Abstract
Promiscuous substrate recognition, the ability to catalyze transformations of chemically diverse compounds, is an evolutionarily advantageous, but poorly understood phenomenon. The promiscuity of DNA repair enzymes is particularly important, because it enables diverse kinds of damage to different nucleotide bases to be repaired in a metabolically parsimonious manner. We present enzymological and crystallographic studies of the mechanisms underlying promiscuous substrate recognition by Escherichia coli AlkB, a DNA repair enzyme that removes methyl adducts and some larger alkylation lesions from endocyclic positions on purine and pyrimidine bases. In vitro Michaelis–Menten analyses on a series of alkylated bases show high activity in repairing N1-methyladenine (m1A) and N3-methylcytosine (m3C), comparatively low activity in repairing 1,N6-ethenoadenine, and no detectable activity in repairing N1-methylguanine or N3-methylthymine. AlkB has a substantially higher kcat and Km for m3C compared with m1A. Therefore, the enzyme maintains similar net activity on the chemically distinct substrates by increasing the turnover rate of the substrate with nominally lower affinity. Cocrystal structures provide insight into the structural basis of this “kcat/Km compensation,” which makes a significant contribution to promiscuous substrate recognition by AlkB. In analyzing a large ensemble of crystal structures solved in the course of these studies, we observed 2 discrete global conformations of AlkB differing in the accessibility of a tunnel hypothesized to control diffusion of the O2 substrate into the active site. Steric interactions between a series of protein loops control this conformational transition and present a plausible mechanism for preventing O2 binding before nucleotide substrate binding.
Keywords: iron 2-oxoglutarate dioxygenase, broad substrate specificity, X-ray crystallography
Promiscuous substrate recognition refers to the ability of an enzyme to catalyze equivalent chemical reactions on structurally diverse substrates (1). This phenomenon is sometimes referred to as “broad substrate specificity” (1), and is distinct from “catalytic promiscuity,” which refers to the ability to perform different types of chemical reactions. Both phenomena represent important properties of many naturally evolved enzymes. Promiscuous substrate recognition is particularly advantageous for DNA-repair enzymes, because it allows a single enzyme to protect against structurally diverse types of DNA damage (1). However, the ability of an enzyme to operate on substrates that differ in size, charge, or hydrogen-bonding capacity is naively at odds with concepts of lock-and-key enzymology (2). The ability to bind distinct chemical structures might also complicate efforts to design specific enzyme inhibitors. The inhibition of DNA repair enzymes has been advocated as a means to improve cancer therapy, because these enzyme systems reverse the effects of DNA damaging compounds, which remain among the most commonly used antitumor agents (3). Understanding the structural basis for promiscuous substrate recognition could facilitate inhibitor development, in addition to providing insight into a fundamentally important enzyme property.
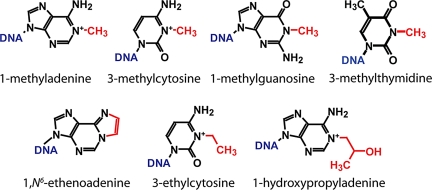
A clear example of promiscuous substrate recognition is provided by AlkB, a direct damage repair enzyme that reverses chemical lesions generated by SN2 alkylation reagents on the endocyclic nitrogen atoms of nucleic acid bases (4, 5). This enzyme has been reported to remove methyl adducts from the N1 atoms of adenine (m1A) and guanine (m1G) and the N3 atoms of cytosine (m3C) and thymine (m3T) (4–8), as well as larger ethyl, propyl, hydroxyalkyl, and exocyclic etheno and ethano adducts from adenine and cytosine bases (Fig. 1) (9–12). Also, AlkB dealkylates bases in both single- and double-stranded DNA and RNA (4, 5, 13, 14). Although there are substantial differences in the methods used in different studies, as well as in the reported specific activities (SI Appendix, Table S1), it is clear that AlkB recognizes a wide variety of chemically distinct substrates.
Fig. 1.
Structures of AlkB substrates. Red lesions are repaired by AlkB.
AlkB hydroxylates alkyl lesions on the carbon atom that is covalently bound to the endocyclic nitrogen atom on the nucleotide base. This reaction results in an unstable bond that spontaneously hydrolyzes to restore the unmodified base. The hydroxylation reaction employs molecular oxygen (O2) as a substrate and 2-oxoglutarate (2OG) as a cosubstrate in addition to an Fe(II) cofactor that catalyzes the redox chemistry. The fold of the catalytic core in AlkB and the central features of its reaction chemistry are shared with a superfamily of enzymes called Fe-2OG dioxygenases (15), which represents the largest known group of nonheme iron enzymes.
Recently, the crystal structure of an anaerobic Michaelis complex of Escherichia coli AlkB was solved (16) with the enzyme bound to Fe(II), 2OG, and an m1A containing DNA substrate, i.e., dT-(1-methyl-dA)-dT (called TmAT in this article). Although the catalytic core of AlkB is homologous to other Fe-2OG dioxygenases (15), a unique nucleotide-recognition lid is formed by residues from the N terminus of the enzyme. Dynamic flexibility in this unique subdomain was hypothesized to have a role in docking substrates of varying size into the active site (16).
To elucidate the enzymological and structural basis of promiscuous substrate recognition by E. coli AlkB, we present here coordinated cocrystallization studies and Michaelis–Menten kinetic analyses on a series of chemically distinct substrates. In the course of this study, we observed that the catalytic core of AlkB undergoes a discrete conformational change (CC) that alters the relative accessibility of a putative O2-diffusion tunnel leading into the active site. This CC may prevent premature generation of reactive oxygen intermediates during the multistep redox reaction cycle of AlkB and other Fe-2OG dioxygenases.
Results and Discussion
Kinetic Analyses of Repair of Model Nucleotide Substrates in Vitro.
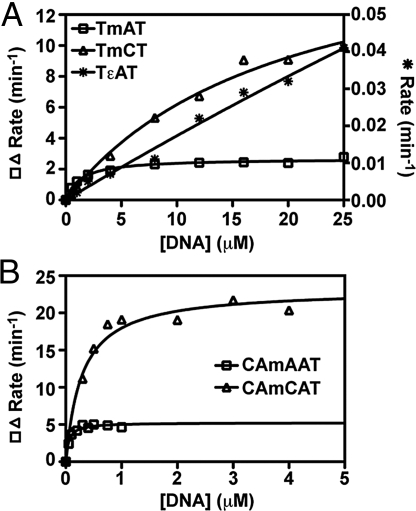
We performed Michaelis–Menten analyses on a series of synthetic trinucleotides with thymine bases flanking a central alkylated base. Oxidative dealkylation by AlkB-ΔN11 was quantified using HPLC methods (10, 16). In addition to the previously characterized TmAT and dT-(1,N6-etheno-dA)-dT (TεAT) substrates (10, 11, 16), we assayed activity on dT-(3-methyl-dC)-dT (TmCT), dT-(1-methyl-dG)-dT (TmGT), and dT-(3-methyl-dT)-dT (TmTT). Assaying repair of lesions in a conserved oligonucleotide background enabled activity to be compared without interference from variations in substrate length and sequence identity. To ensure that conclusions did not depend on backbone structure, we also assayed repair of m1A and m3C in the pentanucleotides dC-dA-(1-methyl-dA)-dA-dT (CAmAAT) and dC-dA-(3-methyl-dC)-dA-dT (CAmCAT). Reaction times were optimized to ensure accurate measurement of initial enzyme velocity without interference from substrate depletion (SI Appendix, Fig. S1). These assays yielded kcat and Km values for the TmAT, TmCT, TεAT, CAmAAT, and CAmCAT substrates (Fig. 2 and Table 1).
Fig. 2.
Michaelis–Menten analysis of AlkB-ΔN11 repair activity on model substrates. (A) DNA trimers and (B) pentamers were incubated at 37 °C under standard reaction conditions. AlkB was used at a concentration of 0.1 μM for the TmAT (□) and TmCT (△), 0.002 μM for CAmAAT (□) and CAmCAT (△), and 1.0 μM for TεAT (*). Reactions quenched by addition of EDTA were analyzed by reversed-phase HPLC, and the dealkylation rate was determined by quantifying substrate and product peak areas in the resulting chromatogram. Curves represent nonlinear regressions to the noncooperative Michaelis–Menten equation. Due to its substantially slower turnover rate, initial velocity of TεAT repair is plotted on the right y axis in A.
Table 1.
Enzyme kinetic parameters for AlkB
| Substrate | kcat, min−1 | Km, μM | kcat/Km, min−1μM−1 |
|---|---|---|---|
| TmAT | 2.7 ± 0.8 | 1.4 ± 0.9 | 1.9 |
| TmCT | 21 ± 4 | 24 ± 5 | 0.9 |
| TεAT | 0.13 ± 0.05 | 60 ± 14 | 0.002 |
| CAmAAT | 5.4 ± 1.3 | 0.06 ± 0.01 | 97.0 |
| CAmCAT | 23 ± 10 | 0.29 ± 0.03 | 78.3 |
Initial reaction velocities were analyzed by nonlinear regression curve fitting using Prism software to obtain the kcat, Km, and SE.
The kcat (2.7 min−1) and Km (1.4 μM) values measured for TmAT are similar to those previously determined for AlkB-ΔN11 or full-length AlkB (SI Appendix, Table S1) (10, 16). However, substantially higher kcat and Km values of 21 min−1 and 23 μM, respectively, were determined for TmCT. Note that the Km for TmCT is so high that assays could not be conducted at saturating substrate concentrations, introducing substantial uncertainty into the estimates of its kcat and Km (Table 1). However, the kcat/Km ratio is determined accurately even under these circumstances, because it is given directly by the slope of initial velocity vs. substrate concentration in the limit of zero substrate concentration (as can be seen mathematically by differentiation of the Michaelis–Menten equation). It is noteworthy that the kcat/Km ratio is very similar for both TmAT and TmCT substrates (1.9 vs. 0.9 min−1μM−1). The failure to achieve enzyme saturation with TmCT, at concentrations much higher than those required to achieve saturation with TmAT, demonstrates that the Km must be substantially higher for TmCT. Futherfore, because kcat/Km is accurately measured for both substrates, we can conclude that TmCT must also have a higher kcat, which is also qualitatively clear from the initial-velocity data (Fig. 1). Therefore, the conclusion is robust that both kcat and Km are elevated for TmCT, despite uncertainty in the individual values.
The same basic pattern was observed in assays on CAmAAT and CAmCAT (Table 1), where full saturation of the enzyme could be achieved with both substrates. Irrespective of backbone structure, the m3C substrate with lower affinity was repaired faster than the m1A substrate with higher affinity (Table 1). The kcat value for each pentamer substrate was very similar to that of the timer containing the same lesion (Table 1), as expected for identical alkylated bases if the polynucleotide backbone does not influence active-site stereochemistry. However, the Km values were 20- to 80-fold lower for the pentamer substrates compared with the trimer substrates (Table 1), indicating that AlkB bound the longer substrates substantially better. Although surprising relative to previously reported observations on some longer DNA substrates (10, 17), higher affinity for pentamer vs. trimer substrates is consistent with the observed backbone interactions in the crystal structures of AlkB cross-linked to double-stranded DNA 13-mers, which show protein contacts to all of the phosphates corresponding to our pentameric substrates (but no phosphates further removed from the alkylated target base) (18).
As noted above, despite the differences in kcat and Km for the m1A vs. m3C lesions, the net catalytic efficiency (the kcat/Km ratio) is very similar for both lesions when assayed in a consistent polynucleotide background (1.9 vs. 0.9 min−1·μM−1 for the trimers, and 97 vs. 78 min−1·μM−1 for the pentamers). This conclusion is consistent with all previously published studies (SI Appendix, Table S1) (4, 5, 7, 13, 14, 17). [A discrepancy in our values for m3C compared with those reported in one study (17) is discussed in SI Appendix.] Therefore, offsetting changes in kcat and Km yield similar net turnover efficiency for the chemically distinct m1A and m3C substrates. This phenomenon, which we call “kcat-Km compensation,” makes an important contribution to promiscuous substrate recognition by AlkB.
Consistent with previous literature (9, 11), the TεAT substrate was repaired much less efficiently, with very approximate kcat and Km values of 0.13 min−1 and 60 μM, respectively (Table 1). After 60 min, only ≈20% of TεAT was repaired in a reaction containing 1 μM enzyme and 10 μM substrate (SI Appendix and SI Appendix, Fig. S2.) The kcat/Km ratio (0.002 min−1·μM−1) is 3 orders of magnitude lower for TεAT than for the equivalent trinucleotide substrates with m1A or m3C bases, indicating substantially lower repair efficiency.
Despite clear separation between methylated and control nonmethylated trimers during HPLC analysis, no detectable repair of the TmGT or TmTT substrates was observed in our assays using enzyme concentrations up to 10 μM in buffers ranging from pH 6 to 8 (data not shown). Previous studies have consistently shown that E. coli AlkB repairs m1G or m3T with substantially lower efficiency than m1A or m3C (6–9) (SI Appendix, Table S1). However, in contrast to our results, detectable repair of m1G and m3T has been observed both in vitro and in vivo (6, 7). Differences in nucleic acid substrate length (as observed here for trimeric vs. pentameric substrates and in published work on DNA glycosylases; see ref. 19) could account for the varying results observed in vitro, whereas accessory factors could potentially influence recognition of damaged bases in vivo.
Crystal Structures of E. coli AlkB Bound to Different Trinucleotide Substrates.
To characterize the structural mechanisms underlying promiscuous substrate recognition, E. coli AlkB-ΔN11 was cocrystallized with TmCT or TεAT substrates. Crystal structures were determined for AlkB bound to TmCT, 2OG, and either an Fe(II) cofactor under anaerobic conditions (1.6 Å) or a Mn(II) cofactor under aerobic conditions (1.5 Å) (SI Appendix, Tables S2 and S3). There are no significant differences between these 2 structures, which show a rmsd of 0.29 Å for least-squares superposition of 200 Cα atoms. Cocrystallization with 12 mM TεAT under aerobic conditions yielded a crystal structure at 1.4-Å resolution with 2OG and Mn(II) bound in the active site, but no evidence of electron density for the trinucleotide substrate (SI Appendix, Table S2). To our knowledge, this structure is the first determined for AlkB bound to a metal cofactor and 2OG without a nucleotide substrate, although we previously reported a nucleotide-free structure of the enzyme bound to Fe(II) and succinate, the product of 2OG decarboxylation (16). Also, a higher resolution structure was determined for AlkB cocrystallized with Fe(II), 2OG, and the TmAT substrate under anaerobic conditions (SI Appendix, Table S2). This structure at 1.7-Å resolution is nearly identical to the published 2.3-Å structure of the same complex (16), giving an rmsd of 0.21 Å for least-squares superposition of 199 Cα atoms. Comparing the higher resolution Fe-2OG-TmAT structure, which is used in all analyses presented below, to the equivalent Fe-2OG-TmCT structure shows much larger conformational differences, giving an rmsd of 0.71 Å for least-squares superposition of 198 Cα atoms. The origin and significance of these differences are dissected in detail below.
Two Global AlkB Conformations Differing in Accessibility of the Putative O2-Diffusion Tunnel.
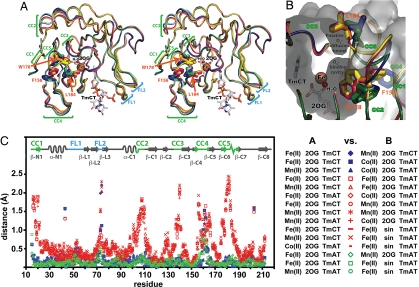
Structural superposition of the complete ensemble of available crystal structures of E. coli AlkB revealed substantial conformational differences in the double-stranded β-helix comprising the catalytic core of this enzyme and homologous Fe-2OG-dioxygenases. Detailed structural analyses showed that 64 of 131 residues in the conserved core adopt an equivalent backbone conformation in all structures, but that most of the remaining residues adopt 1 of 2 discrete alternative conformations (Fig. 3; SI Appendix, Figs. S3–S5).
Fig. 3.
The ensemble of AlkB-ΔN11 crystal structures reveals a discrete change in global protein conformation. (A) Nucleotide-bound structures after least-squares superposition of core β-strands in the Fe-2OG dioxygenase domain [with 2OG, Fe(II), and TmCT shown from just a single structure]. Discrete local CCs are observed in 5 protein segments (CC1–CC5). FLs (FL1 and FL2), which contact the backbone of the trinucleotide substrate, exhibit elevated backbone B-factors as well as conformational variability not correlated with the conformational state of the enzyme (16). The water molecule replaced by O2 to initiate the oxidation reaction occupies the sixth site on the octahedrally coordinated Fe ion. AlkB-Fe(II)-2OG-TmCT is shown in orange, AlkB-Mn(II)-2OG-TmCT in yellow, AlkB-Fe(II)-2OG-TmAT (at 1.7 Å) in green, AlkB-Mn(II)-2OG-TmAT (16) in blue, AlkB-Co(II)-2OG-TmAT (16) in magenta, and AlkB-Fe(II)-succinate-TmAT (16) in cyan. (B) The putative O2-diffusion channel and O2-binding cavity with structures colored as in A. The molecular surface of AlkB-Fe(II)-2OG-TmAT is shown in gray. (C) Plot of Cα displacements between all nucleotide-bound crystal structures after least-squares superposition of core β-strands. The legend indicates the color/symbol used for pairwise structural comparisons. Blue and green are used for comparisons between pairs of structures both in the closed or open conformational state, respectively, whereas red is used for comparisons between structures in different states. For stereo view of B, see SI Appendix, Fig. S3.
When the conserved cores are aligned, concerted conformational changes (CC1-5) are observed involving 5 different polypeptide segments (residues 14–22, 104–111, 136–141, 155–166, and 175–188) that make extensive packing interactions with one another (Fig. 3). Backbone Cα atoms in these polypeptide segments move up to 2.5 Å relative to the conformationally invariant segments (Fig. 3C). Although the loop spanning residues 158–164 (part of CC4) exhibits the most variable backbone conformation of any segment involved in the global CC (Fig. 3C; SI Appendix, Fig. S5B), there is only minimal overlap in the positions it adopts in the 2 conformations (Fig. 3A; SI Appendix, Fig. S5). Also, the backbone B-factors in this segment are elevated consistently in only 1 conformation (blue and green in SI Appendix, Fig. S6). Therefore, the conformation of the 158–164 loop is clearly influenced by the discrete global CC in AlkB. In contrast, 2 loops in the nucleotide-recognition lid occupy a continuum of different positions, leading to their designation as flexible loops (FL1 and FL2 in Fig. 3).
A central feature of the discrete global CC is a coupled rotation of the side chains of aromatic residues F156 and W178 (Fig. 3 A and B; SI Appendix, Figs. S3–S5), which are invariant in AlkB orthologs (16), and make van der Waals contacts to one another in both conformations. In one conformation, these residues line the wall of the O2-binding cavity, whereas in the other, they both flip into the cavity, substantially reducing its volume. When flipped in, the side chain of W178 pushes directly on the 310-helix between strands β-C6 and β-C7, and the resulting movement of this structure pushes the side chain of L184 into a putative O2-diffusion tunnel that leads from the surface of the protein to a water-filled O2-binding cavity adjoining the catalytic iron site. This movement effectively closes the tunnel (Fig. 3B; SI Appendix, Figs. S3 and S4). Therefore, we designate the 2 conformations as “closed” (magenta, yellow, and orange in Fig. 3 A and B) and “open” (blue, green, and cyan). Seven of the 11 structures that we have determined are in the open conformation, whereas the remainder are in the closed conformation (Table 2), as are the 3 DNA cross-linked structures determined by another group (18).
Table 2.
Summary of E. coli AlkB crystal structures
| PDB ID | Ligands | Space group | Conformation | Ref. | ||
|---|---|---|---|---|---|---|
| 3I2O | Fe(II) | 2OG | TmAT | P43 | Open | This study |
| 2FD8 | Fe(II) | 2OG | TmAT | P43 | Open | 16 |
| 3I49 | Fe(II) | 2OG | TmCT | P43 | Closed | This study |
| 3I3M | Mn(II) | 2OG | TmCT | P43 | Closed | This study |
| 3I3Q | Mn(II) | 2OG | — | P1† | Closed | This study |
| 2FDH | Mn(II) | 2OG | TmAT | P43 | Open | 16 |
| 2FDF | Co(II) | 2OG | TmAT | P43 | Closed | 16 |
| 2FDG | Fe(II) | sin | TmAT | P43 | Open | 16 |
| 2FDJ | Fe(II) | sin | — | P1 | Open | 16 |
| 2FDI | Fe(II) | 2OG | TmAT | P43 | Open | 16 |
| 2FDK | Fe(II) | 2OG | TmAT | P43 | Open | 16 |
| 3BKZ | Mn(II) | 2OG | DNA1‡ | P21 | Closed | 18 |
| 3BI3 | Mn(II) | 2OG | DNA5‡ | P21 | Closed | 18 |
| 3BIE | Mn(II) | 2OG | DNA4‡ | P21 | Closed | 18 |
For expanded version, see Table S3.
†Only crystal form with 2 protomers in the asymmetric unit.
‡Disulfide-tethered cytosine (C*) in double-stranded DNA substrates were cross-linked to the protein. DNA1, TAG GTAAC*ATCGT; DNA4, TAGGTAA(m1A)AC*CGT; DNA5, TAGGTAA(m1A)AC*CGT.
Allosteric Interactions Coupled to Opening/Closing of the O2-Diffusion Tunnel.
The consistency of the backbone conformation observed in the open vs. closed structures of AlkB (SI Appendix, Fig. S5) suggests that the transition between these discrete global conformational states is an intrinsic property of the protein. Also, the fact that both conformations are observed in crystals with the same DNA substrate that grew in the same space group from equivalent solution conditions (Table 2) suggests that this transition involves a relatively modest free energy change, as observed for functional allosteric CCs in some other proteins (20). In the crystal lattice, the transition appears to be controlled by interprotomer packing interactions, because the observed conformational state of the protein correlates perfectly with the occurrence of alternative packing contacts near the 158–164 loop (in CC4) and the entrance of the O2-diffusion tunnel (spanning CC3 and CC5) (SI Appendix, Fig. S7). However, there are several mechanisms by which this transition could have a functional role in the catalytic reaction cycle of AlkB outside of the confines of a crystal lattice. It couples stereochemical changes near the catalytic Fe ion (SI Appendix, Table S4) to CCs in the 158–164 loop (part of CC5), which interacts weakly with the trinucleotide in our crystal structures (Fig. 4 and Fig. S8), but more strongly with the larger nucleotide substrate in the DNA cross-linked structures (18). This pathway of allosteric communication could conceivably couple binding of the DNA substrate to changes in the redox properties of the Fe ion that activate it for catalysis. DNA binding could also potentially trigger opening of the O2-diffusion tunnel, which could prevent gratuitous turnover of 2OG and release of reactive oxygen species before the binding of DNA. Additional work will be required to critically evaluate these possibilities. More detailed discussions of this material are presented in SI Appendix.
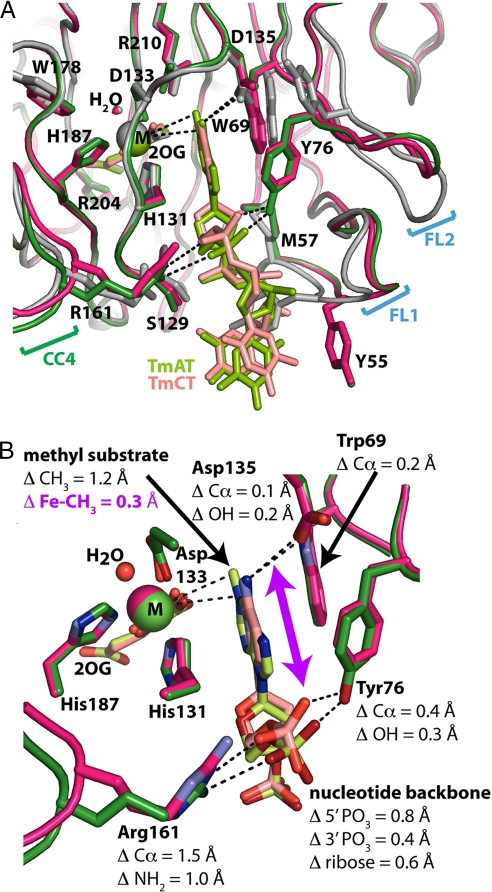
Fig. 4.
Stereochemical comparison of 1-methyadenosine vs. 3-methylcytosine-bound structures. (A) Active site stereochemistry in the Co(II)-2OG-TmAT (green), Fe(II)-2OG-TmCT (magenta), and Mn(II)-2OG (gray) structures. Invariant residues H131, D133, and H187 chelate the metal ion cofactor (M), whereas conserved residues R204 and R210 interact with 2OG. Residue R210 also makes van der Waals contacts to the target methyl group. The partially disordered phosphates on the 5′ terminal thymine are omitted for clarity, as are water molecules other than that occupying the sixth coordination site on the metal. (B) Magnified view of the methylated DNA bases in the Co(II)-2OG-TmAT (green) and Fe(II)-2OG-TmCT (magenta) structures. Nitrogen and oxygen atoms are colored blue and red, respectively, whereas carbon atoms are colored like the protein backbone. Only the methylated bases and flanking phosphates of the nucleotide substrates are shown. The purple arrow is aligned approximately parallel to the 0.6-Å displacement of the ribose group of the alkylated nucleotide in the m3C vs. m1A structures. This displacement effectively slides the alkylated base deeper into a binding slot that is approximately coplanar with the sidechain rings of W69 and H131 and terminates at the catalytic iron center. For stereo views, see SI Appendix, Fig. S8.
The discrete global conformational transition that we have characterized crystallographically seems to be substantially different from the transition recently observed in solution NMR studies of AlkB bound to 2OG vs. succinate (21). Most of the residues perturbed in these studies surround the 2OG-binding cavity and are located in regions of AlkB that do not show significant conformational differences in the ensemble of available crystal structures (SI Appendix, Fig. S9). Residue L184, which gates opening/closing of the putative O2-diffusion tunnel, was observed to experience chemical exchange broadening in the succinate-bound, but not 2OG-bound conformations of AlkB in solution. However, most other residues involved in the crystallographically observed transition do not show any comparable effects, so it is unclear whether the dynamics of tunnel opening are enhanced on conversion of 2OG to succinate.
Stereochemistry of 1-Methyladenosine Vs. 3-Methylcytosine Binding.
The original crystal structures of E. coli AlkB bound to TmAT (16) showed that the alkylated base is sandwiched between the side chains of invariant residues W69 and H131 in a deep binding slot terminating at the catalytic iron center (Fig. 4). The polynucleotide backbone is held in place primarily by contacts to side chains on 3 flexible regions in the protein (FL1, FL2, and R161 in CC4). It was suggested that the flexibility of these loops would enable alkylated bases of variable structures to be repositioned at the catalytic center while maintaining equivalent contacts to the invariant polynucleotide backbone (i.e., by sliding the base into or out of the slot, as necessary) (16). The crystal structures of AlkB bound to TmCT reported in this article confirm this prediction, showing partial compensation for the change in base dimensions via this mechanism. A substrate-binding slot with equivalent geometric properties and base-stacking interactions is observed in the DNA-cross-linked crystal structure of the human AlkB homolog ABH2 (18), but not in the nucleotide-free structure of ABH3 (SI Appendix and SI Appendix, Fig. S10) (22).
Detailed stereochemical analyses must be performed on structures in the same global conformational state to avoid confusing effects related to opening the O2-diffusion tunnel with those involved in accommodating alternative nucleotide substrates. Therefore, Fig. 4 shows a superposition of the Fe(II)-TmCT and Co(II)-TmAT structures, both of which are in the closed conformation. These structures contain different metal cofactors, but this difference in cofactors is not a significant complication, because detailed comparisons of structures containing the same nucleotide substrate in the same global conformational state show only minimal changes in ligand binding geometry with Fe(II), Co(II), or Mn(II) cofactors (16). After least-squares alignment of the conformationally invariant Fe-2OG core, the ribose ring of the target nucleotide in the TmCT structure is positioned 0.6 Å closer to the metal cofactor than in the TmAT structure, even though the polynucleotide backbone makes nearly identical contacts to the protein in both cases (Fig. 4). This displacement partially compensates for the smaller size of the modified cytosine base by moving the target methyl atom closer to its position when attached to a larger m1A base. The protein segments mediating this displacement exhibit much larger movements in crystal structures of AlkB without a bound nucleotide (Fig. 4A; SI Appendix, Fig. S5A). Therefore, the flexibility of these protein segments, which undergo an induce-fit CC on DNA substrate binding, directly contributes to accommodating structurally diverse substrates.
This displacement only compensates for ≈1/3 of the difference in the dimensions of the 2 substrates. The target methyl substituents are 1.7 Å apart after backbone alignment of the otherwise equivalent trinucleotide substrates (SI Appendix, Fig. S8A), but the repositioning mediated by the flexible protein loops moves the substrates only 0.6 Å relative to one another in the active site. There is still a 1.2-Å displacement in their locations after aligning either the conformationally invariant regions of the core β-strands (Fig. 4B) or the metal cofactor and its liganding atoms (SI Appendix, Fig. S11). The direction of this displacement is parallel to the plane of the alkylated base, which is essentially perpendicular to the vector from the metal ion to the target methyl group. Because of this geometrical relationship, the 1.2-Å displacement in the position of the target methyl group produces a more modest 0.3-Å change in its distance from the metal ion (Fig. 4B). Therefore, the geometric architecture of the active site itself contributes to promiscuous substrate recognition by binding the alkylated base with its plane oriented approximately perpendicular to the vector from the catalytic Fe ion to the target oxidation site. Larger displacements in absolute substrate position are thereby converted into smaller changes in the distance from the catalytic Fe ion to the target alkyl group.
Structural Basis of kcat-Km Compensation.
Despite these 2 complementary strategies for accommodation of structurally diverse substrates, there are still significant differences in atomic structure in the active sites of the TmCT vs. TmAT-bound enzymes. The atomic packing density near the catalytic center is substantially lower in the structure with the smaller TmCT substrate, which displays an ≈10-fold faster kcat. In contrast, the atomic packing density will be higher for the bulkier TεAT substrate (SI Appendix and Fig. S12), which displays an ≈20-fold slower kcat (Table 1). Given this correlation, the faster kcat observed for the m3C lesion could be attributable to its lower packing density in the active site. Alternatively, it could be attributable to the 1.2-Å shift in the location of its methyl group compared with that of m1A. One or both of these stereochemical factors is likely to account for the faster kcat of AlkB for m3C vs. m1A, due to either quantum-mechanical effects influencing the rate of electronic rearrangements, or steric effects influencing the rate of movement of reactive atoms or product molecules during the multistep redox reaction.
Given the smaller size of the m3C base compared with m1A base (Fig. 1), its reduced affinity (i.e., higher Km) is likely to be attributable to having less hydrophobic surface area buried in the substrate binding slot. The active site stereochemistry in AlkB seems likely to have evolved to maximize the turnover rate for the smaller m3C substrate to compensate for its lower binding affinity and thereby maintain similar net catalytic efficiency (Table 1) on its 2 predominant physiological substrates (23). These compensating changes in kcat and Km make an important contribution to promiscuous substrate recognition by AlkB.
Conclusions.
The redox reaction cycle of AlkB involves complex local dynamics in the active site, including the diffusional release of CO2 after the initial oxidation of 2OG to succinate, followed by physical migration of the reactive oxygen atom on the resulting “oxyferryl” [Fe(IV) = O] intermediate (16). Even though precise positioning of substrates is required for efficient catalysis via this multistep redox reaction, AlkB is able to repair substrates of different sizes and chemical structures. The structural adaptations described in this study contribute to promiscuous substrate repair by enabling the enzyme to maintain efficient catalytic geometry for chemically diverse substrates.
Broad substrate specificity is an important feature of DNA-repair enzymes (1). However, stereochemical constraints combined with physiological requirements will ultimately impose limits on enzyme performance. Exposure to DNA-alkylating reagents produces over a dozen toxic DNA modifications (23). Although N1-adenine and N3-cytosine account for ≈30% of observed lesions (23), the equivalent damage to N1-guanine and N3-thymine is comparatively rare. This difference in substrate prevalence seems likely to account for the ≈10,000-fold lower efficiency of E. coli AlkB in repairing m1G and m3T compared with m1A and m3C as observed in our assays. AlkB makes 2 weak polar interactions to the exocyclic amino group on adenine/cytosine (via the backbone carbonyl of Q132 and the side-chain carboxylate of D135) (16), which probably enhance affinity for these substrates while reducing affinity for guanine/thymine because they have an exocyclic carbonyl group at the equivalent position (Fig. 1). While AlkB has evolved to promiscuously repair alkylation damage to adenine and cytosine, its 2 most prevalent physiological substrates, it appears to have optimized its affinity for these substrates by diminishing its capacity to repair the comparatively rare guanine and thymine substrates with equivalent damage.
Materials and Methods
Materials.
Trinucleotide substrates (TmAT, TmCT, TmGT, and TεAT) with a central alkylated base flanked by unalkylated thymine bases were synthesized by Biosynthesis, Inc. The TmTT, CAmAAT and CAmCAT nucleotide substrates were synthesized by Midland Certified Reagent Company.
Protein Purification, Crystallization, and X-Ray Structure Determination.
Methods were equivalent to those used for the initial structure determination of AlkB-ΔN11 (16). Briefly, protein expressed from a pET plasmid in E. coli BL21λ(DE3) was purified by Ni-NTA chromatography, repassage through a second Ni-NTA column after cleavage of the C-terminal hexa-histidine tag by TEV protease, and gel filtration. Substrates and metals were added to the protein immediately before crystallization in an anaerobic glovebox (COY Laboratories Products Inc.) via 1:1 hanging-drop vapor diffusion against a well containing 18% PEG 3350, 0.2 M sodium formate. Crystals were passed briefly through 22% PEG 3350, 0.2 M sodium formate, 10% glycerol before freezing in liquid propane for cryogenic data collection. Structures were solved by molecular replacement with the AlkB-ΔN11 structure (16) using COMO (24) and refined using “O” (25), COOT (26), and CNS (27). For additional information on crystallization and refinement, see SI Appendix. The conformationally invariant segments of the core β-strands used for final least-squares alignment comprised residues 19–22, 115–134, 143–158, 166–178, 186–189, and 204–210.
Enzyme Kinetics.
AlkB-ΔN11 (typically 0.1 μM) was mixed with the nucleotide substrate (1–25 μM) in 50 μg/mL BSA, 75 μM Fe(NH4)2(SO4)2, 1 mM 2OG, 2 mM ascorbic acid, and 50 mM K-Hepes, pH 8.0. Reactions were incubated at 37 °C for 2 min and quenched with 10 mM EDTA. Relative substrate and product concentrations were determined by integration of digitized UV absorbance traces from reversed-phase HPLC analyses on a Phenomenex Luna C18 column (16).
Supplementary Material
Acknowledgments.
We thank M. Arbing, J. Benach, B. Gibney, G. Verdon, J. Schwanof, and R. Abramowitz for advice and technical support. This work was supported by a National Institutes of Health (NIH) R01 grant and an American Heart Association Established Investigator award (J.F.H.), as well as by an NIH Protein Structure Initiative grant to the Northeastern Structural Genomics Consortium. B.Y. is a Research Fellow of the Terry Fox Foundation through an award from the National Cancer Institute of Canada, and recipient of a Canadian Institute of Health Research Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812938106/DCSupplemental.
References
- 1.O'Brien PJ. Catalytic promiscuity and the divergent evolution of DNA repair enzymes. Chem Rev. 2006;106:720–752. doi: 10.1021/cr040481v. [DOI] [PubMed] [Google Scholar]
- 2.Jencks WP. Catalysis in Chemistry and Enzymology. New York: Dover; 1987. p. xvi.p. 836. [Google Scholar]
- 3.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat Rev Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 4.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 5.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 6.Koivisto P, Robins P, Lindahl T, Sedgwick B. Demethylation of 3-methylthymine in DNA by bacterial and human DNA dioxygenases. J Biol Chem. 2004;279:40470–40474. doi: 10.1074/jbc.M407960200. [DOI] [PubMed] [Google Scholar]
- 7.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci USA. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falnes PO. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:6260–6267. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delaney JC, et al. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat Struct Mol Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 10.Koivisto P, Duncan T, Lindahl T, Sedgwick B. Minimal methylated substrate and extended substrate range of Escherichia coli AlkB protein, a 1-methyladenine-DNA dioxygenase. J Biol Chem. 2003;278:44348–44354. doi: 10.1074/jbc.M307361200. [DOI] [PubMed] [Google Scholar]
- 11.Mishina Y, Yang CG, He C. Direct Repair of the Exocyclic DNA Adduct 1,N(6)-Ethenoadenine by the DNA Repair AlkB Proteins. J Am Chem Soc. 2005;127:14594–14595. doi: 10.1021/ja055957m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frick LE, Delaney JC, Wong C, Drennan CL, Essigmann JM. Alleviation of 1,N6-ethanoadenine genotoxicity by the Escherichia coli adaptive response protein AlkB. Proc Natl Acad Sci USA. 2007;104:755–760. doi: 10.1073/pnas.0607377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aas PA, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 14.Falnes PO, Bjoras M, Aas PA, Sundheim O, Seeberg E. Substrate specificities of bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:3456–3461. doi: 10.1093/nar/gkh655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clifton IJ, et al. Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J Inorg Biochem. 2006;100:644–669. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 16.Yu B, et al. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature. 2006;439:879–884. doi: 10.1038/nature04561. [DOI] [PubMed] [Google Scholar]
- 17.Roy TW, Bhagwat AS. Kinetic studies of Escherichia coli AlkB using a new fluorescence-based assay for DNA demethylation. Nucleic Acids Res. 2007;35:e147. doi: 10.1093/nar/gkm1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang CG, et al. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452:961–965. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dizdaroglu M. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat Res. 2005;591:45–59. doi: 10.1016/j.mrfmmm.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 20.Tsai CJ, del Sol A, Nussinov R. Allostery: Absence of a change in shape does not imply that allostery is not at play. J Mol Biol. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bleijlevens B, et al. Dynamic states of the DNA repair enzyme AlkB regulate product release. EMBO Rep. 2008;9:872–877. doi: 10.1038/embor.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sundheim O, et al. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J. 2006;25:3389–3397. doi: 10.1038/sj.emboj.7601219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sedgwick B. Repairing DNA-methylation damage. Nat Rev Mol Cell Biol. 2004;5:148–157. doi: 10.1038/nrm1312. [DOI] [PubMed] [Google Scholar]
- 24.Jogl G, Tao X, Xu Y, Tong L. COMO: A program for combined molecular replacement. Acta Crystallogr D. 2001;57:1127–1134. doi: 10.1107/s0907444901006783. [DOI] [PubMed] [Google Scholar]
- 25.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Cowtan EP. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.