doorn HR, et al. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in the Netherlands. J Infect Dis. 2000;182:1788–90. doi: 10.1086/317598. [DOI] [PubMed] [Google Scholar]
- 26.van Doorn HR, de Haas PE, Kremer K, et al. Public health impact of isoniazid-resistant Mycobacterium tuberculosis strains with a mutation at amino-acid position 315 of katG: a decade of experience in The Netherlands. Clin Microbiol Infect. 2006;12:769–75. doi: 10.1111/j.1469-0691.2006.01495.x. [DOI] [PubMed] [Google Scholar]
- 27.Kelley CL, Rouse DA, Morris SL. Analysis of ahpC gene mutations in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41:2057–8. doi: 10.1128/aac.41.9.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman DR, Mdluli K, Hickey MJ, et al. AhpC, oxidative stress and drug resistance in Mycobacterium tuberculosis. Biofactors. 1999;10:211–7. doi: 10.1002/biof.5520100219. [DOI] [PubMed] [Google Scholar]
- 29.Dhandayuthapani S, Zhang Y, Mudd MH, et al. Oxidative stress response and its role in sensitivity to isoniazid in mycobacteria: characterization and inducibility of ahpC by peroxides in Mycobacterium smegmatis and lack of expression in M. aurum and M. tuberculosis. J Bacteriol. 1996;178:3641–9. doi: 10.1128/jb.178.12.3641-3649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson TM, Collins DM. ahpC, a gene involved in isoniazid resistance of the Mycobacterium tuberculosis complex. Mol Microbiol. 1996;19:1025–34. doi: 10.1046/j.1365-2958.1996.449980.x. [DOI] [PubMed] [Google Scholar]
- 31.Sherman DR, Mdluli K, Hickey MJ, et al. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science. 1996;272:1641–3. doi: 10.1126/science.272.5268.1641. [DOI] [PubMed] [Google Scholar]
- 32.Sreevatsan S, Pan X, Zhang Y, et al. Analysis of the oxyR-ahpC region in isoniazid-resistant and -susceptible Mycobacterium tuberculosis complex organisms recovered from diseased humans and animals in diverse localities. Antimicrob Agents Chemother. 1997;41:600–6. doi: 10.1128/aac.41.3.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergval IL, Vijzelaar RN, Dalla Costa ER, et al. Development of multiplex assay for rapid characterization of Mycobacterium tuberculosis. J Clin Microbiol. 2008;46:689–99. doi: 10.1128/JCM.01821-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anthony RM, Schuitema AR, Bergval IL, et al. Acquisition of rifabutin resistance by a rifampicin resistant mutant of Mycobacterium tuberculosis involves an unusual spectrum of mutations and elevated frequency. Ann Clin Microbiol Antimicrob. 2005;4:9. doi: 10.1186/1476-0711-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergval IL, Klatser PR, Schuitema AR, et al. Specific mutations in the Mycobacterium tuberculosis rpoB gene are associated with increased dnaE2 expression. FEMS Microbiol Lett. 2007;275:338–43. doi: 10.1111/j.1574-6968.2007.00905.x. [DOI] [PubMed] [Google Scholar]
- 37.David HL. Probability distribution of drug-resistant mutants in unselected populations of Mycobacterium tuberculosis. Appl Microbiol. 1970;20:810–4. doi: 10.1128/am.20.5.810-814.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manca C, Paul S, Barry CE, III, et al. Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect Immun. 1999;67:74–9. doi: 10.1128/iai.67.1.74-79.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riska PF, Jacobs WR, Alland D. Molecular determinants of drug resistance in tuberculosis. Int J Tuberc Lung Dis. 2000;4:S4–S10. [PubMed] [Google Scholar]
- 40.Zhang Y, Young D. Strain variation in the katG region of Mycobacterium tuberculosis. Mol Microbiol. 1994;14:301–8. doi: 10.1111/j.1365-2958.1994.tb01291.x. [DOI] [PubMed] [Google Scholar]
- 41.Telenti A. Genetics of drug resistance in tuberculosis. Clin Chest Med. 1997;18:55–64. doi: 10.1016/s0272-5231(05)70355-5. [DOI] [PubMed] [Google Scholar]
- 42.Sajduda A, Brzostek A, Poplawska M, et al. Molecular characterization of rifampin- and isoniazid-resistant Mycobacterium tuberculosis strains isolated in Poland. J Clin Microbiol. 2004;42:2425–31. doi: 10.1128/JCM.42.6.2425-2431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 44.Slayden RA, Barry CE., III The genetics and biochemistry of isoniazid resistance in Mycobacterium tuberculosis. Microbes Infect. 2000;2:659–69. doi: 10.1016/s1286-4579(00)00359-2. [DOI] [PubMed] [Google Scholar]
- 45.Mdluli K, Swanson J, Fischer E, et al. Mechanisms involved in the intrinsic isoniazid resistance of Mycobacterium avium. Mol Microbiol. 1998;27:1223–33. doi: 10.1046/j.1365-2958.1998.00774.x. [DOI] [PubMed] [Google Scholar]
- 46.Heym B, Stavropoulos E, Honoré N, et al. Effects of overexpression of the alkyl hydroperoxide reductase AhpC on the virulence and isoniazid resistance of Mycobacterium tuberculosis. Infect Immun. 1997;65:1395–401. doi: 10.1128/iai.65.4.1395-1401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bifani P, Mathema B, Kurepina N, et al. The evolution of drug resistance in Mycobacterium tuberculosis: from a mono-rifampin-resistant cluster into increasingly multidrug-resistant variants in an HIV-seropositive population. J Infect Dis. 2008;198:90–4. doi: 10.1086/588822. [DOI] [PubMed] [Google Scholar]
- 48.Jenkins C, Claxton AP, Shorten RJ, et al. Rifampicin resistance in tuberculosis outbreak, London, England. Emerg Infect Dis. 2005;11:931–4. doi: 10.3201/eid1106.041262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lipin MY, Stepanshina VN, Shemyakin IG, et al. Association of specific mutations in katG, rpoB, rpsL and rrs genes with spoligotypes of multidrug-resistant Mycobacterium tuberculosis isolates in Russia. Clin Microbiol Infect. 2007;13:620–6. doi: 10.1111/j.1469-0691.2007.01711.x. [DOI] [PubMed] [Google Scholar]
- 50.Hillemann D, Kubica T, Rüsch-Gerdes S, et al. Disequilibrium in distribution of resistance mutations among Mycobacterium tuberculosis Beijing and non-Beijing strains isolated from patients in Germany. Antimicrob Agents Chemother. 2005;49:1229–31. doi: 10.1128/AAC.49.3.1229-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rinder H, Dobner P, Feldmann K, et al. Disequilibria in the distribution of rpoB alleles in rifampicin-resistant M. tuberculosis isolates from Germany and Sierra Leone. Microb Drug Resist. 1997;3:195–7. doi: 10.1089/mdr.1997.3.195. [DOI] [PubMed] [Google Scholar]
- 52.O'Sullivan DM, McHugh TD, Gillespie SH. The effect of oxidative stress on the mutation rate of Mycobacterium tuberculosis with impaired catalase/peroxidase function. J Antimicrob Chemother. 2008;62:709–12. doi: 10.1093/jac/dkn259. [DOI] [PubMed] [Google Scholar]
- 53.Chakravorty S, Aladegbami B, Motiwala AS, et al. Rifampin resistance, Beijing-W clade-single nucleotide polymorphism cluster group 2 phylogeny, and the Rv2629 191-C allele in Mycobacterium tuberculosis strains. J Clin Microbiol. 2008;46:2555–60. doi: 10.1128/JCM.00666-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manjunatha UH, Boshoff H, Dowd CS, et al. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:431–6. doi: 10.1073/pnas.0508392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.