Abstract
The quadruplex binding affinities and selectivities of two large π-surface PtII phenanthroimidazole complexes, as well as a smaller π-surface platinum bipyridine complex and a larger RuII complex, were evaluated by electrospray ionization mass spectrometry. Circular dichroism (CD) spectroscopy was used to determine the structures of various quadruplexes and to study the thermal denaturation of the quadruplexes in the absence and presence of the metal complexes. In addition, chemical probe reactions with glyoxal were used to monitor the changes in the quadruplex conformation because of association with the complexes. The platinum phenanthroimidazole complexes show increased affinity for several of the quadruplexes with elongated loops between guanine repeats. Quadruplexes with shorter loops exhibited insubstantial binding to the transition metal complexes. Similarly binding to duplex and single strand oligonucleotides was low overall. Although the ruthenium-based metal complex showed somewhat enhanced quadruplex binding, the PtII complexes had higher quadruplex affinities and selectivities that are attributed to their square planar geometries. The chemical probe reactions using glyoxal indicated increased reactivity when the platinum phenanthroimidazole complexes were bound to the quadruplexes, thus suggesting a conformational change that alters guanine accessibility.
Keywords: mass spectrometry, electrospray ionization, quadruplex, platinum complex
INTRODUCTION
Throughout the development of platinum-centered complexes as DNA-interactive agents, the primary focus has traditionally been on covalent attachment rather than noncovalent binding with one landmark example being the anti-cancer drug cisplatin.1 Other transition metal complexes, such as those based around RuII, have more often been designed to interact noncovalently with DNA, especially via intercalation.2–7 However, recent work has focused on creating noncovalent DNA interactive agents with PtII centers because of the relatively easy synthetic pathways and the different coordination geometry of platinum compared with ruthenium which extends the array of possible metal-ligand architectures.8–10
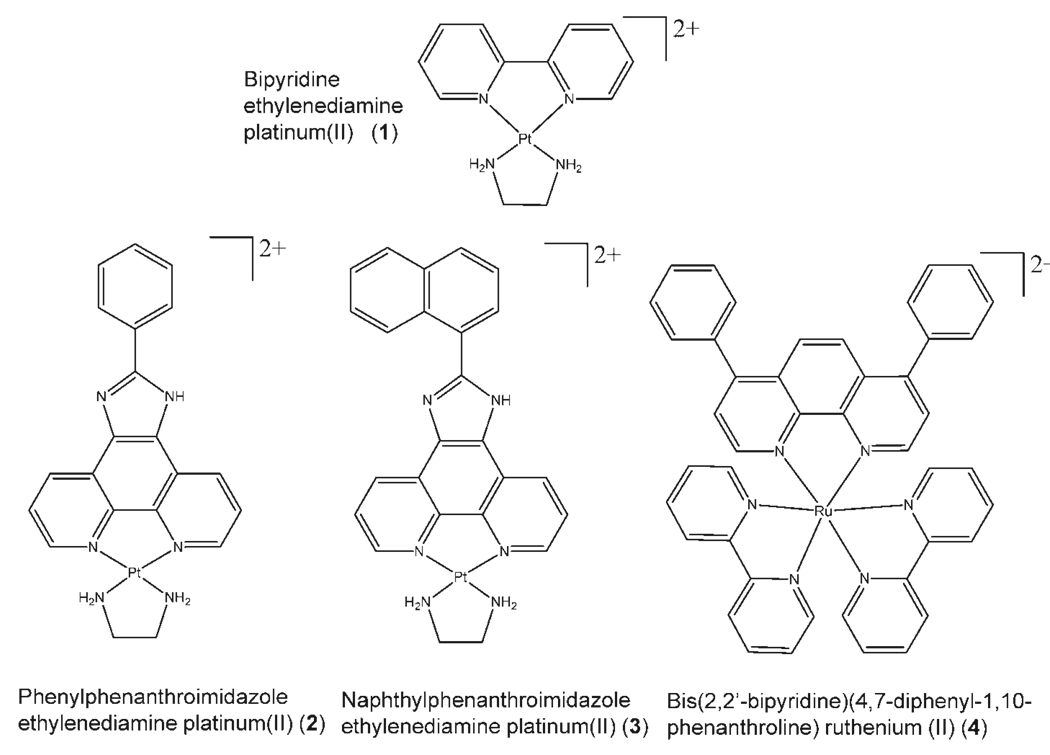
The complexes of interest in the present work, phenylphenanthroimidazole ethylenediamine platinum(II) (2) and naphthylphenanthroimidazole ethylenediamine platinum(II) (3) (see Scheme 1), were developed specifically to interact with quadruplex structures. Quadruplexes are a target of considerable biological interest because of their role in regulating the binding of telomerase.11–16 Telomerase lengthens telomeres, the G-rich sequences at the 3′ ends of DNA that allow replication or transcription to continue along the entire length of the strand. When the telomere arranges into a quadruplex structure rather than remaining as an unfolded single strand, telomerase can neither bind nor lengthen the telomere.15,17,18 This prevents cells, more specifically cancer cells, from becoming immortal.
SCHEME 1.
Structures of metal complexes.
To increase quadruplex specificity, complexes 2 and 3 were designed with an extended aromatic π-surface area.19 Because PtII complexes form a square planar geometry rather than an octahedral geometry, such as that adopted by ruthenium compounds, the π-surface of these transition metal complexes is able to interact more effectively with the planar surface of the G-quadruplex. In comparison to other platinum-based complexes, these complexes have extended π-conjugated surfaces designed for optimal interaction with the G-quadruplex surface through size and electronic complementarity.
In the past decade, electrospray ionization mass spectrometry (ESI-MS) has become a versatile tool for the study of noncovalent interactions. In addition to examination of the interactions between proteins and small molecules or between proteins,20 mass spectrometry has been very useful for studying the noncovalent binding between oligonucleotides,21–30 between DNA and proteins,31–34 between DNA and small molecules,35–42 and between quadruplexes and ligands.22,28,38,43–53 Recently, the Ralph group has used ESI-MS and circular dichroism (CD) spectroscopy to probe the binding of ruthenium and platinum complexes to duplex and quadruplex oligonucleotides.7,51,54 It was determined that the small platinum complexes studied showed low binding affinities to quadruplexes in comparison to duplexes.51 This result is consistent with the prediction that large π-surface areas are necessary to enhance interactions with the G-quartet.
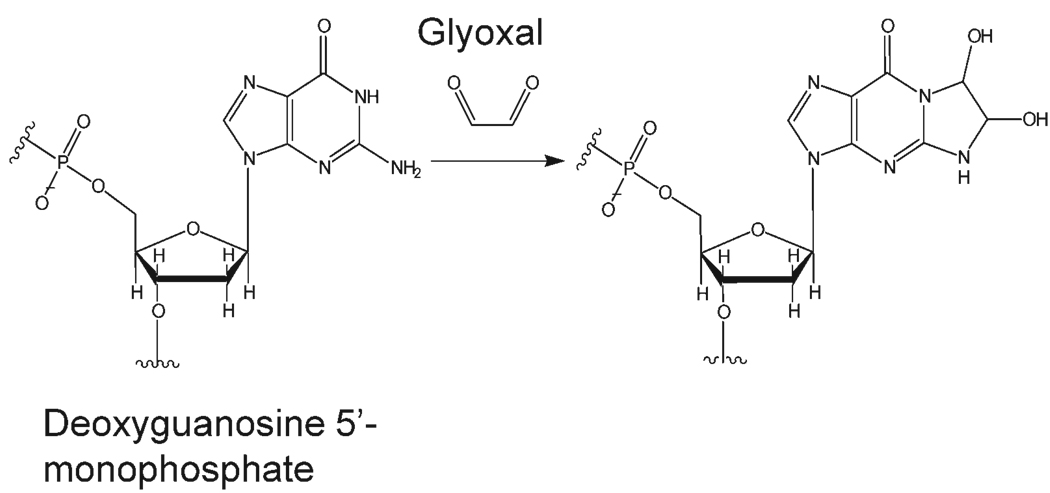
Recently, our group has begun using chemical probes in combination with ESI-MS to study the impact of small molecule binding on oligonucleotide structure.55 Unlike traditional chemical probe techniques in which the outcomes of the reactions are evaluated using gel electrophoresis, our strategy uses mass spectrometry to monitor the extent of reaction with the chemical probe. In addition, we have applied tandem mass spectrometry, including both collisionally induced dissociation (CID) and infrared multiphoton photodissociation (IRMPD), to determine the identity and location of the chemical probe reaction sites. Although our first efforts used potassium permanganate,55 which oxidizes thymine bases, we are extending our mass spectrometric methods to other DNA-reactive chemical probes, specifically glyoxal in the present study. Glyoxal has been used for traditional chemical probe analysis of both duplexes and quadruplexes.56–58 Glyoxal is known to react with guanine bases at the N1 and N2 atoms,58 as illustrated in Scheme 2. As these nitrogen sites are involved in hydrogen bonding between guanine bases in G-quadruplexes, they should exhibit reduced reactivity with glyoxal. Moreover, any change in the secondary structure of a guanine-containing oligonucleotide upon binding of a ligand could change the accessibility of the active sites for reaction with glyoxal.
SCHEME 2.
Reaction between glyoxal and deoxyguanidine.
In this work, the relative DNA affinities of two established duplex intercalators, one a small PtII complex, 1, and the other a large RuII complex, 4, were compared with two novel complexes based on PtII centers, 2 and 3 (Scheme 1). The latter two complexes have extended structures intended to facilitate binding to quadruplex DNA. Although the RuII complex is similar in size to the novel PtII complexes evaluated in the present study, the former has an octahedral geometry rather than square planar. The square planar geometry should increase the π-surface area available for interaction with the quadruplex, thus enhancing binding to the quadruplex. Circular dichroism was used to determine the structure of the quadruplexes and to monitor changes in the melting points upon binding of the metal complexes. ESI-MS was used to evaluate the relative binding affinities of the complexes to various oligonucleotide structures. In addition, reactions with glyoxal were undertaken to assess the variation in reactivities of the guanine bases induced by interaction of the complexes with quadruplexes. We present results that show that the novel complexes 2 and 3 have enhanced quadruplex selectivity over the smaller PtII and RuII complexes.
EXPERIMENTAL
Materials
Oligonucleotides were obtained from IDT DNA, (Coralville, IA) and used without further purification (see Table I). Quadruplexes and duplexes were annealed in 150 mM ammonium acetate by placing the strands in a hot water bath and cooling to room temperature overnight (approximately 12 h). The DNA was annealed at concentrations to produce complexes in the range of 300–500 µM. Thus, DS, Q2, Q3, and Q4 were all annealed at 500 µM whereas Q1 was annealed at 1.2 mM strand concentration. The annealed solutions were stored for at least 24 h in the freezer before use. Although the conformations of the quadruplexes obtained for these annealing conditions are well mapped based on CD measurements, these are not necessarily the same conformations as obtained in vivo. As the SS sequence has the potential to form homodimers or hairpins, nondenaturing gel electrophoresis was used to determine that the majority of the sample is in the single strand conformer under experimental conditions. The PtII complexes were synthesized as discussed previously.19 All three complexes were dissolved to 1 mM in DMSO and stored in the refrigerator when not in use. Glyoxal was purchased from Sigma-Aldrich (St. Louis, MO).
Table I.
DNA Sequences Used in this Work
| DNA | Sequence | Type of DNA Structurea | Number of Tetrads (for Quadruplexes) |
|---|---|---|---|
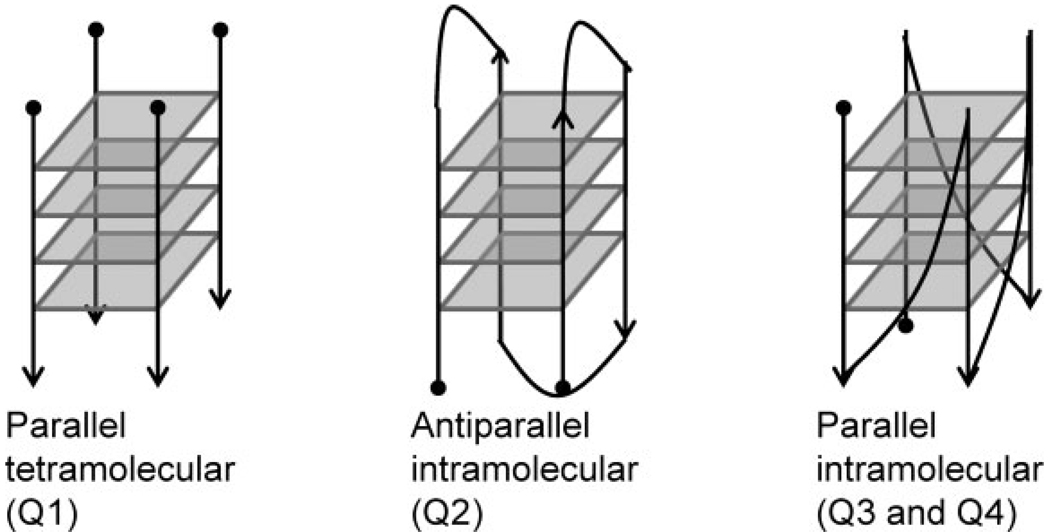
| Q1 | [d(T4G4T4)]4 | Parallel 4-stranded quadruplex | 4 |
| Q2 | dT4G4T4G4T4G4T4G4T4 | Antiparallel intramolecular quadruplex | 4 |
| Q3 | dT2G4T2G4T2G4T2G4 | Parallel intramolecular quadruplex | 4 |
| Q4 | dT2AG3T2AG3T2AG3T2AG3 | Parallel intramolecular quadruplex | 3 |
| DS | dGCGCGGAACCGCGC/dGCGCGGTTCCGCGC | Duplex | – |
| SS | dGCGCGGAACCGCGC | Single strand | – |
Assuming quadruplexes and duplex annealed in ammonium acetate.
The synthesis of bis(2,2′-bipyridine)(4,7-diphenyl-1,10-phenanthroline)ruthenium(II) was performed by first synthesizing bis(2,2′-bipyridine)ruthenium(II) chloride as described previously.59 One equivalent of 4,7-diphenyl-1,10-phenanthroline was added to this precursor in 70% ethanol, and the mixture was refluxed for 30 min.60 TLC was performed to confirm that only one product was present, and the product was air dried. The identity of the product was confirmed using MS. As with the PtII complexes, the product (4) was dissolved in DMSO and stored in the refrigerator.
Methods
ESI-MS
Three Thermo (San Jose, CA) ion trap mass spectrometers, an LTQ, an LCQ-Duo, and an LCQ Deca modified for IRMPD,61 were used to perform ESI-MS. For IRMPD, a 50-W Synrad CO2 laser (Mukilteo, WA) was used to irradiate the samples through a hole in the ring electrode. CID experiments were performed on the LCQ Deca instrument using a collision voltage of 1.0 V and an activation time of 30 ms. IRMPD experiments employed 99% laser power for 1.3 ms. The ligands and oligonucleotides were combined in a 1:1 ratio, unless otherwise indicated, just before analysis. To assist with desolvation and reduction of salt adducts, 20% methanol and 50 mM ammonium acetate was added to the samples immediately before analysis. The DNA concentration for all ESI-MS samples was 10 µM. ESI-MS was performed in negative mode using a spray voltage of 3.5 kV with a heated capillary temperature of 90°C. The tube lens voltage was varied between −150 and −250 V depending on the particular sample. The percentage of bound DNA was calculated from the ESI mass spectra based on:
in which RAn:m indicates the relative abundances of the various DNA:metal complex products and RADNA corresponds to the relative abundance of the unbound DNA in the ESI mass spectra.38,39
CD
A Jasco J-815 circular dichroism system (Easton, MD) was used to collect circular dichroism spectra. For melting point determinations, the CD signal, in mdeg, at 293 nm was monitored as the temperature was increased from 25 to 90°C at a rate of 0.2°C/min. The melting curve was imported into Origin 7.0 (OriginLab, Northampton, MA) for analysis. The melting point, T1/2, was determined by fitting the data with a sigmoidal curve and determining the melting point from the inflection point. Oligonucleotide concentrations for CD were 5 µM.
Chemical Probes
For the glyoxal reactions, 20 µM oligonucleotide complex in water was incubated with 0.5 µL glyoxal for 30 min at 37°C. For the experiments involving the DNA with the metal complexes, the metal complexes were added 10 min prior to glyoxal at a 2:1 complex to DNA ratio. The samples were cleaned up using Pepclean C18 spin columns (Pierce, Rockford, IL) to remove excess glyoxal before analysis. These samples were analyzed by ESI-MS under similar conditions to those described above. Similar calculations to those described above to determine the percentage of bound DNA from ESI mass spectra were used to determine the extent of reaction with glyoxal, but RAn:m indicates the relative abundances of the glyoxal adducts rather than the relative abundances of DNA: metal complexes.
RESULTS
A variety of experiments in both the gas phase and solution were undertaken to map the nature of the DNA/complex interactions. To determine the degree of structure-specific binding of the metal complexes, a variety of DNA structures were studied. Although it was expected that the metal complexes would show preference for quadruplex structures, quadruplexes have a range of orientations and sequences. Four quadruplexes were used in this study: Q1 (a tetramolecular quadruplex), Q2 (an antiparallel intramolecular quadruplex), Q3 (a parallel intramolecular quadruplex), and Q4 (an intramolecular quadruplex with a sequence consistent with the human telomeric repeat) (see representative structures in Scheme 3). One guanine-rich duplex, DS, was studied as well, in addition to a guanine-rich single strand, SS, as representative models of other secondary structures. ESI-MS was used to determine the percentage of bound DNA for the various samples to compare the relative binding affinities of the different metal complexes and, when used in conjunction with glyoxal reactions, to reflect changes in the secondary structure of the oligonucleotides upon binding of the metal complexes. Although ESI-MS results reveal insight into DNA binding (based on relative abundances and stoichiometries), CD results give a broader indication of the overall structure of the quadruplex.
SCHEME 3.
Cartoons of quadruplex structures used in this work.
CD Spectroscopy of Quadruplexes
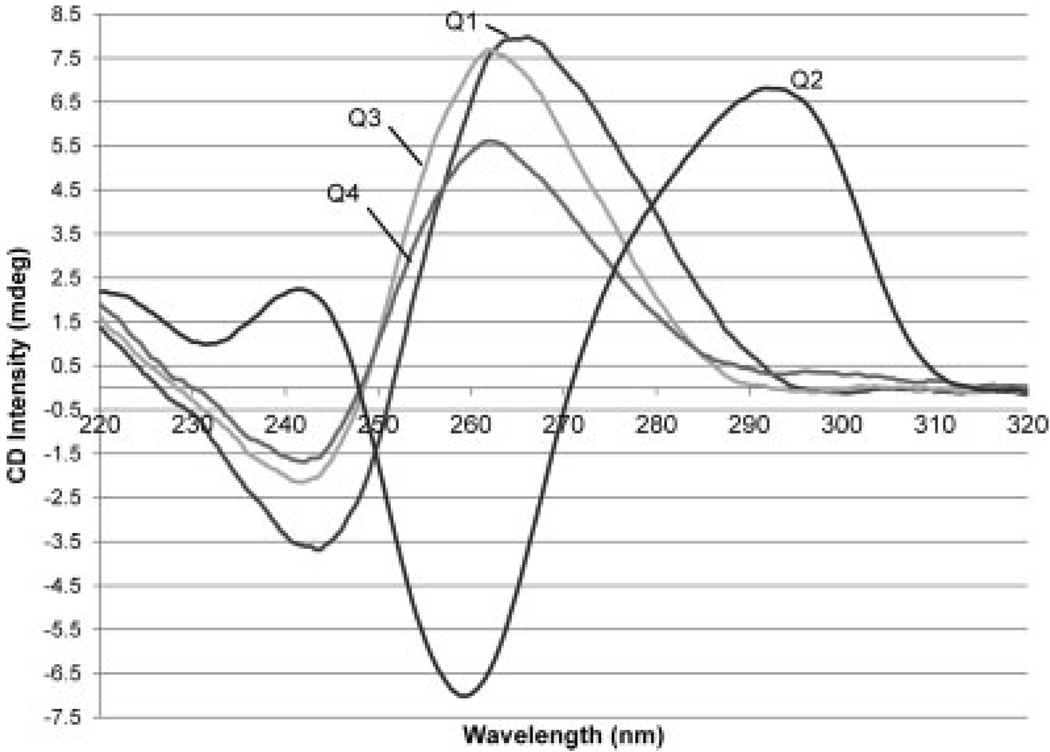
Quadruplexes have specific CD signatures depending on the overall structure of the quadruplex. For instance, an antiparallel intramolecular quadruplex has a peak at 295 nm and a trough at 260 nm whereas a parallel quadruplex has a peak at 265 nm and a trough at 240 nm.62,63 These features can be used to determine the structures of quadruplexes based on the CD spectra. As expected, Q1 exhibits a peak at 266 nm and a trough at 243 nm (see Figure 1) which is consistent with a parallel quadruplex. Both Q3 and Q4 yielded similar spectra to the tetramolecular quadruplex (Q1) indicating parallel conformations with positive peaks at 262 nm and troughs at 242 nm. In the presence of sodium or potassium cations rather than ammonium cations (as used in the present study), the constituent oligonucleotides often result in the formation of antiparallel or mixtures of antiparallel and parallel quadruplexes.64,65 In fact, when these sequences were analyzed without annealing, but were mixed with ammonium acetate added immediately before analysis, signature peaks are present for both parallel and antiparallel quadruplexes (see Supp. Info. Figure S-1). It is only upon annealing these strands in ammonium acetate that the parallel conformation dominates. The parallel conformation of similar quadruplexes, especially human telomeric sequence (Q4), has been observed with other annealing cations66,67 and with ammonium.53 For parallel intramolecular quadruplexes, the loops between guanine repeats run diagonally across the sides of the quadruplex rather than over the top as is seen with antiparallel conformations.68 Q2 provided a very different CD spectrum with positive peaks at 292 and 242 nm and a trough at 259 nm. This spectrum is characteristic of an antiparallel quadruplex structure. The longer thymine loops between guanine repeats allows the loops to form over the top of the quadruplex rather than across the sides as occurs with the other two intramolecular quadruplexes. Because complexes 2 and 3 are designed to endstack with quadruplexes, both antiparallel and parallel intramolecular quadruplexes were included in our study to allow evaluation of the impact of the loop structure on the relative DNA binding affinities of the PtII complexes.
FIGURE 1.
CD spectra of quadruplexes after annealing in 150-mM ammonium acetate.
Relative Binding Affinities and Selectivities of Metal Complexes by ESI-MS
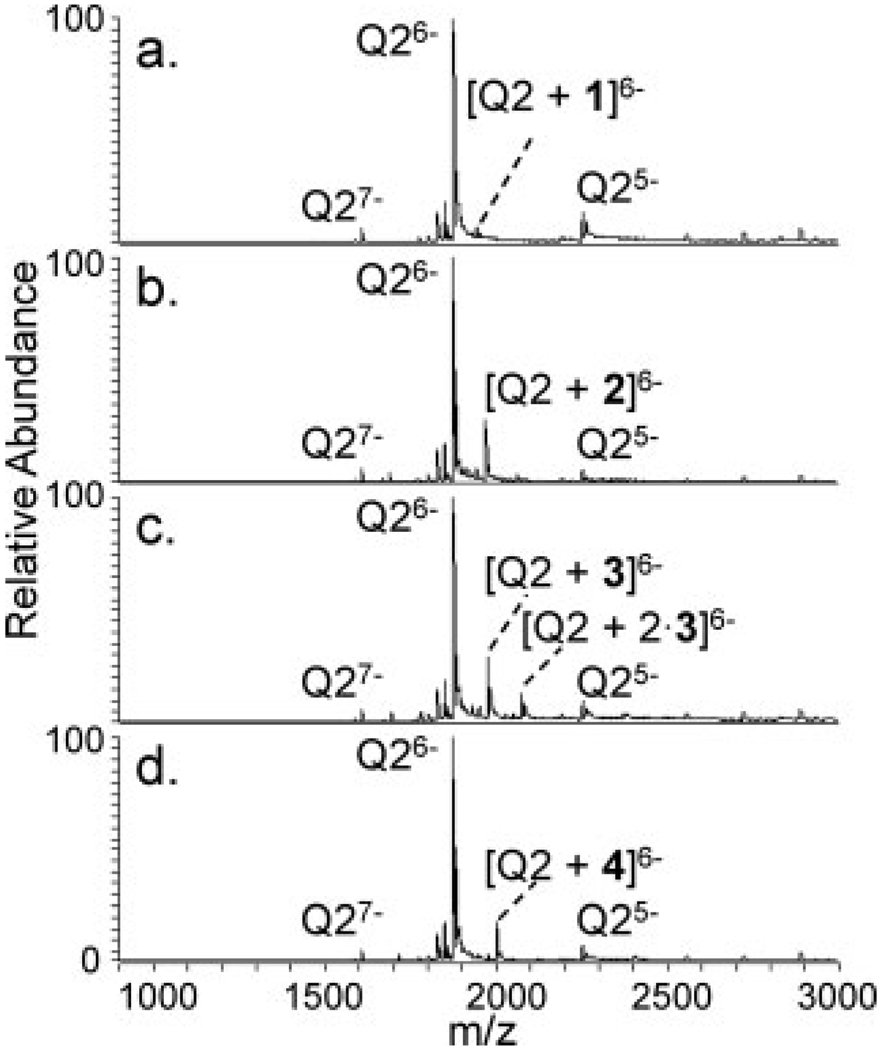
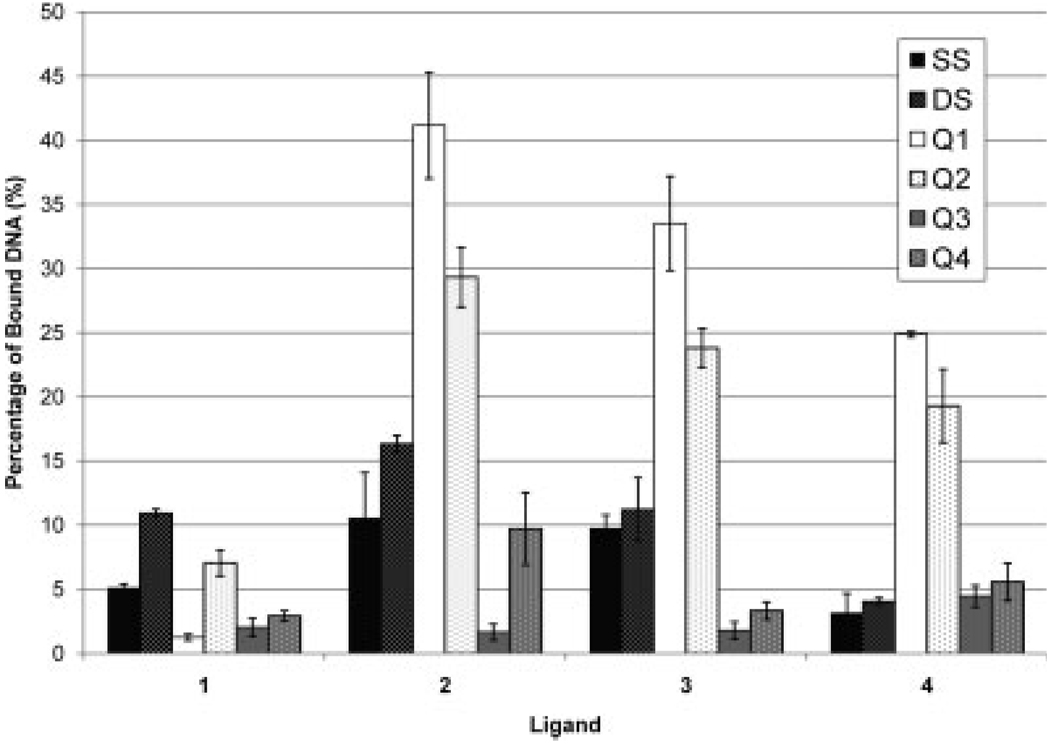
To determine the quadruplex selectivities of the PtII complexes, solutions containing equimolar concentrations of one of the four complexes and one of the four quadruplexes were analyzed by ESI-MS. Examples of the resulting ESI mass spectra are shown in Figure 2, and the results are summarized in bar graph form in Figure 3 (see Supp. Info. Figure S-2 for spectra of the metal complexes with the other quadruplexes). Based on the ESI-MS results and the calculated percentage bound values, Q2 exhibited very low affinity for the small complex, 1. The percentage of bound Q2 increased by over an order of magnitude for the two PtII complexes, 2 and 3, with 2 more favored than 3. The RuII complex, 4, also showed a high degree of binding to Q2. Interestingly, a comparison of the spectra obtained for Q2 with 2 and 3, shown in Figure 2b and 2c respectively, shows that a maximum of two molecules of 3 are bound to the quadruplex whereas only one molecule of 2 was bound. Although it is thought that both PtII complexes bind through the same mode of interaction, the ability of the larger complex to bind in a 2:1 stoichiometry whereas the slightly smaller complex only binds in a 1:1 stoichiometry is intriguing and yet unexplained. Both 1:1 and 2:1 stoichiometries are possible for an end-stacking binding mode as there are two ends at which the complexes can bind. Moreover, the small percentage of bound DNA observed for 1 is consistent with the earlier results of the Ralph group that showed low binding of small platinum-centered complexes to quadruplexes.51 Similar trends in binding affinity were obtained for Q1, which was also used in the previous study reported for these complexes although those results, obtained by UV-vis and equilibrium dialysis, showed greater binding for 3 than for 2.19 The abundances of complexes between Q1 and 2 or 3 were far greater than those of Q1 with 1, with binding to 2 slightly favored.
FIGURE 2.
ESI mass spectra of quadruplex Q2 with each metal complex. The quadruplex:metal complex concentration ratio was 1:1. The ion naming scheme involves denoting Q2 bound to one moiety of 3, for example, as [Q2 +3] while Q2 bound to two moieties of 3 is labeled [Q2 + 2 ⋅ 3].
FIGURE 3.
Percentage of bound DNA for each metal complex. Each bar represents the average of three samples. Refer to Scheme 1 for the structures of compounds 1–4.
Surprisingly, quite different results were obtained for the two parallel intramolecular quadruplexes (Q3 and Q4). The percentage of bound DNA for both Q3 and Q4 was found to be 10% or less for each of the four complexes. These results suggest that the geometry of the loop regions with these quadruplexes defines the potential binding interactions with the complexes. The free thymine bases on the ends of the intermolecular quadruplex (Q1) and the arching loops of the antiparallel intramolecular quadruplex (Q2), both facilitate favorable binding between the complexes and the quadruplexes. However, the shorter loops that result in a parallel conformation for the intramolecular quadruplexes (Q3 and Q4) restrict binding with the complexes and result in low abundances for the quadruplex/metal complex species. These loops are suggested to run diagonally across the sides of the quadruplex rather than over the top68 which may diminish complex binding to the quadruplex. This structure-based suppression of metal complex binding is further supported by the slight increase in binding of 2 to Q4 over Q3 because Q4 has one more nucleotide between guanine repeats which may result in less strained outer tetrads as the extra nucleotide provides greater length to transverse the quadruplex.
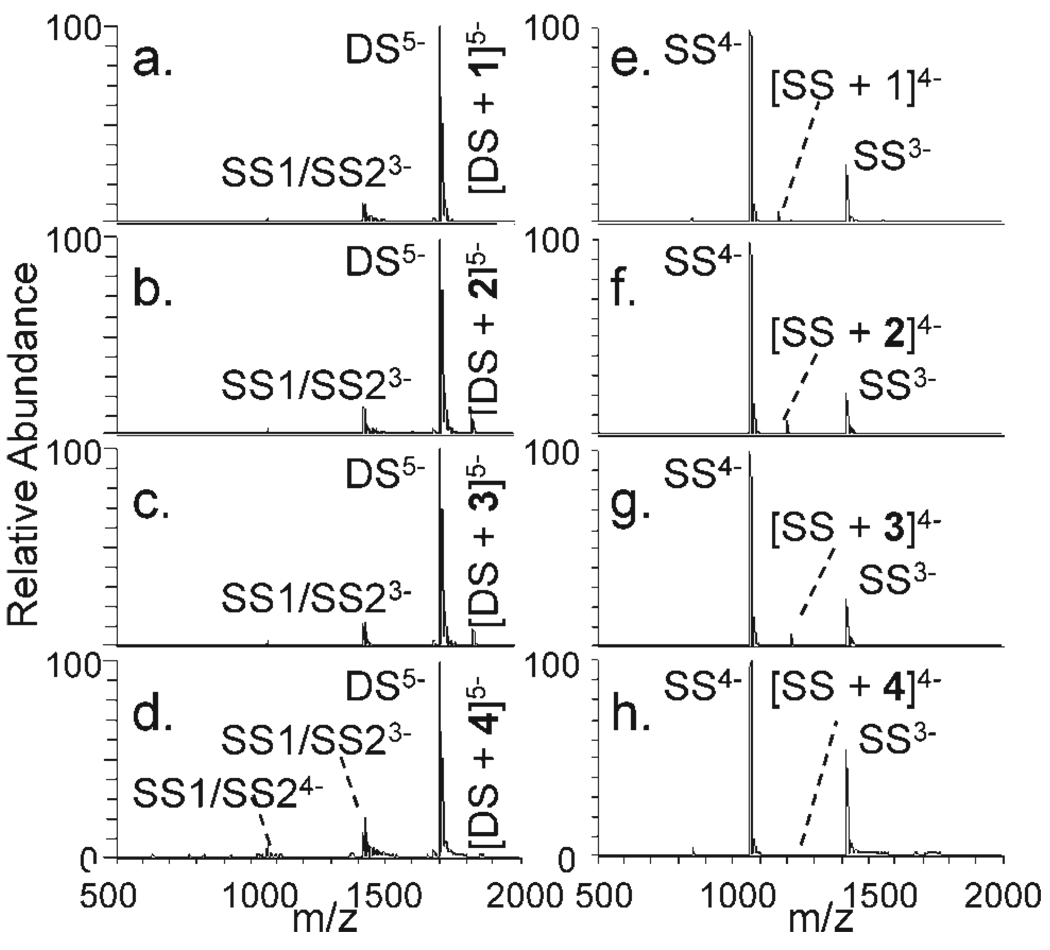
To assess the degree of complex selectivity for quadruplexes versus other DNA structures, ESI mass spectra were acquired for solutions containing an equimolar mixture of a guanine-rich duplex (DS) or guanine-rich single strand (SS) and each of the metal complexes. The resulting mass spectra are shown in Figure 4, and the results are summarized in Figure 3 to allow easier visual comparison to the quadruplex results. There was minimal binding to the single strand for all complexes and slightly greater binding to the duplex. Complex 1 resulted in a higher percentage of bound DNA for the duplex compared with the four quadruplexes or single strand, thus supporting its greater affinity for duplex DNA. The other three complexes, 2, 3, and 4, exhibit selectivity for interaction with quadruplexes over the duplex or single strand DNA.
FIGURE 4.
ESI mass spectra of DS and SS with each metal complex. The concentration ratio of metal complex to DNA is 1:1.
Additional experiments were undertaken in which the concentration ratio of complex to DNA was 2:1 rather than 1:1 to allow a broader assessment of the potential range of stoichiometries of the complex/DNA products. Solutions with even higher concentration ratios were not studied in detail, because of the presence of salt contaminants in the complexes that resulted in overly cluttered, uninterpretable mass spectra. Mass spectra obtained for the 2:1 solutions are shown in Supp. Info. Figure S-3, and a chart of the percentage of bound DNA for these samples is provided in Supp. Info. Figure S-4. With SS and DS, in addition to an overall increase in the percentage of bound DNA, 2 and 3 showed low abundances of 2:1 complexes not seen in the spectra acquired with the 1:1 concentration ratio of complex to DNA. Binding between the metal complexes and Q3 was similar to that observed for the 1:1 solutions. The spectrum obtained for 2 with Q4 showed a marked increase in the percentage of bound DNA and low abundance 2:1 products. The greatest changes in product formation were seen for the Q1 and Q2 quadruplexes. The percentage of bound DNA increased and additional higher order products were observed for both of these quadruplexes with 2 and 3. 2 formed 2:1 products with both Q1 and Q2, whereas 3:1 products were observed with 3 and both Q1 and Q2. The 3:1 products containing 3 with Q1 and Q2 are surprising, because an end-stacking binding mode would only account for association of two molecules of 3 with the quaduplexes. It is possible that the additional molecule of 3 is interacting along the side of the quadruplex or two 3 moieties might stack on one another, but there is no specific evidence supporting either of these scenarios. When analyzed alone by ESI-MS, 3 did not form aggregates, but the planarity of the structure might permit stacking. The other metal complexes, 1 and 4, showed a minimal increase in the percentage of bound DNA for either Q1 or Q2.
Reactions of Glyoxal with DNA and DNA/Metal Complexes
To monitor the perturbation of the DNA structures upon noncovalent binding of the metal complexes, chemical probe reactions were undertaken in conjunction with mass spectrometric analysis. As discussed in the experimental section, solutions containing DNA or DNA with one of the metal complexes were incubated with glyoxal, followed by clean-up with C18 spin columns. Glyoxal reacts at guanine bases, thus serving as a chemical probe of accessible guanines.58 To confirm that thymine bases are unreactive with glyoxal, a 6-mer sequence containing only thymines was reacted under the same conditions as the other strands. The results (not shown) indicate that no reaction occurs. Initial experiments involving the duplexes or quadruplexes in the absence of metal complexes indicated the loss of secondary structure of the DNA after the glyoxal/clean-up procedure based on the detection of only single strand oligonucleotides in the mass spectra. To determine whether this disruption of secondary structure was due to heating during the reactions or to the C18 clean-up procedure, samples of DS and Q1 were analyzed by ESI-MS before heating, after heating at 37°C, and after C18 sample clean up, in each case without the presence of glyoxal. The results (see Supp. Info. Figure S-5) confirmed survival of the duplexes and quadruplexes both before and after heating. However, after the C18 clean-up procedure, intact quadruplexes or duplexes were no longer detected by ESI-MS. This series of results indicates that the loss of secondary structure occurs during the C18 clean-up and is not a confounding result of the glyoxal reactions. This means that ESI-MS can be used to monitor the impact of the metal complexes on the DNA reactivity with glyoxal based on detection of glyoxal adducts of the constituent DNA sequences. Typical ESI mass spectra are shown in Figure 5.
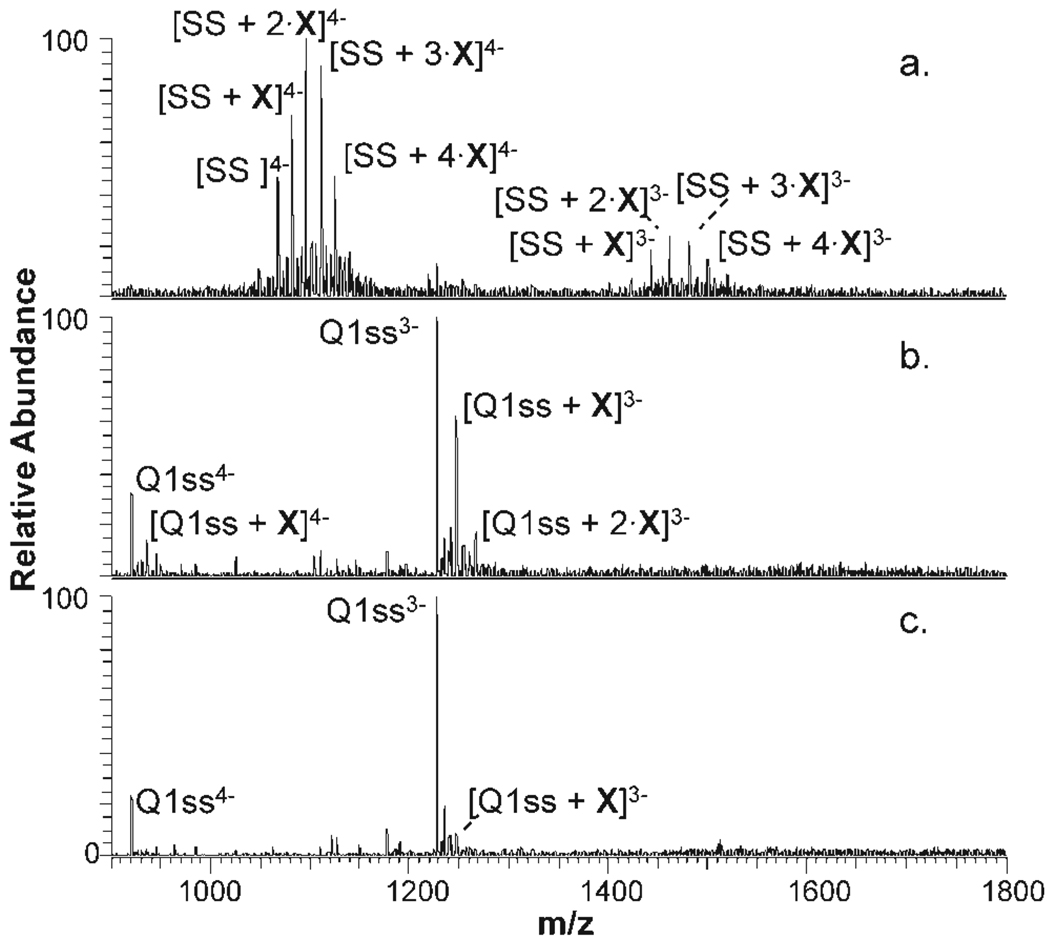
FIGURE 5.
ESI mass spectra of DNA after reaction with glyoxal. Spectra were collected for (a) SS, (b) Q1ss, and (c) and Q1. Q1ss corresponds to the single strand form of Q1 that was not annealed. As discussed in the text, the quadruplex Q1 disassembles during C18 clean-up procedure resulting in detection of the single strand Q1ss. Glyoxal adductions are noted by X.
As expected, SS, with its less rigid secondary structure, proved the most reactive with glyoxal (Figure 5a). The guanine bases are not blocked by intramolecular hydrogen bonds or bonds with other guanines or cytosines, thus leaving them more accessible to reaction with glyoxal. Up to four glyoxal moieties were observed bound to the single strand. The reactivity of DS with glyoxal is approximately two times lower than that of SS (see Supp. Info. Figure S-6), and a maximum of two glyoxal moieties are bound to the strands. Hydrogen bonding between the guanine bases and complementary cytosine bases apparently suppresses the reactions with glyoxal.
Q1 was chosen for the glyoxal studies because, as an intermolecular quadruplex, the reactions of its constituent sequence (Q1ss as a single strand) can be studied in parallel to the reactions of the quadruplex. Comparisons of the glyoxal reactivity of Q1ss and Q1 permit the differentiation of the impact of intrastrand hydrogen bonding and the influence of the bound metal complexes. CD spectra of the intramolecular quadruplexes (Q2, Q3, and Q4) indicated that even without prior annealing these strands fold into quadruplex structures, thus preventing ready comparative studies of the glyoxal reactions of these quadruplexes and their single strand counterparts. The ESI mass spectra obtained for the glyoxal reaction solutions containing Q1 or Q1ss are shown in Figure 5b and 5c. Although Q1ss was approximately half as reactive as SS, the latter which contains six guanine bases compared with four guanines in Q1ss, the substantial reactivity of Q1ss with glyoxal confirmed the significant accessibility of its guanines. The reactivity of quadruplex Q1 with glyoxal decreased by a factor of four relative to that of Q1ss, presumably because the Hoogsteen hydrogen bonds between the guanine bases of Q1 block access to and/or suppress reactions of glyoxal.
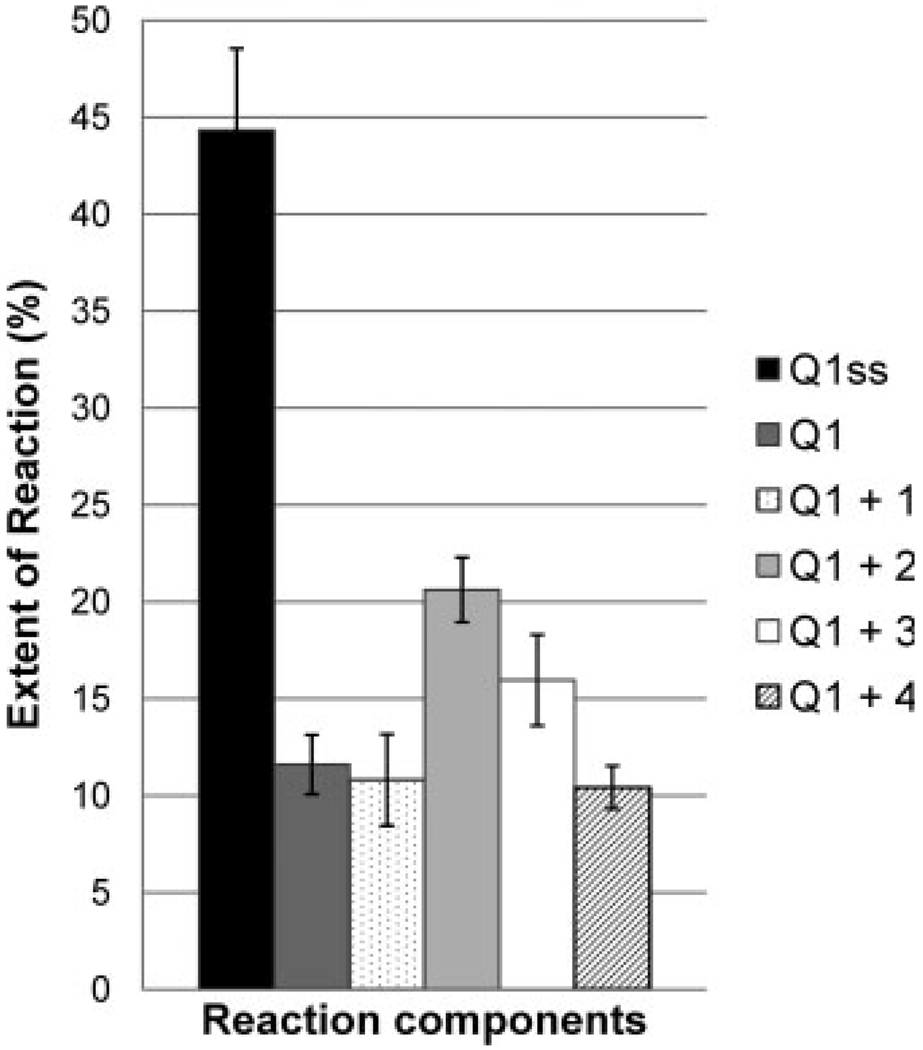
Figure 6 summarizes the results of similar glyoxal reactions performed after Q1 was incubated with each of the metal complexes. The extent of reaction with Q1 is calculated from the ESI mass spectra in a similar manner to that used to quantify the percentage of bound DNA for solutions containing DNA and the metal complexes. In this case, the abundances of all of the glyoxal-adducted DNA ions were summed and divided by the summed abundances of the all DNA species (both unreacted DNA and the DNA/glyoxal adducts) and then converted into a percentage. As mentioned above, the C18 clean-up procedure used to process the glyoxal-containing samples disrupts the secondary structure of the Q1 quadruplexes, causing their conversion to single strand species. Thus, the results summarized in Figure 6 reflect the extent of reaction based on detection of single strand products even when the target DNA was annealed into a quadruplex form before incubation with the metal complexes and subsequent reactions with glyoxal. Interaction of the quadruplex Q1 with 1 caused minimal change in the reactivity of the quadruplex. However, interaction with 2 increased the extent of reaction with glyoxal by a factor of two. This result is an indication that 2 alters the structure of the quadruplex in such a way that increases the accessibility of some of the guanine bases. Similarly, 3 also increase the extent of reaction of the quadruplex with glyoxal, although to a lesser extent than 2. These data are consistent with the trend in relative binding affinities discussed earlier in which a higher percentage of bound DNA was found for 2 over 3. Comparable to 1, 4 did not cause a change in the extent of reactivity of the quadruplex with glyoxal compared with the free quadruplex. Despite the large size of the ligand, the RuII complex 4 itself does not appear to cause the same structural changes upon binding that are seen with the PtII complexes, 2 and 3, suggesting either a weaker interaction or different binding mode between 4 and Q1 than 2 or 3. The possibility of cross-reactivity of glyoxal with the metal complexes themselves, a process that would sequester the complexes and intervene with binding, was also considered. There was no evidence to support the occurrence of this cross-reaction.
FIGURE 6.
Summary of the extent of reaction with glyoxal for Q1ss, Q1, and Q1 in the presence of the metal complexes. The bars represent the average of three reactions.
The reactions of glyoxal with the double strand, DS, both in the absence and presence of the metal complexes, were also evaluated by ESI-MS. A chart comparing the relative reactivities of these samples can be found in Supp. Info. Figure S-6. Based on the abundances of the DNA/glyoxal adducts, the differences in the reactivity of glyoxal with the free duplex or with the duplex bound to the metal complexes were determined to be negligible. This finding suggests that the metal complexes do not cause a significant change in the structure of the duplex, either in terms of blocking reaction sites or by unwinding the duplex.
Tandem Mass Spectrometry of Glyoxalated Quadruplexes
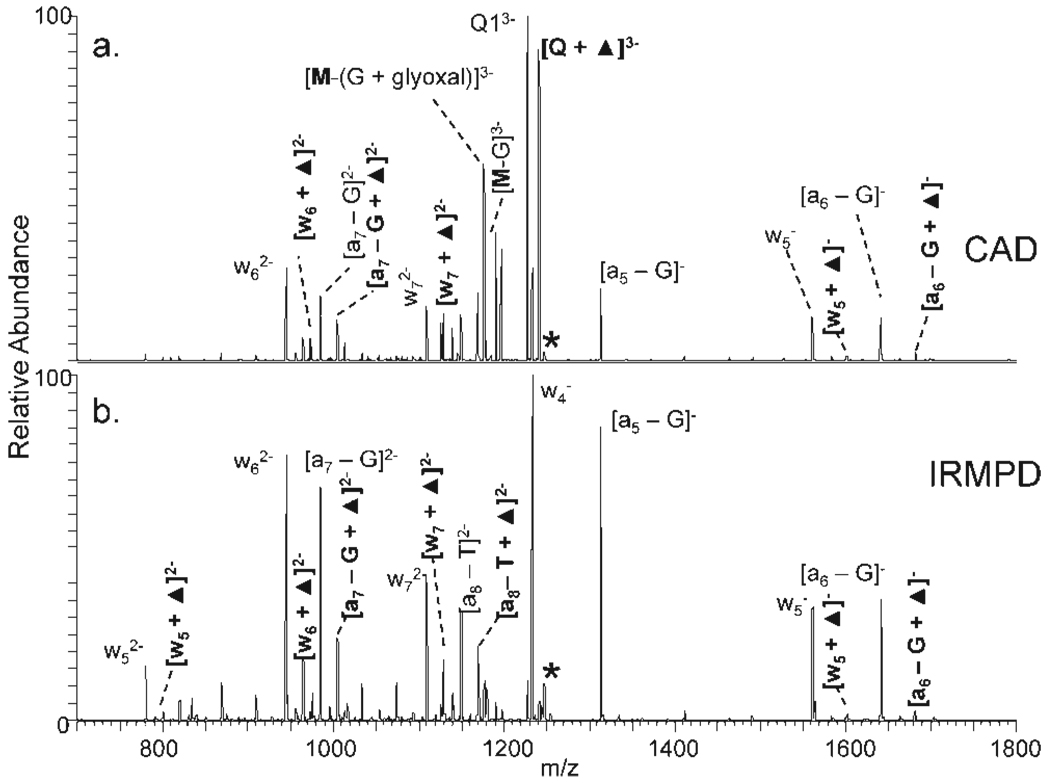
Tandem mass spectrometry via collision induced dissociation (CID) and infrared multiphoton dissociation (IRMPD) was used to examine the structures of the DNA/glyoxal adducts and determine the specificity of the glyoxal reaction sites. Examples of the resulting mass spectra are shown in Figure 7 for the Q1ss/glyoxal adducts in the 3-charge state. Upon CID, the most abundant product ion stemmed from loss of the glyoxal moiety (Figure 7a). Dehydration of the Q1ss/glyoxal adduct, and loss of either a guanine base (e.g., [M–G]3−) or a glyoxal-modified guanine base (e.g., [M–(G + glyoxal)]3−), were also dominant fragmentation pathways. These channels are not useful for identifying the specific location of the glyoxal reaction site. Upon IRMPD, there was more extensive cleavage of the DNA backbone, resulting in both (a and b) and w fragment ions that retained the dehydrated glyoxal modification (Figure 7b). The majority of these (a and b) and w ions stem from cleavages that occur adjacent to guanine bases. Product ions ranging from a5 to a8 and w5 to w7 were found to retain the dehydrated glyoxal moiety. This array of diagnostic sequence ions indicates that the glyoxal reaction occurs nonspecifically at different guanine sites rather than localizing at specific guanine bases. The IRMPD mass spectra acquired from the samples containing the quadruplex Q1 in the presence of each of the metal complexes were similar, indicating that the metal complexes do not appear to activate or suppress the glyoxal reaction at uniquely identifiable guanines.
FIGURE 7.
Dissociation of [Q1ss + glyoxal]3− by (a) CID at 1.0 V collision voltage and (b) IRMPD at 99% laser power for 1.3 ms. Ions that include the dehydrated glyoxal moiety are noted by ▲. The parent ions are indicated by an asterisk.
T1/2 of Q2 and Metal Complexes
Relative comparison of quadruplex melting points (T1/2 values) in the presence and absence of DNA-interactive ligands can be used to assess how the ligands affect the stabilities of the quadruplexes. An increase in the melting point of a quadruplex upon binding of a ligand suggests an increase in stability against thermally stress. The Q2 quadruplex was chosen for melting point analysis because it produced the most abundant complexes with 2 and 3 by the ESI-MS analysis. In this case, Q2 was analyzed in the presence and absence of the four metal complexes (see Supp. Info. Figure S-7). The melting and cooling curves of the quadruplex did not overlap, indicating the quadruplex was not at equilibrium (data not shown). Although this implies that the melting points are not the true equilibrium melting points and should not be used for thermodynamic calculations, the results are still useful for these simple relative comparisons of thermal stability.69 In fact, it is common for quadruplexes in general70 and this sequence in particular71 to have slow association/dissociation kinetics that result in hysteresis between the melting and annealing curves even at very low heating and cooling rates. Absolute quantitative values are not critical for this work, and the values given for T1/2 are useful for relative comparisons.
Without the presence of a metal complex, the Q2 quadruplex yielded a T1/2 value of 50.3°C (Table II). The addition of 1 caused an insignificant increase in the T1/2 (50.5°C). Moreover, the addition of 4 only increased the T1/2 of Q2 by approximately 4°C (to 54.1°C). In comparison, the addition of 2 increased the melting temperature by 18°C, thus indicating a greater degree of stability against thermal denaturation. The addition of 3 caused the greatest increase in the T1/2 to 72°C. As was shown above, in comparison with 2, 3 exhibited slightly lower binding affinities to quadruplexes based on the ESI-MS results. However, the protection of Q2 against thermal denaturation by 3 here is greater than 2. The larger size of the ligand in this metal complex appears to stabilize the quadruplex to a greater extent than 2, perhaps indicating a higher binding avidity, despite the slightly lower binding affinity estimated from the ESI-MS results. It is also possible that 3 forms higher order adducts with the DNA than 2, as is suggested by the ESI-MS results obtained for the 2:1 metal complex:DNA solutions.
Table II.
T1/2 Values for Q2 in the Absence and Presence of All Four Metal Complexes
| Sample | Melting Point ± Standard Deviation (°C) | ΔT1/2 |
|---|---|---|
| Q2 | 50.3 ± 0.2 | – |
| Q2 + 1 | 50.5 ± 0.2 | 0.2 |
| Q2 + 2 | 68 ± 1 | 18 |
| Q2 + 3 | 72 ± 1 | 22 |
| Q2 + 4 | 54.1 ± 0.7 | 3.8 |
CONCLUSIONS
The combination of large, planar ligands attached to the PtII core appears to provide better quadruplex interaction than complexes that have small ligands or are based around a metal with the RuII octahedral geometry. Thermal denaturation studies and reactions with glyoxal in the presence of the four metal complexes also show that 2 and 3 afford increased thermal stability over the other complexes, and induce changes in guanine accessibility in the quadruplexes. Although showing relatively high affinities for Q1 and Q2, 2, 3, and 4 exhibited much lower binding to Q3 and Q4. This striking difference in binding of the complexes highlights the importance of quadruplex structure. As end-stacking binders, these metal complexes are sensitive to the secondary structure surrounding the quadruplex. It should also be reiterated that Q4, in the presence of sodium and potassium, often forms antiparallel or mixed orientation quadruplexes rather than parallel quadruplexes. This could permit binding similar to that seen for Q2 to occur in vivo.
Supplementary Material
Additional Supporting Information may be found in the online version of this article.
Acknowledgments
Contract grant sponsor: Robert A. Welch Foundation
Contract grant number: F-1155
Contract grant sponsor: National Institutes of Health
Contract grant number: RO1 GM65956
We thank TI-3D and Eun Jeong Cho for use of and help with the Jasco J-815 circular dichroism system, and Wendy Marriner for assistance with the gel electrophoresis.
Footnotes
This article was originally published online as an accepted preprint. The ‘Published Online’ date corresponds to the preprint version. You can request a copy of the preprint by emailing the Biopolymers editorial office at biopolymers@wiley.com
REFERENCES
- 1.Roner MR, Carraher CE., Jr . In: Inorganic and Organometallic Macromolecules: Design and Applications. Abd-El-Aziz AS, Carraher CE Jr, Pittman CU Jr, Zeldin M, editors. New York, NY: Springer; 2008. pp. 193–220. [Google Scholar]
- 2.Friedman AE, Chambron JC, Sauvage JP, Turro NJ, Barton JK. J Am Chem Soc. 1990;112:4960–4962. [Google Scholar]
- 3.Pierard F, Kirsch-De Mesmaeker A. Inorg Chem Commun. 2006;9:111–126. [Google Scholar]
- 4.Zeglis BM, Pierre VC, Barton JK. Chem Commun. 2007:4565–4579. doi: 10.1039/b710949k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metcalfe C, Thomas JA. Chem Soc Rev. 2003;32:215–224. doi: 10.1039/b201945k. [DOI] [PubMed] [Google Scholar]
- 6.Ralph SF, Beck JL, Gupta R, Urathamakul T, Sheil MM, Aldrich-Wright JR. J Inorg Biochem. 2003;96 214–214. [Google Scholar]
- 7.Urathamakul T, Beck JL, Sheil MM, Aldrich-Wright JR, Ralph SF. Dalton Trans. 2004:2683–2690. doi: 10.1039/B406889K. [DOI] [PubMed] [Google Scholar]
- 8.Kieltyka R, Englebienne P, Fakhoury J, Autexier C, Moitessier N, Sleiman HF. J Am Chem Soc. 2008;130:10040–10041. doi: 10.1021/ja8014023. [DOI] [PubMed] [Google Scholar]
- 9.Cusumano M, Di Pietro ML, Giannetto A. Inorg Chem. 2006;45:230–235. doi: 10.1021/ic050880o. [DOI] [PubMed] [Google Scholar]
- 10.Cusumano M, Di Pietro ML, Giannetto A. Inorg Chem. 1999;38:1754–1758. doi: 10.1021/ic9809759. [DOI] [PubMed] [Google Scholar]
- 11.Han HY, Hurley LH. Trends Pharmacol Sci. 2000;21:136–142. doi: 10.1016/s0165-6147(00)01457-7. [DOI] [PubMed] [Google Scholar]
- 12.Mergny JL, Helene C. Nat Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 13.Fedoroff OY, Salazar M, Han H, Chemeris VV, Kerwin SM, Hurley LH. Biochemistry. 1998;37:12367–12374. doi: 10.1021/bi981330n. [DOI] [PubMed] [Google Scholar]
- 14.Zahler AM, Williamson JR, Cech TR, Prescott DM. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 15.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fletcher TM, Sun DK, Salazar M, Hurley LH. Biochemistry. 1998;37:5536–5541. doi: 10.1021/bi972681p. [DOI] [PubMed] [Google Scholar]
- 17.Huppert JL, Philos Trans R. Soc London Ser A. 2007;365:2969–2984. doi: 10.1098/rsta.2007.0011. [DOI] [PubMed] [Google Scholar]
- 18.Burger AM, Bibby MC, Double JA. Br J Cancer. 1997;75:516–522. doi: 10.1038/bjc.1997.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kieltyka R, Fakhoury J, Moitessier N, Sleiman HF. Chem Eur J. 2008;14:1145–1154. doi: 10.1002/chem.200700783. [DOI] [PubMed] [Google Scholar]
- 20.Gingras A-C, Aebersold R, Raught B. J Physiol (London) 2005;563:11–21. doi: 10.1113/jphysiol.2004.080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li HH, Yuan G, Du DM. J Am Soc Mass Spectrom. 2008;19:550–559. doi: 10.1016/j.jasms.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Baker Erin S, Bernstein Summer L, Bowers Michael T. J Am Soc Mass Spectrom. 2005;16:989–997. doi: 10.1016/j.jasms.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 23.Gabelica V, De Pauw E. J Mass Spectrom. 2001;36:397–402. doi: 10.1002/jms.141. [DOI] [PubMed] [Google Scholar]
- 24.Gabelica V, De Pauw E. Int J Mass Spectrom. 2002;219:151–159. [Google Scholar]
- 25.Gidden J, Baker ES, Ferzoco A, Bowers MT. International J Mass Spectrom. 2005;240:183–193. [Google Scholar]
- 26.Hofstadler SA, Griffey RH. Chem Rev. 2001;101:377–390. doi: 10.1021/cr990105o. [DOI] [PubMed] [Google Scholar]
- 27.Mazzitelli CL, Wang J, Smith SI, Brodbelt JS. J Am Soc Mass Spectrom. 2007;18:1760–1773. doi: 10.1016/j.jasms.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosu F, Gabelica V, Houssier C, Colson P, De Pauw E. Rapid Commun Mass Spectrom. 2002;16:1729–1736. doi: 10.1002/rcm.778. [DOI] [PubMed] [Google Scholar]
- 29.Veenstra TD. Biophysical Chem. 1999;79:63–79. doi: 10.1016/s0301-4622(99)00037-x. [DOI] [PubMed] [Google Scholar]
- 30.Williams ER, Jurchen JC, Garcia DE, Lemoff AS, Bush MF. Adv Mass Spectrom. 2004;16:79–94. [Google Scholar]
- 31.Akashi S, Osawa R, Nishimura Y. J Am Soc Mass Spectrom. 2005;16:116–125. doi: 10.1016/j.jasms.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 32.Hanson CL, Robinson CV. J Biol Chem. 2004;279:24907–24910. doi: 10.1074/jbc.R300037200. [DOI] [PubMed] [Google Scholar]
- 33.Jensen ON, Kulkarni S, Aldrich JV, Barofsky DF. Nucleic Acids Res. 1996;24:3866–3872. doi: 10.1093/nar/24.19.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapur A, Beck JL, Brown SE, Dixon NE, Sheil MM. Protein Sci. 2002;11:147–157. doi: 10.1110/ps.27702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deterding LJ, Kast J, Przybylski M, Tomer KB. Bioconjugate Chem. 2000;11:335–344. doi: 10.1021/bc990123c. [DOI] [PubMed] [Google Scholar]
- 36.Furlan RLA, Watt SJ, Garrido LM, Amarante-Mendes GP, Nur-E-Alam M, Rohr J, Brana A, Mendez C, Salasd JA, Sheil MM, Beck JL, Padilla G. J Antibiot. 2004;57:647–654. doi: 10.7164/antibiotics.57.647. [DOI] [PubMed] [Google Scholar]
- 37.Iannitti-Tito P, Weimann A, Wickham G, Sheil MM. Analyst. 2000;125:627–633. doi: 10.1039/a908920i. [DOI] [PubMed] [Google Scholar]
- 38.Mazzitelli CL, Brodbelt JS, Kern JT, Rodriguez M, Kerwin SM. J Am Soc Mass Spectrom. 2006;17:593–604. doi: 10.1016/j.jasms.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 39.Mazzitelli CL, Chu YJ, Reczek JJ, Iverson BL, Brodbelt JS. J Am Soc Mass Spectrom. 2007;18:311–321. doi: 10.1016/j.jasms.2006.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazzitelli CL, Rodriguez M, Kerwin SM, Brodbelt JS. J Am Soc Mass Spectrom. 2008;19:209–218. doi: 10.1016/j.jasms.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith SI, Guziec LJ, Guziec FS, Hasinoff BB, Brodbelt JS. J Mass Spectrom. 2007;42:681–688. doi: 10.1002/jms.1205. [DOI] [PubMed] [Google Scholar]
- 42.Beck JL, Colgrave ML, Ralph SF, Sheil MM. Mass Spectrom Rev. 2001;20:61–87. doi: 10.1002/mas.1003. [DOI] [PubMed] [Google Scholar]
- 43.Li W, Zhang M, Zhang JL, Li HQ, Zhang XC, Sun Q, Qiu CM. Febs Letters. 2006;580:4905–4910. doi: 10.1016/j.febslet.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 44.Pierce SE, Sherman CL, Jayawickramarajah J, Lawrence CM, Sessler JL, Brodbelt JS. Analytica Chimica Acta. 2008;627:129–135. doi: 10.1016/j.aca.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker Erin S, Lee Jeong T, Sessler Jonathan L, Bowers Michael T. J Am Chem Soc. 2006;128:2641–2648. doi: 10.1021/ja0564968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.David WM, Brodbelt J, Kerwin SM, Thomas PW. Anal Chem. 2002;74:2029–2033. doi: 10.1021/ac011283w. [DOI] [PubMed] [Google Scholar]
- 47.Evans SE, Mendez MA, Turner KB, Keating LR, Grimes RT, Melchoir S, Szalai VA. JBIC, J Biol Inorg Chem. 2007;12:1235–1249. doi: 10.1007/s00775-007-0292-0. [DOI] [PubMed] [Google Scholar]
- 48.Gornall KC, Samosorn S, Talib J, Bremner JB, Beck JL. Rapid Commun Mass Spectrom. 2007;21:1759–1766. doi: 10.1002/rcm.3019. [DOI] [PubMed] [Google Scholar]
- 49.Rosu F, De Pauw E, Guittat L, Alberti P, Lacroix L, Mailliet P, Riou J-F, Mergny J-L. Biochemistry. 2003;42:10361–10371. doi: 10.1021/bi034531m. [DOI] [PubMed] [Google Scholar]
- 50.Rosu F, Gabelica V, Shin-ya K, De Pauw E. Chemical Commun. 2003:2702–2703. doi: 10.1039/b309394h. [DOI] [PubMed] [Google Scholar]
- 51.Talib J, Green C, Davis KJ, Urathamakul T, Beck JL, Aldrich-Wright JR, Ralph SF. Dalton Trans. 2008:1018–1026. doi: 10.1039/b715458e. [DOI] [PubMed] [Google Scholar]
- 52.Carrasco C, Rosu F, Gabelica V, Houssier C, De Pauw E, Garbay-Jaureguiberry C, Roques B, Wilson WD, Chaires JB, Waring MJ, Bailly C. ChemBioChem. 2002;3:1235–1241. doi: 10.1002/1439-7633(20021202)3:12<1235::AID-CBIC1235>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 53.Zhou J, Yuan G. Chem-Eur J. 2007;13:5018–5023. doi: 10.1002/chem.200601605. [DOI] [PubMed] [Google Scholar]
- 54.Urathamakul T, Waller DJ, Beck JL, Aldrich-Wright JR, Ralph SF. Inorg Chem. 2008;47:6621–6632. doi: 10.1021/ic702179a. [DOI] [PubMed] [Google Scholar]
- 55.Mazzitelli CL, Brodbelt JS. Analytical Chemistry. 2007;79:4636–4647. doi: 10.1021/ac070145p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bui CT, Rees K, Cotton RGH. Curr Pharmacogenomics. 2004;2:325–332. [Google Scholar]
- 57.Kasai H, Iwamoto-Tanaka N, Fukada S. Carcinogenesis. 1998;19:1459–1465. doi: 10.1093/carcin/19.8.1459. [DOI] [PubMed] [Google Scholar]
- 58.Shapiro R, Cohen BI, Shiuey S-J, Maurer H. Biochemistry. 1969;8:238–245. doi: 10.1021/bi00829a034. [DOI] [PubMed] [Google Scholar]
- 59.Sullivan BP, Salmon DJ, Meyer TJ. Inorg Chem. 1978;17:3334–3341. [Google Scholar]
- 60.Pyle AM, Rehmann JP, Meshoyrer R, Kumar CV, Turro NJ, Barton JK. J Am Chem Soc. 1989;111:3051–3058. [Google Scholar]
- 61.Wilson JJ, Brodbelt JS. Anal Chem. 2006;78:6855–6862. doi: 10.1021/ac060760d. [DOI] [PubMed] [Google Scholar]
- 62.Gray DM, Wen JD, Gray CW, Repges R, Repges C, Raabe G, Fleischhauer J. Chirality. 2008;20:431–440. doi: 10.1002/chir.20455. [DOI] [PubMed] [Google Scholar]
- 63.Rachwal PA, Findlow IS, Werner JM, Brown T, Fox KR. Nucleic Acids Res. 2007;35:4214–4222. doi: 10.1093/nar/gkm316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smargiasso N, Rosu F, Hsia W, Colson P, Baker ES, Bowers MT, De Pauw E, Gabelica V. J Am Chem Soc. 2008;130:10208–10216. doi: 10.1021/ja801535e. [DOI] [PubMed] [Google Scholar]
- 65.Rujan IN, Meleney JC, Bolton PH. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee JY, Yoon J, Kihm HW, Kim DS. Biochemistry. 2008;47:3389–3396. doi: 10.1021/bi702013d. [DOI] [PubMed] [Google Scholar]
- 67.Pedroso IM, Duarte LF, Yanez G, Baker AM, Fletcher TM. Biochem Biophys Res Commun. 2007;358:298–303. doi: 10.1016/j.bbrc.2007.04.126. [DOI] [PubMed] [Google Scholar]
- 68.Bugaut A, Balasubramanian S. Biochemistry. 2008;47:689–697. doi: 10.1021/bi701873c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mergny J-L, De Cian A, Ghelab A, Sacca B, Lacroix L. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gros J, Rosu F, Amrane S, De Cian A, Gabelica V, Lacroix L, Mergny J-L. Nucleic Acids Res. 2007;35:3064–3075. doi: 10.1093/nar/gkm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brown NM, Rachwal PA, Brown T, Fox KR. Org Biomol Chem. 2005;3:4153–4157. doi: 10.1039/b511706b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article.