Abstract
Human PC4 and the yeast ortholog Sub1 have multiple functions in RNA polymerase II transcription. Genome-wide mapping revealed that Sub1 is present on Pol III-transcribed genes. Sub1 was found to interact with components of the Pol III transcription system and to stimulate the initiation and reinitiation steps in a system reconstituted with all recombinant factors. Sub1 was required for optimal Pol III gene transcription in exponentially growing cells.
Keywords: Chip chip, PC4, reinitiation, TFIIIB
PC4 plays an important role in various cellular processes, including transcription, DNA repair, and replication (1–3). First identified as a RNA polymerase (Pol) II coactivator (2, 4), PC4 was shown to interact with activators and components of the Pol II basal transcription machinery (2, 5) and to enhance activator-dependent transcription, stimulating both initiation and promoter escape (6). However, in the absence of TFIIH and TFIID, PC4 represses basal transcription (5, 7). Sub1, the yeast ortholog of PC4, was characterized biochemically as a coactivator required for activated transcription in vitro (8) and genetically as a suppressor of certain TFIIB mutations (9). Further investigations extended the role of PC4/Sub1 to transcription elongation and mRNA processing. Sub1 was shown to regulate enzymes modifying the CTD of the largest subunit of Pol II and might therefore enhance elongation (10). Functional interactions between not only PC4/Sub1 and Cstf-64/Rna15 (11) but also Pta1 and Sub1 (12) established additional connections between Sub1/PC4 and mRNA processing. PC4 has also been copurified with human TFIIIC and was found to stimulate RNA Pol III transcription in vitro (13). Recently, PC4 was found to be associated with chromatin and to be important for chromatin organization, suggesting a more general role in transcription regulation (14).
Apart from its role in transcription, PC4 was implicated in other cellular processes, through its capacity to bind tightly to melted DNA and to single-stranded DNA (ssDNA, ref. 7). A direct interaction between PC4 and XPG, a subunit of the nucleotide excision factor, was correlated with the genetic interaction between their yeast counterparts, Sub1 and Rad2, suggesting a role for PC4/Sub1 in the repair of oxydative DNA damage (1). Furthermore, PC4 can form complexes with human HSSB protein on ssDNA and influences its replication function in vitro (3).
In this study, we identified the gene targets of Sub1 in exponentially growing yeast cells. Analysis of the genome-wide localization of Sub1 revealed its association to all of the genes transcribed by Pol III. We focused on the role of Sub1 in Pol III transcription. We present evidence that Sub1 is involved in Pol III transcription initiation and reinitiation processes in vitro and is required for optimal Pol III transcription in vivo.
Results
Genome-Wide Analysis of Sub1 Occupancy.
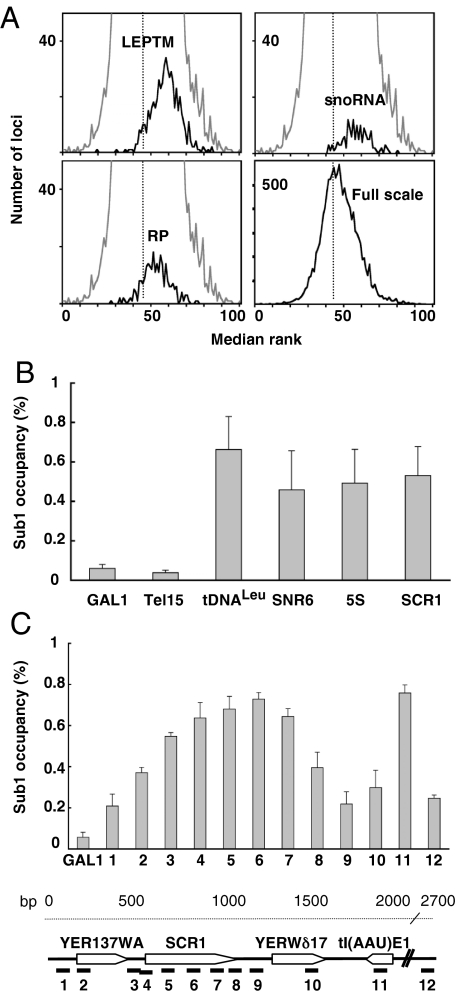
To define Sub1 gene targets in vivo, we performed a genome-wide analysis of Sub1 occupancy in exponentially growing cells. Chromatin immunoprecipitation (ChIP) assays were performed on epitope-tagged Sub1–3HA cross-linked chromatin and analyzed by hybridization to DNA microarrays harboring ORFs and intergenic regions similar to those that we used in a previous work to analyze the genomic location of the Pol III transcription machinery (15). A defined number of loci were significantly enriched (991 loci with a P value <0.01) under active growth conditions. Approximately one-fourth of the enriched loci were located within ORF; the others corresponded to intergenic regions and to genes encoding nontranslated RNAs. We noted that the ACT1, PMA1, PYK1, ADH1, and snoRNA genes previously identified as DNA targets of Sub1 by ChIP and PCR amplification (10, 16, 17) were indeed enriched in our data. The distribution of the loci corresponding to snoRNA genes shown in Fig. 1A extended the binding of Sub1 to all of the H/ACA box or C/D box snoRNA genes. To further identify Pol II gene targets of Sub1, the enriched loci with a P value <0.01 were analyzed by using the GoTermFinder software. Three overrepresented GO categories (P value <10−5) were identified, indicating that Sub1 was preferentially bound to a subset of Pol II-transcribed genes encoding constituents of the cell wall (30 genes), the nucleosome (H2A, H2B, H3, and H4 histone genes) and the ribosome (10% of the enriched loci, corresponding to genes encoding constituents of the translational apparatus, including TEF1, TEF2, and 50 ribosomal protein (RP) genes). The distribution of the loci corresponding to RP genes is shown in Fig. 1A. Because RP and histone genes are among the most highly transcribed genes in exponentially growing cells, we determined the relationship between transcription rates and Sub1 occupancy. Interestingly, although Sub1 was also localized on very poorly transcribed ORFs, Sub1-associated Pol II genes globally tended to be highly transcribed [supporting information (SI) Fig. S1]. Remarkably, one-third of Sub1 enriched loci corresponded to Pol III-transcribed genes present on the arrays or to the intergenic regions and ORFs adjacent to these genes. As shown in Fig. 1A, the Loci Enriched by the Pol III Transcription Machinery (LEPTM, ref. 15), comprising tRNA genes and other genes transcribed by Pol III, like 5S RNA gene, SNR6, RPR1, and SCR1 were significantly overrepresented among the most enriched DNA regions, suggesting the association of Sub1 to all Pol III-transcribed genes in conditions of active growth.
Fig. 1.
Sub1 is present on Pol I, Pol II and Pol III-transcribed genes. (A) Genome-wide binding of Sub1. The distribution of medium ranks of binding ratios is represented as histograms. Loci corresponding to the Loci Enriched by the Pol III Transcription Machinery (LEPTM), RP genes and snoRNA genes are represented as indicated (black curves) within the global distribution of loci (gray curve). The full-scale shows the distribution of all of the loci. (B) Quantitative ChIP analysis on Pol III selected genes or all along the SCR1 gene locus (C). The amounts of immunoprecipitated DNA from Tap-tagged Sub1 cells expressed as a value relative to that of the input are shown as histograms. The GAL1 promoter or Tel15 DNA region were used as controls. Error bars represent the standard deviation between at least 3 independent replicates. A schematic organization of the SCR1 locus and the positions of the 12 DNA fragments amplified by PCR are represented.
To confirm the results obtained by microarray hybridizations, we performed conventional ChIP assays on a set of selected Pol III-transcribed genes. Using real-time PCR on DNA fragments immunopurified from a Tap-tagged Sub1 strain, we quantified the binding of Sub1 to the tRNALeu, 5S RNA, SNR6 and SCR1 genes that were significantly enriched (6- to 13-fold) compared with background signals measured on the GAL1 gene promoter or on a telomeric region of chromosome XV (Fig. 1B). Note that the binding of PC4 to one tRNA gene (but not to the U6 gene) has been recently detected by ChIP experiments (18).
Previous works provided evidence that Sub1 was present at the promoter, the downstream region and across the entire length of several Pol II-transcribed genes (10, 17). The SCR1 gene (522 bp) is the only Pol III-transcribed gene long enough to allow a spatial resolution by ChIP of the distribution of a binding protein. A ChIP experiment was thus performed analyzing 12 real-time PCR amplicons distributed along a 2.7-kb DNA region of chromosome V that encompassed SCR1 and the tI(AAU)E1 (tRNAIleu) gene. As shown in Fig. 1C, Sub1 was detected significantly all along the SCR1 gene and over the tRNAIleu gene, but not on the intergenic regions that separate these loci. A comparison of the enrichment profile of Sub1 across the SCR1 gene with that of TFIIIC, TFIIIB and Pol III (19) suggested a colocalization of Sub1 with Pol III.
The DNA arrays used in our studies had a poor coverage of the rDNA gene locus. Nevertheless, we found a significant enrichment of all loci corresponding to that region, suggesting that Sub1 associates at many locations throughout the rDNA gene. A conventional ChIP experiment on the rDNA locus (Fig. S2) confirmed that Sub1 (but not TFIIIC) bound all along the rDNA gene locus but preferentially to the Pol I-transcribed region, an enrichment profile similar to that of TFIIS, a protein involved in Pol II transcription that has been shown recently to play a role in Pol III transcription (19).
We wondered whether Sub1 genome-wide location could be modified under conditions where a global decrease of transcription was observed. To address this question, we performed ChIP on chip experiments from cells grown to stationary phase where a severe shutdown of transcription by Pol II has been described (20). The comparison of Sub1 binding in exponential or stationary phase grown cells showed that Sub1 occupancy went down substantially in stationary phase as illustrated in Fig. S3 for a subset of its Pol II targets or for LEPTM where the decrease in Sub1 binding was highly similar (Fig. S3B) to those published (21) for TFIIIB or Pol III.
The binding of Sub1 to the transcribed sequences of genes that tend to be highly transcribed raised the possibility that part of the binding signals were because of nonspecific interactions of Sub1 with RNA transcripts (22). Although this hypothesis could not be ruled out, all in vitro data presented below strongly suggested that ChIP binding signals resulted from Sub1 association with DNA.
All together, our results suggested that Sub1 could be involved in all 3 transcription systems. In the following experiments, we focused on the role of Sub1 in Pol III transcription both in vitro and in vivo.
Sub1 Stimulates a Reconstituted Pol III Transcription System.
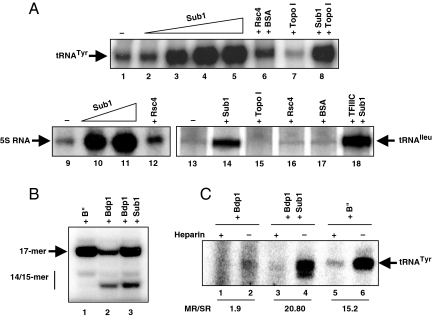
The association of Sub1 to all Pol III-transcribed genes in vivo and the previous observation of a stimulatory effect of PC4 on a human Pol III transcription system in vitro (13) prompted us to test the effect of Sub1 on in vitro transcription. A transcription system reconstituted with all recombinant factors and highly purified Pol III directs a low level of RNA synthesis and needs to be supplemented with partially purified fractions like B′′ (23) or TFIIIE (24, 25) to restore strong transcription rates (26), suggesting the existence of positive auxiliary factors enhancing Pol III transcription. As shown in Fig. 2A, the presence of recombinant Sub1 (Fig. S4A) in a transcription system reconstituted with recombinant factors strongly stimulated (4- to 6-fold) SUP4 tRNATyr, 5S RNA or tRNAIleu gene expression in contrast to any of the control proteins assayed (Rsc4, Topo I or BSA). Only specific Pol III transcription was stimulated because the presence of Sub1 did not change the nonspecific transcription activity of Pol III from DNA plasmid templates (Fig. S4C). Interestingly, Western blot analysis revealed the presence of Sub1 in the B″ fraction but not in the most purified TFIIIE fractions (Fig. S4B and ref. 27) showing that Sub1 could not account for TFIIIE activity.
Fig. 2.
Sub1 is a Pol III transcription activator in vitro. (A) Sub1 stimulates the minimal Pol III transcription system. In vitro transcription of the SUP4-tDNATyr gene, 5S RNA gene or tDNAIleu gene containing a TATA-box was carried out in the presence of rTBP, rBrf1, rBdp1, rTFIIIC (except for lanes 13–17), rTFIIIA (lanes 9–12 only), purified Pol III, varying amounts of purified rSub1 (lanes 2–5: 8, 24, 80, 240 ng; lane 8: 240 ng; lanes 10–11: 40, 160 ng; lanes 14, 18: 200 ng), or 200 ng of control proteins Rsc4, a recombinant protein purified under the same conditions than rSub1, BSA or Topo I, a single-stranded DNA binding protein like Sub1 that may play a role in human Pol III transcription (13). (B) Sub1 stimulates transcription initiation. Synthesis of the 17-mer RNA from the SUP4-tDNATyr template was carried out by using rTFIIIC, rTBP, rBrf1 and purified Pol III in the presence of rBdp1, B′′ or rSub1 (100 ng). The positions of the 14/15-mer and 17-mer transcripts are indicated. (C) Sub1 enhances Pol III transcription reinitiation. Preinitiation complexes were assembled on the SUP4-tDNATyr gene for 20 min in the presence of rTFIIIC, rTBP, rBrf1 and rBdp1 (lanes 1–4), or B′′ (lanes 5–6). rSub1 (100 ng) was added in lanes 3–4. Purified Pol III (10 ng) was then added together with a mixture lacking CTP and the incubation was continued for 20 min. Transcription was then resumed by the addition of CTP, either in the presence (+) or in the absence (−) of heparin, and the incubation was continued for 5 min. The ratios of multiple round (MR) versus single round (SR) of transcription are indicated.
A factor-independent in vitro transcription assay where Pol III autonomously initiates transcription at a 3′-end overhang of a linear DNA was then carried out, as described (28), to test whether Sub1 stimulation required the presence of transcription factors. Sub1 was able to stimulate Pol III transcription in this TFIIIB/IIIC-independent assay but only when Pol III was in limiting amounts (Fig. S5), suggesting that Sub1 stimulation of Pol III transcription might take place, at least in part, through its ability to directly facilitate the productive interaction of Pol III with DNA. This stimulation effect could not account, by itself, for the extent of stimulation by Sub1 in factor-dependent multiple round of transcription (Fig. 2A) where all the components were in saturating amounts.
Sub1 Stimulates Pol III Transcription Initiation in Vitro.
We examined the effect of Sub1 on transcription initiation in single round of transcription experiments using a SUP4 tDNA template under conditions that yield a 17-mer RNA (Fig. 2B). As described, several transcription products were observed by using rTFIIIB. The 17- and 14-mer correspond to different start sites whereas the 15-mer results from the cleavage of the 17-mer product (29, 30). In the presence of B′′ fraction, only the 17-mer RNA was observed and the transcription level was stimulated up to 3 times. In the presence of rSub1, a similar stimulation of the transcription was observed but Sub1 had no effect on start site selection, indicating that Sub1 directly affected the initiation efficiency of Pol III transcription.
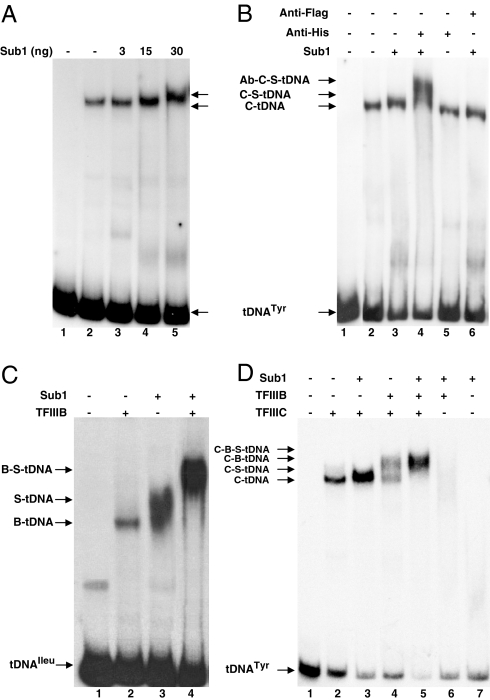
PC4 was found to enhance and extend the interactions of TFIIIC with tDNA (13, 18). We explored a possible role of Sub1 in the assembly of preinitiation complexes using gel shift assays. The addition of 3 or 15 ng of Sub1 was sufficient to promote the binding of a limiting amount of rTFIIIC (1–2 ng) to the SUP4 tRNATyr gene, as evidenced by the clear increase in signal intensity (Fig. 3A). With larger amounts of Sub1, the mobility of the TFIIIC-DNA complexes decreased. Because the amounts of unbound DNA remained largely unchanged, a specific association between TFIIIC and at least one molecule of Sub1 was indicated. As shown in Fig. 3B, the addition of antibodies raised against the histidine epitope further decreased the mobility of the DNA complexes, showing the presence of histidine-tagged Sub1 in the supershifted TFIIIC-DNA complexes.
Fig. 3.
Sub1 enhances the assembly of TFIIIC-TFIIIB-tDNA complexes. The positions of protein-tDNA complexes visualized by gel shift assays are indicated. Lane 1: control without protein. (A) Sub1 promotes the binding of TFIIIC to tDNA. Limiting amounts of rTFIIIC (C, 2 ng) were incubated in the presence of the indicated amounts of Sub1 (S) with the SUP4-tDNATyr probe. (B) Sub1 is recruited with TFIIIC on tDNA. Endogenous yeast TFIIIC was preincubated with the SUP4-tDNATyr probe for 10 min at 25 °C, alone or in the presence of 30 ng of Sub1 and then further incubated for 20 min at 25 °C with anti-histidine or anti-Flag monoclonal antibodies. (C) Sub1 stimulates the assembly of TFIIIB on tDNAIleu. rTFIIIB (B, 10 ng) and Sub1 (100 ng) were incubated as indicated with the tDNAIleu probe containing a TATA-box. (D) Sub1 helps TFIIIC to recruit TFIIIB on tDNA. rTFIIIC (10 ng), rTFIIIB (10 ng) or Sub1 (100 ng) were incubated as indicated with the SUP4-tDNATyr probe.
To test whether Sub1 could also influence the assembly of rTFIIIB into preinitiation complexes, we took advantage of the tRNAIleu (TAT) gene that can be transcribed in a TFIIIC-independent manner (31) and bound by rTFIIIB in gel shift assays (Fig. 3C). We also studied the recruitment of TFIIIB by TFIIIC to the SUP4 tRNATyr gene that results in the formation of TFIIIB-TFIIIC-DNA complexes of slower electrophoretic mobility as shown in Fig. 3D. In both cases, the addition of Sub1 strongly stimulated the binding of TFIIIB resulting in the formation of a larger amount of slow-migrating complexes clearly distinct from the nonspecific protein-DNA complexes obtained with Sub1 alone. The stimulation of TFIIIB assembly on tDNAIleu by Sub1 (Fig. 3C) was correlated with an at least 4-fold increase in the transcription levels observed in the presence of Sub1 using a minimal transcription system composed of rTFIIIB and Pol III (Fig. 2A, lanes 13–14). Furthermore, TFIIIC and Sub1 had additive stimulatory effects on transcription (Fig. 2A, lanes 14 and 18).
Sub1 Stimulates Pol III Transcription Reinitiation in Vitro.
We next wondered whether the effects of Sub1 on Pol III transcription resulted only from the stimulation of preinitiation complexes assembly. To address this question, we carried out a time course analysis of tRNAIleu synthesis (Fig. S4E) using our recombinant system in the presence or absence of Sub1. In both conditions, similar amounts of full-length transcripts could be visualized very rapidly (5 min), suggesting that the effects of Sub1 on transcription factors recruitment were not sufficient to explain the strong stimulation observed in multiple round of transcription (Fig. 2A). After 30 min of incubation, a strong accumulation of tRNAIleu transcripts was observed only when Sub1 was present. Under these conditions, both de novo complex formation and reinitiation were combined. Because the high levels of Pol III transcription come mainly from reinitiation efficiency (32), we thus examined whether Sub1 could promote Pol III transcription reinitiation.
The transcription initiation frequency was determined for the SUP4 tRNATyr gene (Fig. 2C) by comparing the output of multiple round versus single round transcription cycles performed with limiting amounts of Pol III, as described (33). The basal transcription system reconstituted with rTFIIIC could support only 1.9 cycle of transcription in 5 min. In contrast, more than 20 transcription cycles were obtained within 5 min in the presence of Sub1 or B′′ fraction. Similar results were obtained with the tRNAIleu gene (ratio of 1.2 and 19, Fig. S4D) showing that Sub1 played a critical role in the transcription reinitiation processes. Note that under limiting amounts of Pol III, part of the transcription stimulation could be due to a factor-independent effect of Sub1 on Pol III (Fig. S5).
Sub1 Interacts with TFIIIB and TFIIIC Components.
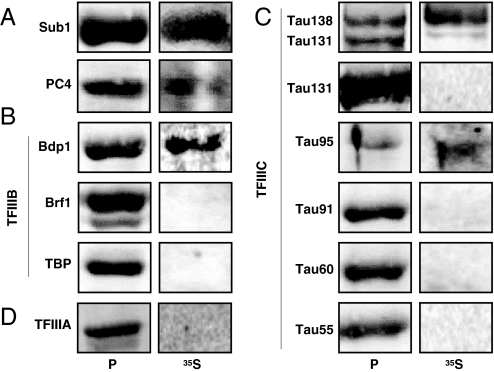
Based on gel shift assays (Fig. 3), our results suggested that Sub1 helps TFIIIB and TFIIIC to assemble on tRNA genes, possibly through direct protein–protein interactions with basal factors, at least in a DNA-dependent manner. Far Western experiments were performed as described (34) using 35S-labeled Sub1 as a probe (Fig. 4). As expected, because PC4 forms tightly associated homodimers (35), we found that Sub1 interacted with itself and with PC4 (Fig. 4A). Sub1 was also found to interact with Bdp1, a component of TFIIIB and with τ138 and τ95, 2 subunits of TFIIIC, in good agreement with the coimmunopurification of PC4 with human TFIIIC (13). No interaction was detected with TFIIIA. According to these data, the shifts seen in EMSA experiments (Fig. 3) after the addition of Sub1 may likely reflect both the binding of Sub1 to DNA in addition to direct interactions with TFIIIB and TFIIIC.
Fig. 4.
Sub1 interacts with components of the Pol III transcription machinery. rSub1 or rPC4 (A), subunits of rTFIIIB (B) or of rTFIIIC (C), or rTFIIIA (D) were subjected to SDS/PAGE, transferred onto a membrane, stained with Ponceau S (lanes P) and then probed with 35S-labeled Sub1 (lanes 35S). Labeled protein complexes were revealed by autoradiography.
Sub1 Is Required for Optimal Pol III Transcription in Vivo.
Because Sub1 acts as an activator of Pol III transcription in vitro, we next investigated whether the presence of Sub1 was important for Pol III transcription in vivo. There was no difference in the cell growth rate of exponentially growing sub1Δ cells as compared with wild type strain (8). Consistently, we could not detect any significant difference between both strains in the steady state levels of Pol III transcripts in exponentially growing cells (Fig. S6B Right). However, we did observe that sub1Δ crude extracts were systematically less efficient than wild type extracts in Pol III transcription as exemplified in Fig. S6A for the SUP4 tRNATyr gene (≈20% less RNA transcripts). The addition of Sub1 stimulated the transcription in both sub1Δ and wild type extracts. However, the 20% lower activity of the sub1Δ extracts was too modest for further analysis.
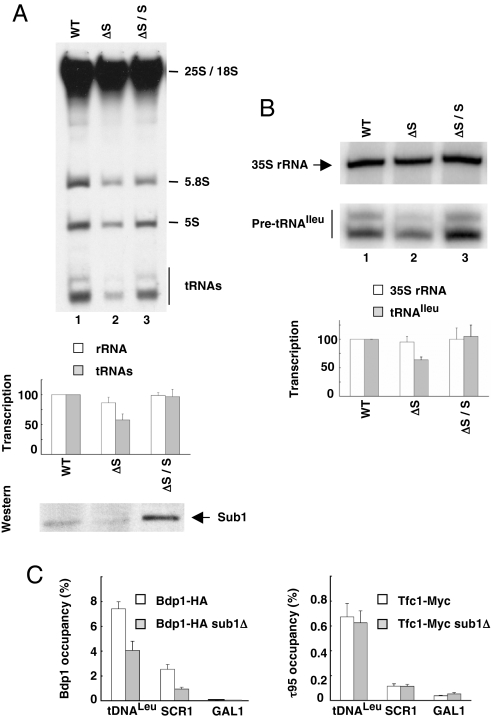
In good accordance with these results, pulse labeling experiments performed on exponentially growing cells revealed that Pol III transcription was decreased in vivo in the absence of Sub1. The level of tRNA gene transcription was ≈40% lower in sub1Δ cells as compared with wild type whereas the level of Pol I transcription measured by the neosynthesis of 25S and 18S RNA was not significantly changed (Figs. 5A and S6B Left). The defect in Pol III transcription was due to the absence of Sub1, because wild type levels of Pol III transcription were restored when Sub1 was expressed in sub1Δ cells from a centromeric plasmid (Fig. 5A Bottom). Primer extension analysis was performed on total RNA prepared from sub1Δ cells (Fig. 5B). Once again, whereas the levels of 35S RNA neosynthesis were not significantly altered, a ≈35% decrease of tRNAIleu (TAT) neosynthesis was observed in sub1Δ cells and wild type transcription levels were restored in sub1Δ cells expressing Sub1 from a centromeric plasmid.
Fig. 5.
Sub1 is an activator of Pol III transcription in exponentially growing cells. (A) Pol III transcription is less efficient in sub1Δ strain. Three micrograms of total labeled RNA prepared from WT, sub1Δ (ΔS) or sub1Δ/pCM-SUB1 cells (sub1Δ transformed with a centromeric plasmid harbouring SUB1, ΔS/S) were analyzed by electrophoresis and fluorography. (Top) The positions of labeled 25S, 18S and 5.8S RNA transcribed by Pol I, and of 5S RNA and tRNAs transcribed by Pol III are indicated. (Middle) Quantitation of Pol I (25S + 18S rRNA) and Pol III (tRNAs) labeled transcripts in ΔS or ΔS/S cells are represented as histograms and as a percentage of their transcription in WT cells. Error bars represent the standard deviation between three independent experiments. (Bottom) The amounts of Sub1 in crude extracts (5 μg) were analyzed by Western blot analysis with antibodies directed to Sub1. (B) The neosynthesis of tRNAIleu is decreased in sub1Δ cells. (Upper) The amounts of neosynthesized 35S rRNA or pretRNAIleu were determined by primer extension analysis on 4 μg of total RNA. (Lower) Quantitation of neosynthesized 35S rRNA or pretRNAIleu in sub1Δ or sub1Δ/pCM-SUB1 cells is represented as histograms and as a percentage of their transcription in WT cells. Error bars represent the standard deviation between three independent experiments. (C) Less TFIIIB factor is associated to its gene targets in sub1Δ cells. ChIP assays were performed on WT or sub1Δ cells by using antibodies against HA (for Bdp1–3HA) or Myc (for Tfc1–13Myc) epitope and analyzed by real-time PCR on selected genes. Error bars represent the standard deviation between at least three independent replicates.
We next wondered whether the decrease in Pol III transcription in sub1Δ cells was correlated to a lower occupancy of Pol III-transcribed genes by the transcription machinery. To address this question, cross-linked chromatin extracts were prepared from sub1Δ strains expressing τ95–13myc or Bdp1–3HA. ChIP experiments were performed by using antibodies directed to the epitopes and analyzed by real-time PCR on selected Pol III-transcribed genes. As shown Fig. 5C, whereas TFIIIC binding was unchanged, less TFIIIB was present on the SCR1 or tRNALeu genes when SUB1 was deleted. In good agreement with our in vitro data, these results suggested that Sub1 may help TFIIIC to efficiently recruit TFIIIB on DNA.
Discussion
In this work, we demonstrate that Sub1 is present on Pol III-transcribed genes. We find that Sub1 stimulates in vitro Pol III transcription at two discrete steps of the transcription cycle. Although Sub1 is not an essential protein in exponentially growing cells, we show that its absence in vivo correlates with a decrease in Pol III transcription efficiency and with lower levels of TFIIIB associated with two of its target genes.
Sub1 Stimulates Two Steps of the Pol III Transcription Cycle.
In this work, we provide biochemical evidence that Sub1 strongly stimulates in vitro transcription by Pol III, in agreement with the stimulation of human Pol III transcription by PC4 (13, 18). Using a transcription system reconstituted with all recombinant factors and highly purified Pol III, we showed that recombinant Sub1 stimulated both the initiation and reinitiation steps of transcription. Sub1 was determined to promote the binding of both TFIIIB and TFIIIC to their cognate sites, likely through direct TFIIIB and TFIIIC interactions, resulting in increased levels of Pol III transcription. However, the main effect of Sub1 was to relieve the Pol III reinitiation defect observed when using recombinant factors. Therefore, Sub1 can be considered as a Pol III reinitiation factor. Although its exact function in that transcription step remains to be determined, one may hypothesize that Sub1 could help Pol III to be directly transferred from the terminator to the start site of transcription as it has been proposed in the facilitated recycling pathway (36).
In all our in vitro assays, optimal stimulations required higher molar amounts of Sub1 relative to that of TFIIIC, TFIIIB or Pol III. Several characteristics of Sub1 might contribute to this phenomenon. First, incomplete or inappropriate modification of Sub1 residues because of its expression in insect cells might decrease its activity (for example, acetylation of PC4 has been shown to interfere with its DNA binding activity, ref. 37). Second, because Sub1 was found to bind both single- and double-stranded DNA (8), a substantial amount of the protein could be sequestered by the nonspecific DNA present in the reaction mixtures. Last, Sub1 might bind to DNA as a dimer or even multimerize along the DNA as suggested for PC4 (35), as a prerequisite for productive interaction with the components of the Pol III transcription machinery on DNA.
Sub1 Is Required for Optimal Pol III Transcription in Exponentially Growing Cells.
Our data imply that Sub1 is a novel Pol III transcriptional activator in vivo. The modest but reproducible decreased efficiency of Pol III transcription in sub1Δ cells suggests that Sub1 is necessary to sustain wild type levels of Pol III transcription under normal growth conditions. The lower transcription efficiency was correlated to a reduced association of Bdp1, a TFIIIB subunit, with Pol III-transcribed genes. These data are in good agreements with a direct role of Sub1 on transcription initiation in vivo through the efficient recruitment of TFIIIB by TFIIIC on DNA.
However, we could not detect any significant difference in the steady-state levels of Pol III transcripts in the absence of Sub1 (Fig. S6B). Consistently, SUB1 is a nonessential gene and its deletion does not interfere with the cell growth rate in exponentially growing cells (1, 8). All these results suggest the existence of regulatory mechanisms that compensate forthe decreased neosynthesis of Pol III transcripts caused by the absence of Sub1 to maintain a wild type pool of tRNA in the cell.
Our genome-wide occupancy studies suggest that Sub1 could play a more general role in transcription than anticipated. Our data demonstrated that Sub1 is not restricted to the Pol II transcription system, but is also involved in Pol III transcription. A function of Sub1 in Pol I transcription remains an open possibility but our preliminary in vivo pulse labeling and primer extension analysis provided no evidence for such a role, at least in exponentially growing cells.
The role of Sub1 is also likely to be more complex than a straightforward activation function. Both PC4 and Sub1 were recently shown to play a negative role in Pol II transcription in vivo. Knocking down PC4 expression using siRNA altered the expression of less than 200 genes, most of them being up-regulated (14), whereas disruption of SUB1 increased IMD2 transcription (38) showing that PC4/Sub1 could also repress the expression of some genes under normal growth conditions. Like the NC2 cofactor (39) or Mot1 (40), Sub1 may thus play both a positive and a negative role in transcription.
Finally, previous studies suggested that Sub1 may have some physiological importance under suboptimal growth conditions. For instance, it has been reported that the expression of Sub1 mRNA rapidly increases after transfer of quiescent yeast cells in rich medium (20). It would be interesting to analyze the possible role of Sub1 in reactivating transcription during recovery from poor growth conditions that were found to repress transcription (nutrients or serum starvation, mitotic repression, secretory pathway defects, oxydative stress, DNA damages, chemical treatments with diverse drugs). Furthermore, Sub1 has been shown to be required for resistance to the oxidizing agents tert-butyl hydroperoxide or hydrogen peroxide (1, 41) suggesting a role of Sub1 in the stress response. Further investigations should be performed to characterize conditions where Sub1 is necessary for cellular growth and to determine whether and by which molecular mechanisms Pol III transcription is regulated by Sub1 in such conditions.
Materials and Methods
Yeast Strains.
All strains used in this study are described in SI Text and Table S1.
ChIP, Microarray Hybridization, and Data Analysis.
ChIP and PCR were performed as described (15). The sequences of the oligonucleotides used in this study are available upon request. For ChIP on chip experiments, DNA from wild type or 3HA-Sub1 strains were competitively hybridized to DNA microarrays harbouring 14,172 yeast ORF and intergenic regions (15, 21). Data from 3 independent experiments were compiled. Data analysis are described in SI Text. The complete raw dataset and analyzed data are available at www.ncbi.nlm.nih.gov/geo (GEO accession no. GSE11054) in SI, Datasets S1 and S2.
In Vitro Transcription Assays.
Transcriptions were performed as described in Ducrot et al. (26) with 10 ng of rTFIIIC, 20 ng of rTBP, 10 ng of rBrf1, 10 ng of rBdp1 or 0.5 μg of partially purified B“ fraction, 100 ng of highly purified Pol III and 40 ng of rTFIIIA when 5S DNA template was used. Facilitated transcription reinitiation assays were performed as described (33). Specific transcripts were visualized with a Typhoon 9200 Imager (Amersham Biosciences). Quantitation was performed by using the Quantity one software (Bio-Rad). See also SI Text.
Supplementary Material
Acknowledgments.
We are grateful to A. Sentenac for helpful discussions and for improving the manuscript and N. Caudy for technical assistance. We thank M. Teichmann (Institut Européen de Chimie et Biologie, Université Bordeaux, Bordeaux, France) for the generous gift of purified PC4 and the pet-PC4 expression plasmid, Franck Amiot, Peggy Maltere, Amélie Robert for providing the DNA chip (CEA, IRCM) and N. Alic for help in 17-mer assay. This work was funded by grant ANR-07-BLAN-0039–01 from the French National Research Agency and by grant 1078 from the Association pour la Recherche contre le Cancer. A.T. was supported by the International PhD Program of the Commissariat à l'Energie Atomique..
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900162106/DCSupplemental.
References
- 1.Wang JY, Sarker AH, Cooper PK, Volkert MR. The single-strand DNA binding activity of human PC4 prevents mutagenesis and killing by oxidative DNA damage. Mol Cell Biol. 2004;24:6084–6093. doi: 10.1128/MCB.24.13.6084-6093.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ge H, Roeder RG. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 3.Pan ZQ, Ge H, Amin AA, Hurwitz J. Transcription-positive cofactor 4 forms complexes with HSSB (RPA) on single-stranded DNA and influences HSSB-dependent enzymatic synthesis of simian virus 40 DNA. J Biol Chem. 1996;271:22111–22116. doi: 10.1074/jbc.271.36.22111. [DOI] [PubMed] [Google Scholar]
- 4.Kretzschmar M, Kaiser K, Lottspeich F, Meisterernst M. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 1994;78:525–534. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- 5.Malik S, Guermah M, Roeder RG. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proc Natl Acad Sci USA. 1998;95:2192–2197. doi: 10.1073/pnas.95.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuda A, et al. Transcriptional coactivator PC4 stimulates promoter escape and facilitates transcriptional synergy by GAL4-VP16. Mol Cell Biol. 2004;24:6525–6535. doi: 10.1128/MCB.24.14.6525-6535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Werten S, et al. Interaction of PC4 with melted DNA inhibits transcription. EMBO J. 1998;17:5103–5111. doi: 10.1093/emboj/17.17.5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henry NL, Bushnell DA, Kornberg RD. A yeast transcriptional stimulatory protein similar to human PC4. J Biol Chem. 1996;271:21842–21847. doi: 10.1074/jbc.271.36.21842. [DOI] [PubMed] [Google Scholar]
- 9.Knaus R, Pollock R, Guarente L. Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J. 1996;15:1933–1940. [PMC free article] [PubMed] [Google Scholar]
- 10.Calvo O, Manley JL. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 2005;24:1009–1020. doi: 10.1038/sj.emboj.7600575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvo O, Manley JL. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol Cell. 2001;7:1013–1023. doi: 10.1016/s1097-2765(01)00236-2. [DOI] [PubMed] [Google Scholar]
- 12.He X, et al. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 2003;17:1030–1042. doi: 10.1101/gad.1075203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Roeder RG. DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol Cell. 1998;1:749–757. doi: 10.1016/s1097-2765(00)80074-x. [DOI] [PubMed] [Google Scholar]
- 14.Das C, et al. Transcriptional coactivator PC4, a chromatin-associated protein, induces chromatin condensation. Mol Cell Biol. 2006;26:8303–8315. doi: 10.1128/MCB.00887-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harismendy O, et al. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 2003;22:4738–4747. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nedea E, et al. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem. 2003;278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- 17.Yang PK, et al. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol Cell Biol. 2005;25:3295–3304. doi: 10.1128/MCB.25.8.3295-3304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mertens C, Roeder RG. Different functional modes of p300 in activation of RNA polymerase III transcription from chromatin templates. Mol Cell Biol. 2008;28:5764–5776. doi: 10.1128/MCB.01262-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghavi-Helm Y, et al. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev. 2008;22:1934–1947. doi: 10.1101/gad.471908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radonjic M, et al. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Oficjalska-Pham D, et al. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol Cell. 2006;22:623–632. doi: 10.1016/j.molcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kassavetis GA, et al. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992;71:1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- 24.Dieci G, et al. A novel RNA polymerase III transcription factor fraction that is not required for template commitment. J Biol Chem. 1993;268:11199–11207. [PubMed] [Google Scholar]
- 25.Ruth J, et al. A suppressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J. 1996;15:1941–1949. [PMC free article] [PubMed] [Google Scholar]
- 26.Ducrot C, et al. Reconstitution of the yeast RNA polymerase III transcription system with all recombinant factors. J Biol Chem. 2006;281:11685–11692. doi: 10.1074/jbc.M600101200. [DOI] [PubMed] [Google Scholar]
- 27.Dieci G, et al. Positive modulation of RNA polymerase III transcription by ribosomal proteins. Biochem Biophys Res Commun. 2009;379:489–493. doi: 10.1016/j.bbrc.2008.12.097. [DOI] [PubMed] [Google Scholar]
- 28.Ferrari R, Dieci G. The transcription reinitiation properties of RNA polymerase III in the absence of transcription factors. Cell Mol Biol Lett. 2008;13:112–118. doi: 10.2478/s11658-007-0041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrau JC, Werner M. B′′ associated factor (s) involved in RNA polymerase III preinitiation complex formation and start-site selection. Eur J Biochem. 2001;268:5167–5175. doi: 10.1046/j.0014-2956.2001.02445.x. [DOI] [PubMed] [Google Scholar]
- 30.Chedin S, et al. The yeast RNA polymerase III transcription machinery: A paradigm for eukaryotic gene activation. Cold Spring Harb Symp Quant Biol. 1998;63:381–389. doi: 10.1101/sqb.1998.63.381. [DOI] [PubMed] [Google Scholar]
- 31.Dieci G, et al. TFIIIC-independent in vitro transcription of yeast tRNA genes. J Mol Biol. 2000;299:601–613. doi: 10.1006/jmbi.2000.3783. [DOI] [PubMed] [Google Scholar]
- 32.Dieci G, Sentenac A. Facilitated recycling pathway for RNA polymerase III. Cell. 1996;84:245–252. doi: 10.1016/s0092-8674(00)80979-4. [DOI] [PubMed] [Google Scholar]
- 33.Ferrari R, Rivetti C, Acker J, Dieci G. Distinct roles of transcription factors TFIIIB and TFIIIC in RNA polymerase III transcription reinitiation. Proc Natl Acad Sci USA. 2004;101:13442–13447. doi: 10.1073/pnas.0403851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaussivert N, Conesa C, Shaaban S, Sentenac A. Complex interactions between yeast TFIIIB and TFIIIC. J Biol Chem. 1995;270:15353–15358. doi: 10.1074/jbc.270.25.15353. [DOI] [PubMed] [Google Scholar]
- 35.Werten S, Moras D. A global transcription cofactor bound to juxtaposed strands of unwound DNA. Nat Struct Mol Biol. 2006;13:181–182. doi: 10.1038/nsmb1044. [DOI] [PubMed] [Google Scholar]
- 36.Dieci G, Sentenac A. Detours and shortcuts to transcription reinitiation. Trends Biochem Sci. 2003;28:202–209. doi: 10.1016/S0968-0004(03)00054-9. [DOI] [PubMed] [Google Scholar]
- 37.Kumar BR, Swaminathan V, Banerjee S, Kundu TK. p300-mediated acetylation of human transcriptional coactivator PC4 is inhibited by phosphorylation. J Biol Chem. 2001;276:16804–16809. doi: 10.1074/jbc.M100934200. [DOI] [PubMed] [Google Scholar]
- 38.Koyama H, et al. Transcriptional repression of the IMD2 gene mediated by the transcriptional co-activator Sub1. Genes Cells. 2008;13:1113–1126. doi: 10.1111/j.1365-2443.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- 39.Willy PJ, Kobayashi R, Kadonaga JT. A basal transcription factor that activates or represses transcription. Science. 2000;290:982–985. doi: 10.1126/science.290.5493.982. [DOI] [PubMed] [Google Scholar]
- 40.Dasgupta A, et al. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc Natl Acad Sci USA. 2002;99:2666–2671. doi: 10.1073/pnas.052397899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol Cancer Res. 2002;1:103–112. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.