Summary
PPARα is activated by drugs to treat human disorders of lipid metabolism. Its endogenous ligand is unknown. PPARα-dependent gene expression is impaired with inactivation of fatty acid synthase (FAS), suggesting that FAS is involved in generation of a PPARα ligand. Here we demonstrate the FAS-dependent presence of a phospholipid bound to PPARα isolated from mouse liver. Binding was increased under conditions that induce FAS activity and displaced by systemic injection of a PPARα agonist. Mass spectrometry identified the species as 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (16:0/18:1-GPC). Knockdown of CEPT1, required for phosphatidylcholine synthesis, suppressed PPARα-dependent gene expression. Interaction of 16:0/18:1-GPC with the PPARα ligand binding domain and co-activator peptide motifs was comparable to PPARα agonists, but interactions with PPARδ were weak and none were detected with PPARγ. Portal vein infusion of 16:0/18:1-GPC induced PPARα-dependent gene expression and decreased hepatic steatosis. These data suggest that 16:0/18:1-GPC is a physiologically relevant endogenous PPARα ligand.
Introduction
Peroxisome proliferator-activated receptors (PPARs) constitute a subfamily of nuclear receptors with three current members: PPARα, PPARγ, and PPARδ. Each receptor appears to modulate pathways at the interface between intermediary metabolism and inflammation, making them physiologically and clinically relevant (Bensinger and Tontonoz, 2008). PPARα is activated by fibrate drugs to lower triglycerides and raise HDL (Barter and Rye, 2008), PPARγ is targeted by glitazones to treat diabetes (Yki-Jarvinen, 2004), and pharmacological activation of PPARδ appears to improve several metabolic parameters in humans (Riserus et al., 2008). PPARs are ligand-activated receptors that heterodimerize with RXR, bind to response elements in target genes, and alter co-activator/co-repressor dynamics to induce transcription. Fatty acids, especially if polyunsaturated, are thought to be preferred PPAR ligands, but a wide variety of lipids (Forman et al., 1997; Kliewer et al., 1997; Krey et al., 1997; Yu et al., 1995) have been implicated in PPAR activation including saturated fatty acids, fatty acyl-CoA species, eicosanoids (prostaglandins, leukotrienes, and HETEs), oxidized fatty acids, and oxidized phospholipids. No endogenous PPAR ligand has been identified.
PPARα is expressed at high levels in liver where it promotes fatty acid oxidation, ketogenesis, lipid transport, and gluconeogenesis (Bernal-Mizrachi et al., 2003; Reddy and Hashimoto, 2001). Systemic levels of free fatty acids change with nutritional status, making PPARα an attractive candidate sensor of energy balance that might respond to fatty acids by accelerating their metabolism. This would require that PPARα, a nuclear protein, be exposed to concentrations of fatty acids that reflect those outside the cell. A simple diffusion gradient for these lipids in the hepatocyte is unlikely to exist since fatty acids crossing the plasma membrane undergo addition of an acyl-CoA group, leading to a myriad of potential fates including storage in lipid droplets, synthesis of phospholipids, incorporation into intracellular organelles or the external plasma membrane, transport into mitochondria for beta oxidation, association with the ER/Golgi for lipoprotein assembly, and others. This scheme suggests that fatty acids are chaperoned from the extracellular environment to the nucleus to activate PPARα, or that another entity reflecting nutritional status is involved in generating the endogenous ligand.
No chaperone has emerged for the PPARα ligand. FABP4, which binds fatty acids, has been implicated in shuttling ligand to another family member, PPARγ (Ayers et al., 2007). FABP1, an abundant protein in liver that binds a broad range of fatty acids, might serve a similar function for PPARα. But PPARα-dependent genes are expressed appropriately in FABP1 null mice (Newberry et al., 2003), indicating that FABP1 probably does not present ligand to PPARα.
There is evidence that a nutritionally responsive entity may generate the endogenous PPARα ligand. Fatty acid synthase (FAS) catalyzes the first committed step in fatty acid biosynthesis, utilizing acetyl-CoA, malonyl-CoA and NADPH to generate mostly the saturated fatty acid palmitate (Semenkovich, 1997). Liver-specific inactivation of FAS results in mice with decreased PPARα-dependent gene expression and a phenotype resembling PPARα deficiency (Chakravarthy et al., 2005). The phenotype is reversed and gene expression rescued after pharmacologic activation of PPARα. This phenomenon is not limited to the liver. Selective inactivation of FAS in the hypothalamus impairs PPARα-dependent gene expression and alters feeding behavior (Chakravarthy et al., 2007). Both are corrected after hypothalamic infusion of a PPARα activator. It thus appears that FAS, known to be regulated by nutrition, is required in some tissues to generate the endogenous ligand for PPARα.
To characterize that ligand, we developed a strategy based on purifying a tagged PPARα molecule from the livers of mice with or without expression of FAS. This strategy was used to test the hypothesis that de novo lipid biosynthesis generates a physiologically relevant endogenous ligand for PPARα.
Results
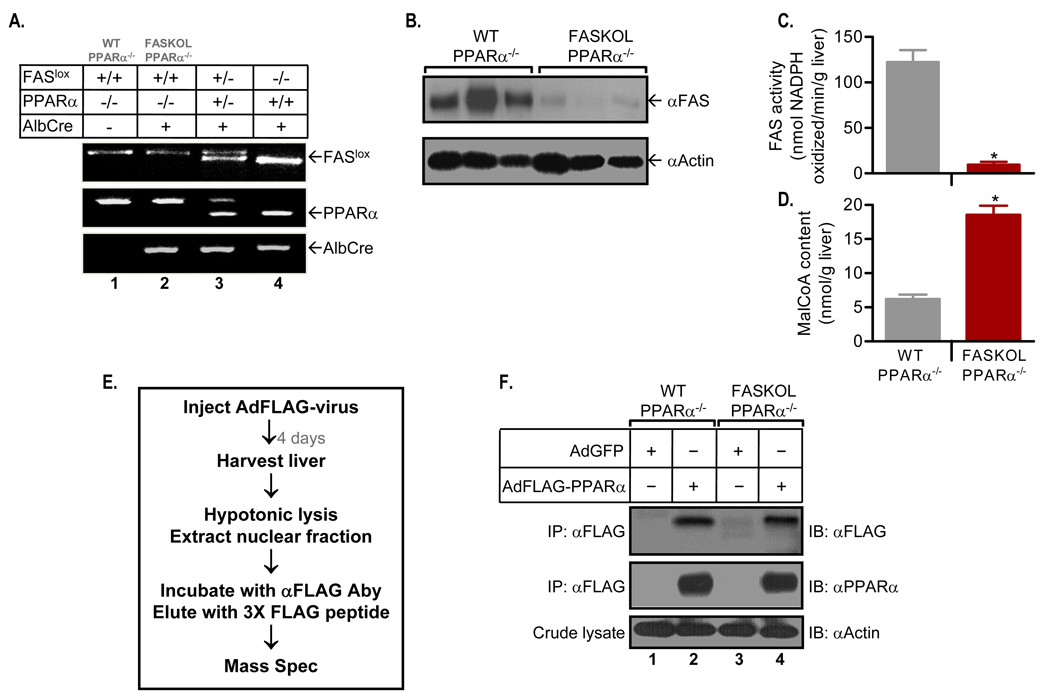
Fatty Acid Synthase KnockOut in Liver (FASKOL) mice have impaired PPARα-dependent gene expression that is rescued after pharmacological activation of PPARα (Chakravarthy et al., 2005). These mice were crossed with PPARα null mice to eliminate the possibility of ligand competition between adenovirally transduced PPARα and endogenous PPARα. Figure 1A shows representative PCR genotyping assays for mice wild type at the PPARα locus (lane 4), PPARα heterozygotes (lane 3), and PPARα-deficient mice used for subsequent experiments with FAS expression (lane 1, WT PPARα−/− ) or without FAS expression (lane 2, FASKOL PPARα−/− ). The fidelity of the FAS knockout in the PPARα background was verified by demonstrating decreased FAS protein (Figure 1B) and enzyme activity (Figure 1C) in the livers of FASKOL mice. The FAS substrate malonyl-CoA accumulated in FASKOL livers (Figure 1D), confirming decreased enzyme activity.
Figure 1. Generation of Liver-specific FAS knockout (FASKOL) Mice on a PPARα null Background and Reconstitution of Liver PPARα Expression.
(A) PCR analysis. Liver DNA was amplified using primer sets for the FAS floxed allele (top panel), PPARα (middle panel), and Cre (bottom panel).
(B) Immunoblot analysis of liver lysates for wild type (WT) and FASKOL mice on a PPARα null background using FAS (top panel) and actin (bottom panel) antibodies.
(C and D) FAS activity (C) and malonyl-CoA content (D). Liver homogenates from overnight-fasted 12 h chow-refed male WT and FASKOL mice on a PPARα null background were assayed. Each bar represents mean ± SEM of 6–8 mice of each genotype. *, P < 0.05.
(E) Diagram for isolation of FLAG-tagged PPARα.
(F) Immunoprecipitation (IP) and immunoblot (IB) analysis in livers of WT and FASKOL mice on a PPARα null background infected with adenoviruses encoding GFP alone (AdGFP) or FLAG-tagged PPARα (AdFLAG-PPARα). Nuclear fractions were immunoprecipitated with anti-FLAG antibodies and immunoblotted with either anti-FLAG antibody (top panel) or anti-PPARα antibody (middle panel). Crude liver lysates were immunoblotted with anti-actin antibody (bottom panel). Blots are representative of >12 independent experiments.
Figure 1E depicts our strategy for detecting the endogenous PPARα ligand. Mice were treated with an adenovirus directing expression of a FLAG-tagged PPARα under conditions that do not induce appreciable liver inflammation as reflected by normal liver function tests (Supplemental Table 1). Our reconstitution method has been shown to achieve levels of PPARα comparable to wild type mice (Bernal-Mizrachi et al., 2003). Reconstitution was followed by affinity-capture (utilizing an antibody recognizing the FLAG epitope) of PPARα under conditions (no detergent or high salt elution buffers) unlikely to disrupt the ligand/nuclear factor interaction. This yielded a dominant PPARα band on protein-stained gels (not shown). Figure 1F shows affinity matrix eluates subjected to immunoprecipitation followed by immunoblotting. These results confirm the absence of endogenous PPARα (lanes 1 and 3) and indicate that the yield of tagged PPARα from liver was similar in mice with and without expression of FAS (lanes 2 and 4).
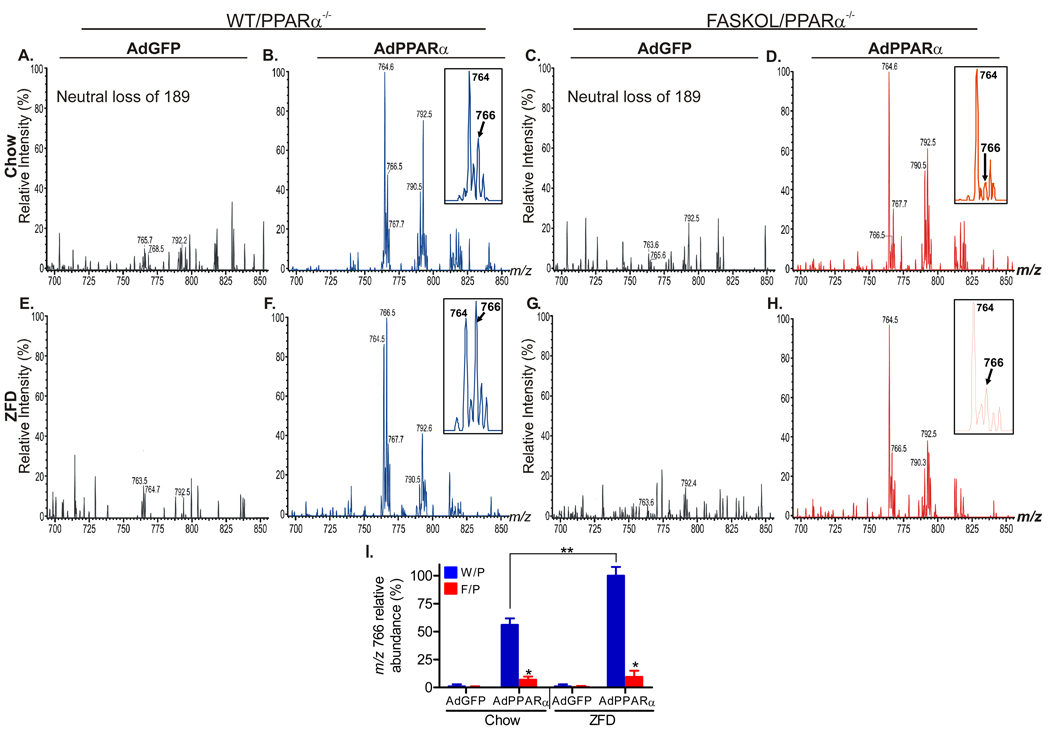
Affinity matrix eluates (with equal protein content) of nuclear fractions from mice treated with AdGFP (as a control) or AdPPARα in the presence (WT/ PPARα−/− ) or absence (FASKOL/ PPARα−/− ) of FAS were subjected to mass spectrometric analysis of extracted lipids. No appreciable lipid signal was detected from GFP eluates (Figure 2A, C, E, G for a portion of the phospholipid spectra; triglyceride and fatty acid signals were also essentially absent, data not shown), suggesting that nonspecific binding of lipids to GFP protein was minimal.
Figure 2. Identification of a Glycerophosphocholine (GPC) Species in FLAG-eluted Hepatic Nuclear Extracts.
Positive ion ESI/MS analyses of lithiated adducts of hepatic nuclear phospholipids were performed to monitor neutral loss of 189 [LiPO4(CH2)2N(CH3)3], which identifies parent [MLi+ ]ions that contain the phosphocholine head-group in lipid mixtures.
(A–D) Representative profiles of GPC species in chow fed WT and FASKOL mice on a PPARα null background infected with AdGFP (A and C) or AdFLAG-PPARα (B and D).
(E–H) Representative profiles of GPC species in zero fat diet (ZFD) fed WT and FASKOL mice on a PPARα null background infected with AdGFP (E and G) or AdFLAG-PPARα (F and H). Insets in B, D, F, and H depict the fragment ion at mass-to-charge ratio (m/z) 766 as the specific GPC species that is both PPARα and FAS dependent.
(I) Quantification of the relative abundance of the m/z 766 ion with respect to genotype (W/P, WT on PPARα null−/− background; F/P, FASKOL on PPARα null−/− background) and diet (chow and ZFD). Each bar represents mean ± SEM from 3 independent experiments with 4–6 mice in each group per experiment. *, P < 0.05 vs. corresponding W/P control; **, P < 0.05.
We did not detect fatty acids or triglycerides bound to PPARα that were consistent with an FAS-dependent ligand. FAS dependence was assessed by isolating PPARα from mice with and without FAS deficiency and from animals fed chow as well as a high carbohydrate, zero fat diet (ZFD), since carbohydrates induce FAS expression. PPARα triglyceride binding was increased in both wild type and FAS-deficient livers (Supplemental Figure 1), and this was more pronounced in the latter, consistent with the hepatic steatosis that occurs with zero fat diet feeding in FASKOL mice. There were no genotype or diet effects on binding of PPARα to any molecular species of phosphatidylethanolamine or phosphatidylinositol (Supplemental Figure 2), common fatty acids (Supplemental Figure 3), or lysophosphatidylcholine (Supplemental Figure 4).
Lipid analyses revealed only one peak that was FAS-dependent. It was observed in positive ion phospholipid spectra. Material represented by a peak with mass to charge ratio (m/z) of 766.5 bound to PPARα purified from livers expressing FAS (arrow, Figure 2B) and its abundance was significantly decreased in PPARα purified from FAS-deficient livers (arrow, Figure 2D). The abundance of m/z 766.5 increased in PPARα purified from WT mice fed a diet high in carbohydrates (ZFD) (arrow, Figure 2F, compare m/z 766 to m/z 764 in Figure 2F and Figure 2B) but remained low with FAS deficiency (arrow, Figure 2H). Quantitation of relative peak abundance in independent experiments is presented in Figure 2I. Increased association of the material represented by the ion at m/z 766 with PPARα after high carbohydrate feeding and its decreased detection with FAS deficiency indicate that this material is FAS-related.
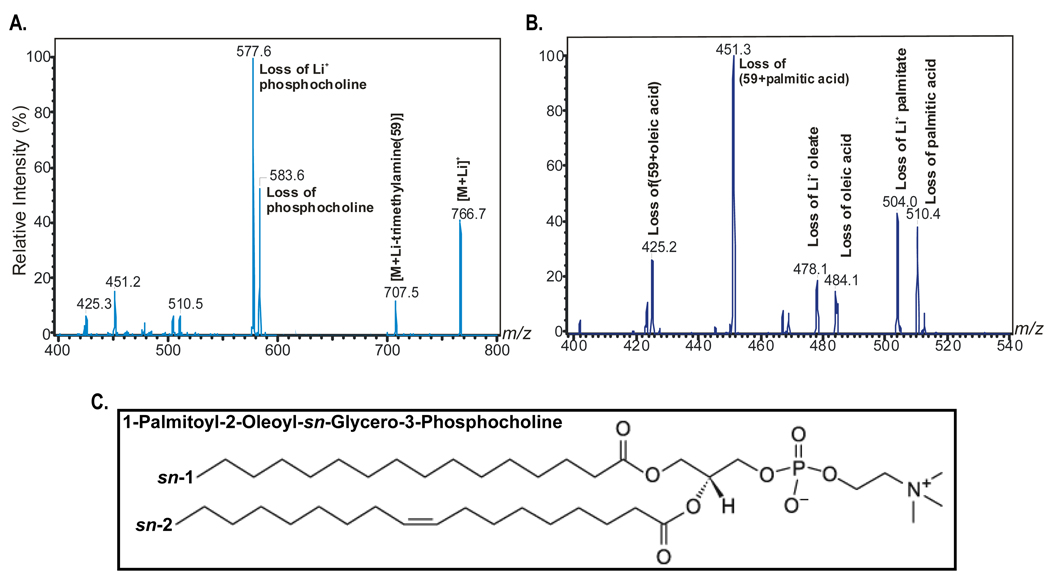
Tandem mass spectrometry (Figure 3) identified this material as the phospholipid molecular species 1-palmitoyl-2-oleoly-sn-glycerol-3-phosphocholine (16:0/18:1-GPC). Figure 3A illustrates the fragmentation pattern upon collisionally-activated dissociation of the ion of m/z 766, which corresponds to the lithiated adduct [MLi+ ] of 16:0/18:1-GPC Li+ permits the formation of complexes with informative fragmentation patterns). Neutral loss of trimethylamine [MLi+ - 59] yields an ion at m/z 707. Ions at m/z 583 [MLi+ - 183] and m/z 577 [MLi+ - 189] reflect net loss of [HPO4(CH2)2N(CH3)3] and of [LiPO4(CH2)2N(CH3)3], respectively. Losses of 59, 183, and 189 are common to the tandem spectra of all GPC-Li+ species, so these ions identify the phosphocholine head-group. The spectrum in Figure 3B contains ions at m/z 510, m/z 504, and m/z 451 that reflect neutral losses of palmitic acid [MLi+ - 256], the lithium salt of palmitate [MLi+ - 262], and trimethylamine plus palmitic acid [MLi+ - 315], respectively. The spectra also contain ions reflecting losses of oleic acid (m/z 484), the lithium salt of oleate (m/z 478), and trimethylamine plus oleic acid (m/z 425). Relative abundances of the ions at m/z 425 [MLi+ - (59 oleic acid)] and 451 [MLi+ - (59 + palmitic acid)] in Figure 3B indicate that palmitate and oleate are the sn-1 and sn-2 substituents, respectively, because the abundance of the ion reflecting loss of trimethylamine plus the sn-1 substituent always exceeds that of the ion reflecting loss of trimethylamine plus the sn-2 substituent (Hsu et al., 1998). Figure 3C shows a diagram of the putative PPARα ligand based on these mass spectra.
Figure 3. Tandem Mass Spectrometry Identifies the GPC species as 1-palmitoyl-2-oleoly-sn-glycerol-3-phosphocholine (16:0/18:1-GPC).
(A) Fragmentation pattern upon collisionally-activated dissociation of the ion of m/z 766, which corresponds to the lithiated adduct [MLi+] of 16:0/18:1-GPC.
(B) Expansion of the mass spectrum in A from m/z 400 to m/z 540 to illustrate relative abundances of ions that represent losses of fatty acid substitutents. The data indicate that palmitate and oleate are the sn-1 and sn-2 substituents, respectively.
(C) Structure of the putative PPARα ligand.
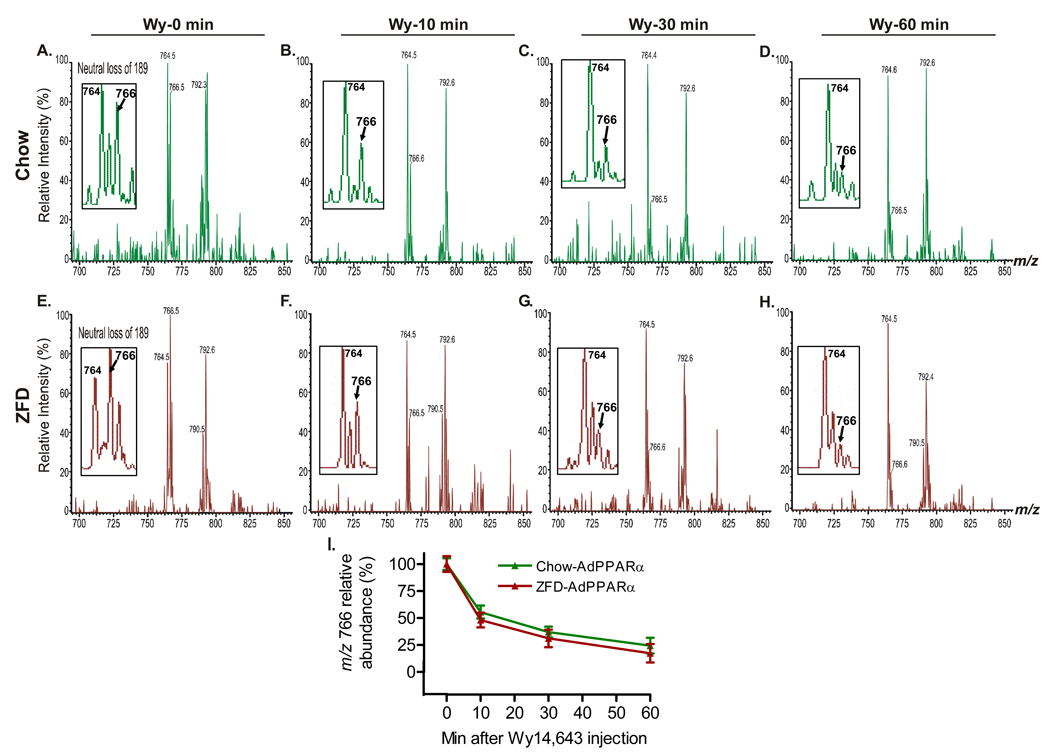
If 16:0/18:1-GPC is an endogenous PPARα ligand, it should be possible to competitively inhibit its binding with a known ligand in living mice. This was demonstrated by administering 50 µg/g of Wy14,643, a dose known to rapidly activate PPARα (Chakravarthy et al., 2005), at time 0, followed by isolation of PPARα-bound lipids from the mouse livers. The putative ligand was displaced from PPARα within minutes of treatment with the known agonist, both in the setting of chow (Figure 4A–D) and zero fat diet feeding (Figure 4E–H). Note the rapid decrease in abundance for m/z 766 (representing 16:0/18:1-GPC) relative to the invariant m/z 764 (16:0/18:2-GPC) and the increased abundance of m/z 766 at 0 minutes with zero fat diet compared to chow diet. Quantitation of relative peak abundance for independent experiments is presented in Figure 4I.
Figure 4. In vivo Displacement of the Endogenous PPARα Ligand with a PPARα Agonist Li+ adducts of GPC molecular species in excess FLAG-eluted hepatic nuclear extracts obtained from Wy14,643 (Wy)-treated mice were analyzed by positive ion ESI/MS/MS scans monitoring neutral loss of 189, which reflects elimination of lithiated phosphocholine from the parent [MLi+] ion.
(A–D) Representative ESI/MS/MS scans of GPC species at baseline (time 0) (A), 10 min (B), 30 min (C) and 60 min (D) after an intraperitoneal injection of 50 µg/g Wy14,643 in chow fed WT mice on a PPARα null background injected with AdFLAG-PPARα adenovirus.
(E–H) Representative ESI/MS/MS scans of GPC species at baseline (time 0) (E), 10 min (F), 30 min (G) and 60 min (H) following the same treatment in ZFD fed mice. Insets in A–H depict the ion at m/z 766 (16:0/18:1-GPC) that is displaced in a time-dependent manner by Wy14,643.
(I) Quantification of the relative abundance of the m/z 766 ion in response to Wy14,643. Graphs represent mean ± SEM from two independent experiments with 4–5 mice in each group per experiment.
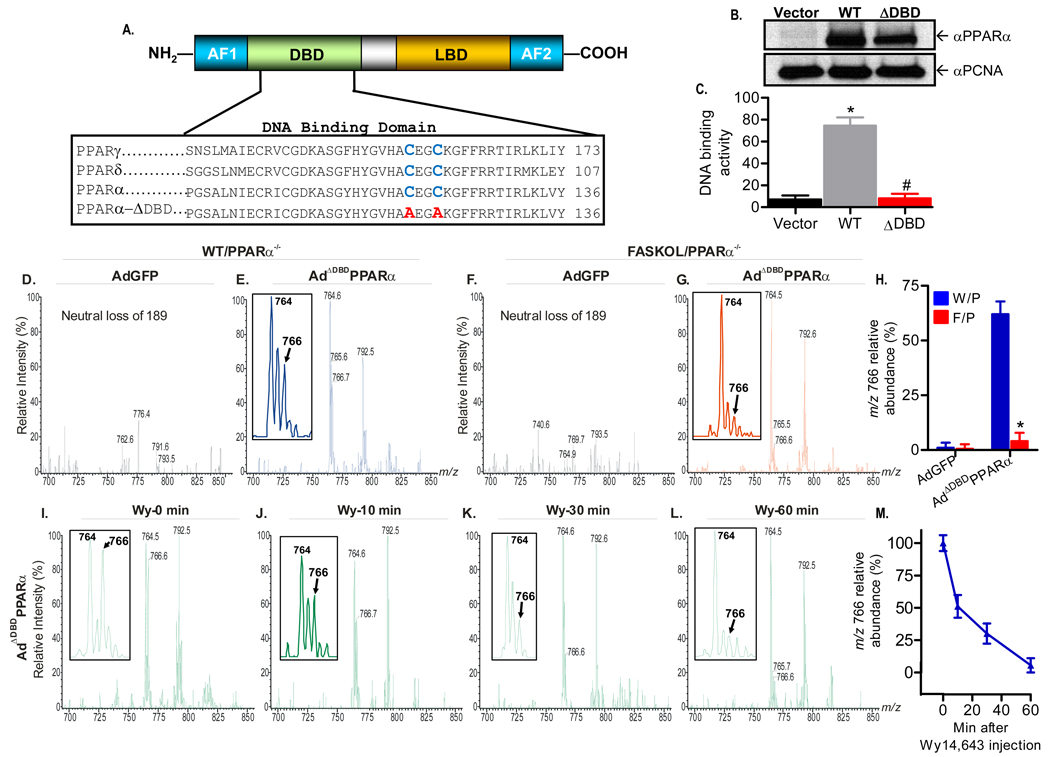
Although the rapid decline in signal with Wy14,643 treatment could represent displacement of endogenous ligand from its binding site on PPARα, it is also possible that lipid loss could represent accelerated metabolism because Wy14,643 activates lipid oxidation. To distinguish between these two possibilities, we repeated the in vivo competitive inhibition experiment using a PPARα molecule with a defective DNA binding domain (Figure 5). This protein, ADBD-PPARα, should retain ligand binding yet be incapable of increasing transcriptional programs promoting lipid metabolism. Two cysteine residues conserved in all PPAR family members were mutated to alanines in PPARα (Figure 5A). Mutation of these residues in PPARδ abolishes DNA binding activity (Shi et al., 2002). Adenoviral expression of this mutated protein resulted in similar levels of protein as wild type PPARα (Figure 5B), and DNA binding was verified to be impaired with this mutant (ΔDBD in Figure 5C). Mice were treated with AdGFP (as a control) or Ad-ΔDBD-PPARα in the presence (WT/PPARα−/−) or absence (FASKOL/ PPARα−/−) of FAS and lipids from affity-purified nuclear extracts were then analyzed by mass spectrometry (Figure 5D–H). Nonspecific binding was minimal (Figure 5D, F), and m/z 766 (representing 16:0/18:1-GPC) was detected using the mutant PPARα in the presence of FAS (Figure 5E, arrow) with a substantial decrease noted in the absence of FAS (Figure 5G, arrow). Quantitation of relative peak abundance for independent experiments is presented in Figure 5H. As seen with wild type PPARα, association of the 16:0/18:1-GPC represented by the ion at m/z 766 with ΔDBD-PPARα was reduced within minutes of administration of Wy14,643 to mice (Figure 5I–L). Competitive inhibition data from independent experiments are presented in Figure 5M. These results confirm that 16:0/18:1-GPC binding to PPARα is FAS-dependent and involves a binding site that is also occupied by a known PPARα activator.
Figure 5. Generation of a PPARα DNA Binding Domain (DBD) Mutant Adenovirus and Mass Spectrometric Analysis.
(A) Schematic of the modular domain structure of PPARα (top panel). AF, activating function; LBD, ligand binding domain. The two highly conserved cysteine residues (blue) within the DBD of the PPAR family (bottom panel) were mutated to alanine (red).
(B) Immunoblot analysis of Cos-7 cells transfected with empty vector, wild type (WT), or DBD-mutant (ADBD) PPARα plasmids using anti-PPARα and proliferating cell nuclear antigen (PCNA) antibodies. Gels are representative of three independent experiments.
(C) Mutation C119A, C122A disrupts PPARα DNA binding activity. Cos-7 cells were transfected and DNA binding activity was assayed. Graphs represent mean ± SEM of experiments performed in triplicate. *, P < 0.05 vs. empty vector. #, P < 0.05 vs. WT control.
(D–G) Representative positive ion ESI/MS/MS scans monitoring neural loss of 189 from lithiated adducts of GPC species in FLAG-eluted hepatic nuclear extracts obtained from chow fed WT and FASKOL mice on a PPARα null background infected with AdGFP (D and F) or AdΔDBDPPARα
(E and G) adenoviruses. Insets in E and G indicate that the material represented by the ion at m/z 766 is the FAS-dependent phospholipid molecular species. A tandem spectrum of this ion (Figure 3) establishes its identity as [MLi+] of 16:0/18:1-GPC
(H) Quantification of the relative abundance of the m/z 766 ion in response to control and mutant adenoviral injections in W/P (WT on PPARα null background) and F/P (FASKOL on PPARα null− background) mice. Each bar represents the mean ± SEM from three independent experiments with 4–6 mice in each group per experiment. *, P < 0.05 vs. corresponding W/P control.
(I–L) Representative neutral loss of 189 ESI/MS/MS scans of GPC species at baseline (time 0) (I), 10 min (J), 30 min (K) and 60 min (L) following intraperitoneal injection of 50 µg/g Wy14,643 in chow fed WT mice on a PPARα null−/− background injected with AdΔDBDPPARα adenovirus. Insets in I–L indicate that the ion at m/z 766 (16:0/18:1-GPC) is displaced from the DBD defective PPARα in a time-dependent manner by Wy14,643.
(M) Quantification of the relative abundance of the m/z 766 ion in response to Wy14,643 administration in WT mice on a PPARα null background injected with AdΔDBDPPARα adenovirus. Graphs represent mean ± SEM from two separate experiments with 3–4 mice in each group per experiment.
Rapid displacement of 16:0/18:1-GPC by the synthetic ligand Wy14,643 might reflect a relatively lower affinity of 16:0/18:1-GPC for PPARα as compared to other receptor-associated species that were not displaced, such as 16:0/18:2-GPC (m/z 764) and 18:1/18:1-GPC (m/z 793). To address this possibility, we performed luminescent proximity (AlphaScreen) assays as well as scintillation proximity assays (Nichols et al., 1998) involving concentration-dependent displacement of radiolabelled Wy14,643. Binding constants of 16:0/18:1-GPC, 16:0/18:2-GPC and 18:1/18:1-GPC for PPARα were identical and, as expected, indicated a lower affinity of these phospholipids for PPARα than Wy14,643 (Supplemental Figure 5). These results are consistent with the notion that the in vivo displacement of 16:0/18:1-GPC by Wy14,643 represents competition of a higher affinity synthetic ligand for an endogenous ligand.
The binding of 16:0/18:1-GPC to PPARα was decreased in the absence of FAS and significantly increased under conditions (high carbohydrate feeding) that induce FAS activity (see Figure 2I), suggesting that FAS is involved in the generation of this ligand. Total hepatic nuclear concentrations of 16:0/18:1-GPC, 16:0/18:2-GPC and 18:1/18:1-GPC were assayed by mass spectrometry under fed and fasting conditions (Supplemental Table 2). 16:0/18:1-GPC and 16:0/18:2-GPC tended to increase with fasting, but these differences were not significant. These assays represent measurements of the total content of a particular phospholipid in the nucleus, not phospholipid bound to PPARα or phospholipid available for signaling. Relevant endogenous ligands should fluctuate during metabolic transitions but current techniques require use of the entire nucleus, which includes a considerable mass of structural GPCs comprising nuclear membranes. The predominant mass of structural GPCs likely overwhelms any changes in the smaller mass of GPCs that might serve a signaling role by liganding nuclear receptors. Assaying the entire nucleus did however allow the determination of the relative abundance of 16:0/18:1-GPC. The contribution of 16:0/18:1-GPC to total nuclear phosphatidylcholine in mouse liver was 11.4 ± 0.6% and its contribution to total nuclear phospholipids was 4.4 ± 1.2%.
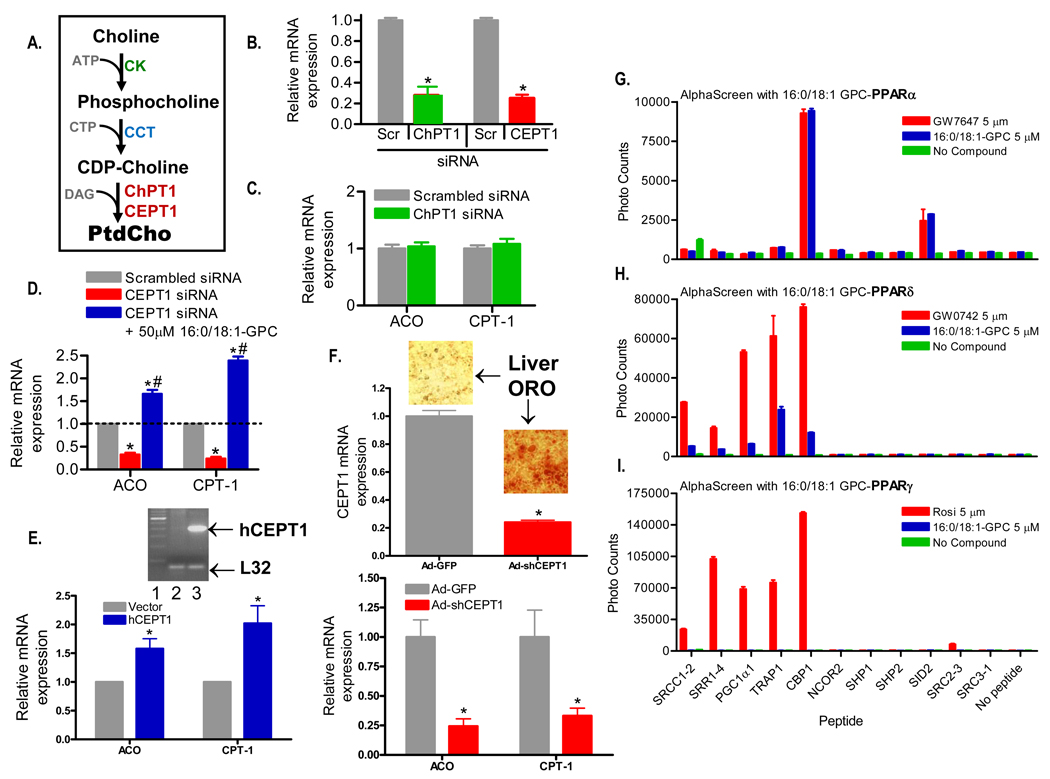
Ligand-dependent activation of PPARα induces expression of genes involved in fatty acid metabolism such as acyl-CoA oxidase and the liver isoform of carnitine palmitoyl transferase 1 (ACO and CPT-1). Incubation of cultured mouse hepatoma cells with exogenous 16:0/18:1-GPC (the FAS-dependent phosphatidylcholine species) increased expression of both ACO and CPT-1 to a similar degree as equimolar amounts of the known PPARα activator Wy14,643 (Supplemental Figure 6). However, the link between FAS and PPARα is most likely to be mediated by effects on endogenous phosphatidylcholine synthesis, which occurs mostly through the Kennedy pathway (Figure 6A). Endogenous synthesis involves the successive action of choline kinase (CK) and CTP:phosphocholine cytidylyltransferase (CCT) to yield CDP-choline (Kent, 2005). This substrate reacts with diacyglycerol (DAG) to yield phosphatidylcholine (PtdCho) through the action of one of two enzymes, choline phosphotransferase 1 (ChPT1), found in the Golgi, and choline-ethanolamine phosphotransferase-1 (CEPT1), found in the nucleus as well as the ER (Henneberry et al., 2002). siRNA-mediated knockdown of these enzymes was achieved in cultured mouse hepatoma cells (Figure 6B), followed by assessment of PPARα-dependent genes. Inactivation of ChPT1, the Golgi enzyme, had no effect on ACO or CPT-1 (Figure 6C). However, knockdown of CEPT1, the nuclear/ER enzyme, decreased PPARα-dependent genes, an effect that was rescued by exogenous 16:0/18:1-GPC (Figure 6D), consistent with the notion that endogenous 16:0/18:1-GPC activates PPARα and that FAS-dependent 16:0/18:1-GPC is an endogenous PPARα ligand.
Figure 6. Gene Expression and Binding Assays.
(A) Schematic of the Kennedy pathway to generate phosphatidylcholine (PtdCho). CK, choline kinase; CTP, cytosine triphosphate; CCT, CTP phosphocholine citidyltransferase; DAG, diacyl glycerol; ChPT1, choline phosphotransferase 1; CEPT1, choline-ethanolamine phosphotransferase 1.
(B) Effect on ChPT1 and CEPT1 mRNA levels normalized to L32 ribosomal mRNA in response to 72 h treatment with corresponding siRNAs and scrambled (Scr) controls in Hepa 1–6 cells.
(C) Effect of 72 h treatment with scrambled and ChPT1 siRNAs on PPARα target genes (ACO and CPT-1) by RT-PCR normalized to L32 ribosomal mRNA in Hepa 1–6 cells.
(D) Effect of 72 h treatment with scrambled and CEPT1 siRNAs on ACO and CPT-1 message levels in Hepa 1–6 cells. Expression of ACO and CPT-1 was also assessed 24 h after addition of 50 µM 16:0/18:1-GPC in a subset of Hepa 1–6 cells previously treated with CEPT1 siRNA. mRNA levels are normalized to control L32 ribosomal mRNA. For C–E, graphs represent mean ± SEM of three separate experiments with each group in triplicate. *, P < 0.05 compared to scrambled controls. #, P < 0.05 compared to CEPT1 siRNA treated cells.
(E) Effect of CEPT1 overexpression on ACO and CPT-1 message levels in Hepa 1–6 cells. Cells were transfected with a human CEPT1 expression vector, and expression was documented by the RT-PCR reaction shown in the inset (lane 1-ladder, lane 2-cells transfected with empty vector, lane 3-cells transfected with the human CEPT1 vector). Graphical results are normalized to L32 mRNA. *, P < 0.05 compared to vector.
(F) Effect of CEPT1 knockdown in living mice. C57BL/6 mice were treated with an shRNA adenovirus for CEPT1 or a control virus expressing GFP. shRNA treatment resulted in decreased expression of CEPT1 (top panel) and livers showed increased staining by Oil Red O (ORO). shRNA-treated livers also showed decreased expression of ACO and CPT-1 (bottom panel). Results are normalized to L32. *, P < 0.05 compared to GFP treatment.
(G–I) Binding of various peptide motifs to the purified PPARα (G), PPARδ (H), and PPARγ (I) LBD in the presence of 5 µM of the corresponding PPAR agonist or 16:0/18:1-GPC as measured by AlphaScreen assays. The background signals of either the respective LBDs or the peptides alone, or without addition of the ligand/agonist (no compound), are all less than 800. Data represent mean ± SEM from three separate experiments.
Two additional pieces of evidence support the link between CEPT1 and induction of PPARα-dependent genes. Overexpression of CEPT1 in hepatoma cells increased expression of ACO and CPT-1 (Figure 6E). We also generated an adenovirus expressing an shRNA for CEPT1. Treatment of living mice with this virus decreased CEPT1 expression in liver (Figure 6F, top panel). Livers with decreased CEPT1 expression were pale-appearing and had increased Oil Red O staining (insets). This intervention resulted in decreased expression of the PPARα-dependent genes ACO and CPT-1 (Figure 6F, bottom panel).
Luminescent proximity (AlphaScreen) assays showed that equimolar amounts of 16:0/18:1-GPC and the known potent PPARα activator GW7647 exhibited similar patterns of interaction with the PPARα ligand binding domain (LBD) and a series of co-activator peptides (Figure 6G). Nearly identical interaction signals with the PPARα LBD were also seen when equimolar amounts of Wy14,643 and 16:0/18:1-GPC were compared (data not shown). As compared to the PPARδ activator GW0742 (Figure 6H), interactions between 16:0/18:1-GPC and the PPARδ LBD were weak, and no signal was detected with the PPARγ LBD as compared to the positive control rosiglitazone (Figure 6I).
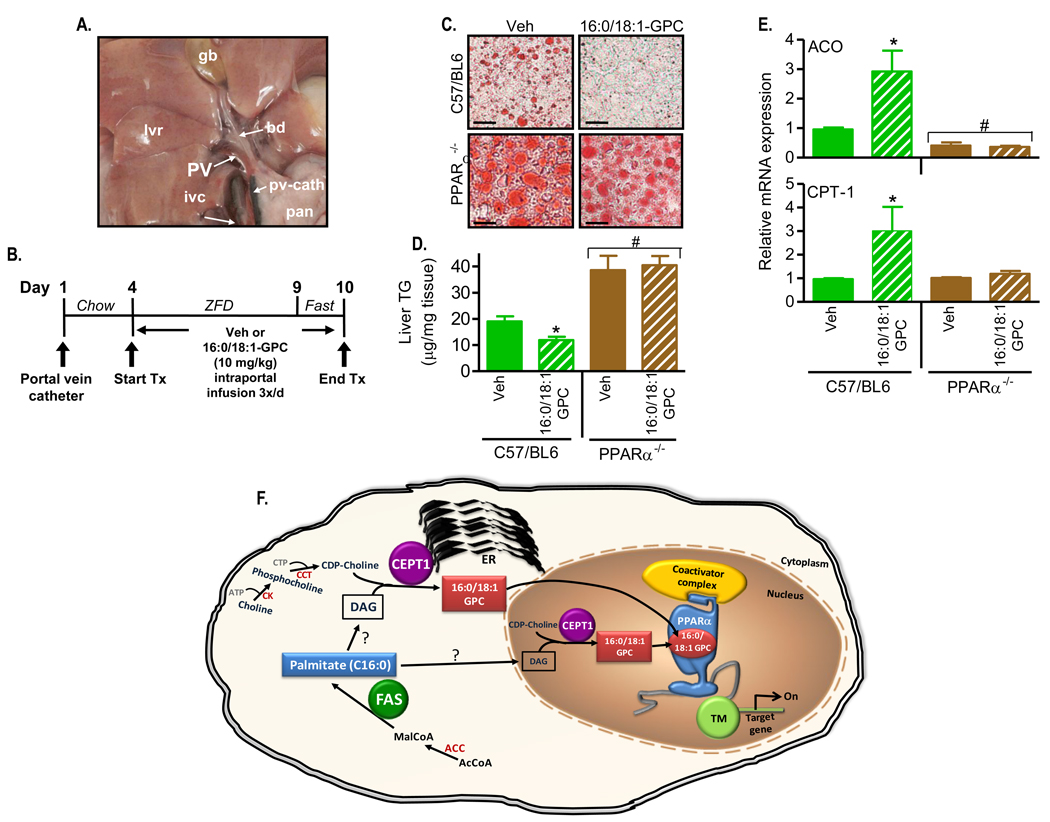
To provide in vivo evidence that 16:0/18:1-GPC serves as a PPARα endogenous hepatic ligand, we implanted catheters in the portal veins of mice, infused this phosphatidylcholine species or vehicle over several days, fasted the animals (while continuing the infusions), then isolated their livers for assays of fat content as well as gene expression. Direct portal vein infusion was prompted by preliminary results showing that intraperitoneal administration of 16:0/18:1-GPC had no effect (data not shown). In additional preliminary experiments, kinetic analyses of radiolabelled phosphatidylcholine showed selective nuclear enrichment within minutes of portal vein administration (Supplemental Figure 7). The appearance of the catheter in the portal vein (pv-cath) is shown in Figure 7A. Our treatment protocol is shown in Figure 7B. After recovering from catheter placement, mice were started on a zero fat diet (ZFD) and treated with thrice-daily infusions of phosphatidylcholine or vehicle between days 4 and 9 followed by a prolonged fast (while continuing the infusions). Fasting causes fatty liver in mice, an effect that is amplified in PPARα null mice. Fat staining of liver is shown in Figure 7C and liver triglyceride quantified in Figure 7D. Lipid content of PPARα-deficient mice was increased as compared to controls and unaffected by phosphatidylcholine infusion. Fat content was decreased in control (C57/BL6) mice with infusion of 16:0/18:1-GPC as compared to vehicle. The PPARα-dependent genes ACO and CPT-1 were increased by 16:0/18:1-GPC in control mice but not in PPARα-deficient mice (Figure 7E).
Figure 7. Portal Vein Infusion of 16:0/18:1-GPC Rescues Hepatic Steatosis in a PPARα-dependent manner.
(A) Operative field depicting the portal vein (PV) cannulated with a catheter (pv-cath) positioned at the entry site into the liver (lvr). The catheter is intentionally marked in black ink at its proximal tip to enhance visualization. Labels indicate gall bladder (gb), bile duct (bd), inferior vena cava (ivc), and pancreas (pan).
(B) Intraportal 16:0/18:1-GPC treatment protocol. After insertion of the portal vein catheter, C57BL/6 mice (wild type for FAS and either wild type or null for PPARα) were allowed to recover. On day 4, chow was changed to a zero fat diet (ZFD) and the mice received 3 intraportal injections/day of 10 mg/kg 16:0/18:1-GPC sonicated in normal saline/0.5% ethanol/0.5% fatty acid-free BSA or vehicle alone. On the last day before the end of treatment (day 9), mice were fasted for 24 h.
(C) At the end of the treatment protocol, liver histological sections were stained with oil red O to visualize neutral lipids (x40 magnification) from wild type (C57/BL6) and PPARα−/− mice treated with 16:0/18:1-GPC or vehicle (Veh). Sections are representative of several animals for each condition.
(D) Quantification of hepatic triglyceride content per unit mass of tissue from vehicle and 16:0/18:1-GPC treated C57/BL6 and PPARα−/− mice. Bars represent mean ± SEM of two separate infusion experiments with 5–8 animals per group in each experiment. *, P < 0.05 vs. corresponding Veh. #, P < 0.05 vs. C57/BL6 controls.
(E) Expression of hepatic ACO (top panel) and CPT-1 (bottom panel) mRNA by RT-PCR normalized to control L32 ribosomal mRNA following the 16:0/18:1-GPC injections. Data represent mean ± SEM of two independent RT-PCR experiments for each gene with 4 mice per genotype per group. *, P < 0.05 vs. corresponding Veh. #, P < 0.05 vs. C57/BL6 controls. (F) Proposed model for the generation of the endogenous PPARα ligand in liver. FAS yields palmitate (C16:0), and 16:0/18:1-GPC is likely generated through the diacylglycerol (DAG) intermediate and the enzymatic activity of CEPT1 either in the ER or the nucleus. Binding of 16:0/18:1-GPC to PPARα in the nucleus activates transcription machinery (TM) turning on PPARα-dependent genes and affecting hepatic lipid metabolism. ACC, acetyl CoA carboxylase; ER, endoplasmic reticulum.
Discussion
PPARs are targets of drugs in use and in development to treat disease, and they modulate metabolic and inflammatory pathways by responding to nutritional signals through ligand activation of transcription. No authentic endogenous PPAR ligand, the molecule occupying the nuclear receptor binding site in vivo while the receptor is actively driving transcription, has been identified. Here we demonstrate PPARα binding of a discrete phospholipid, 16:0/18:1-GPC, in mammalian liver in the presence of FAS, when PPARα is active, but not in the absence of FAS, when PPARα is not. This molecule was displaced from PPARα in vivo with a pharmacological ligand, inhibiting its biosynthesis in cells and living mice disrupted PPARα-dependent gene expression, and infusing it directly into mouse liver altered hepatic lipid metabolism in a PPARα-dependent fashion. These results suggest that this particular phosphatidylcholine species is a physiologically relevant endogenous PPARα ligand in liver.
There is precedent for the interaction of phospholipids with nuclear receptors. The crystal structure of the insect homologue of mammalian RXR includes co-purified phospholipid (Billas et al., 2001; Clayton et al., 2001). Structures of the ligand binding domains of two orphan receptors, SF-1 and LRH-1, include electron density patterns representing phospholipids (Krylova et al., 2005; Li et al., 2005; Ortlund et al., 2005). In contrast to PPARα, which forms heterodimers with RXR and is clearly ligand-activated, SF-1 and LRH-1 bind to DNA as monomers and it is not known whether the ligands for these receptors are involved in dynamic regulation of their activity (Forman, 2005). Oxidized phospholipids, especially those derived from modified low density lipoprotein particles through the action of phospholipases or lipoxygenases, have been implicated in the activation of PPARα and PPARγ (Davies et al., 2001; Delerive et al., 2000; Lee et al., 2000).
Our current results show that the phosphatidylcholine molecular species 16:0/18:1-GPC binds PPARα at an activating ligand binding site (reflected by its displacement with a synthetic PPARα ligand in vivo) when FAS enzyme activity is present. 16:0/18:1-GPC is the only FAS-dependent phosphatidylcholine species we identified bound to PPARα in vivo. However, two other phosphatidylcholine species, 16:0/18:2-GPC and 18:1/18:1-GPC, were also co-purified with tagged PPARα from liver and their interactions with the PPARα ligand binding domain in vitro (Supplemental Figure 5) were indistinguishable from those of 16:0/18:1-GPC. The failure to discriminate between these three GPCs in vitro may be due to differential interactions between ligands and ligand binding domains in vitro as opposed to ligands and full length receptors in vivo. 16:0/18:2-GPC and 18:1/18:1-GPC, but not 16:0/18:1-GPC, remained bound to PPARα in the absence of hepatic FAS, a condition characterized by a striking decrease in PPARα-dependent gene expression. Their association with the receptor in the absence of appropriate activation of gene expression suggests that the binding of 16:0/18:2-GPC and 18:1/18:1-GPC is not sufficient for receptor activation, but we cannot exclude the possibility that the presence of these additional species may be necessary for receptor activation.
In AlphaScreen assays, 16:0/18:1-GPC interacted with PPARα but not PPARγ, suggesting that this species does not bind nonspecifically to PPARs and consistent with data showing that 16:0/18:1-GPC does not interact with the ligand binding domains of PPARγ, SF-1 or LRH-1 (Krylova et al., 2005). One interpretation of our findings is that FAS, which synthesizes predominantly the saturated fatty acid palmitate (16:0), preferentially channels newly synthesized palmitate through diacylglycerol to the site of phosphatidylcholine synthesis for generation of the PPARα ligand. Previous data also link fatty acid synthesis and phosphatidylcholine synthesis. In cultured cells, SREBPs stimulate the synthesis of phosphatidylcholine (but not other phospholipids), and this effect is attenuated by the FAS- inhibitor cerulenin (Ridgway and Lagace, 2003). Figure 7F shows how fatty acid synthesis, phosphatidylcholine synthesis, and PPARα signaling appear to be related based on the current findings.
In the current work, addition of 16:0/18:1-GPC to cultured cells increased PPARα-dependent gene expression (Supplemental Figure 6), its addition rescued PPARα-dependent gene expression when endogenous synthesis of phosphatidylcholine was interrupted (Figure 6D), and its infusion directly into the portal vein of living mice increased PPARα-dependent genes and corrected fasting-induced steatosis only in the presence of PPARα (Figure 7A–E). These experiments support the concept that this GPC species activates PPARα, but do not show that a circulating form of this lipid reports nutritional status. In fact, how altering extracellular concentrations of a charged phospholipid species might affect nuclear events is unknown, although there is evidence that certain extracellular phospholipids have direct access to the nuclear receptor PPARγ (Davies et al., 2001).
Our results showing that portal vein infusion results in nuclear before cytoplasmic accumulation (Supplemental Fig. 7) are consistent with an uncharacterized conduit for phospholipids to the nucleus. Since the portal vein drains the intestine, the anatomic origin of nutrients, the results also suggest that providing a GPC ligand in the diet, if it were able to access the portal vein at sufficient concentrations, could activate hepatic PPARα. The method of infusion, utilizing the portal vein, is likely important. FASKOL mice have elevated circulating levels of fatty acids (in the peripheral venous circulation) that contribute to the hepatic steatosis of these animals (Chakravarthy et al., 2005). Curiously, these fatty acids (so abundant that they cause fatty liver) from the periphery (that we have referred to as “old ” fat as opposed to “new” fat from de novo lipogenesis or diet) are unable to activate PPARα, suggesting (based on the current work) that they are unavailable for GPC synthesis. Lipids from the periphery enter the liver via the hepatic artery while those from the diet enter via the portal vein. The hepatic artery and portal vein comprise anatomically distinct regions of the portal triad, and it is possible that hepatocytes in those different segments respond differently to lipid signals.
While the infusion studies provide proof of principle that 16:0/18:1-GPC can affect liver biology in a PPARα-dependent manner, they are not directly germane to the central finding of this paper that the intracellular enzyme FAS is linked to the generation of a physiologically relevant endogenous ligand for PPARα. However, the infusion studies represent grounds for future research involving portal vein infusions as well as dietary supplementation with GPCs labeled at different substituents to further pursue the notion that an externally provided GPC can reach PPARα intact.
Modulating phospholipids is also complicated by remodeling, a process identified by Lands (Lands, 1960) that results in the rearrangement of fatty acid substituents. Not all of the responsible enzymes have been identified, but PPARα activation induces the expression of a recently discovered lysophosphatidylcholine acyltransferase (LPCAT3) in liver that remodels the sn-1 and sn-2 fatty acid substituents (Zhao et al., 2008) and might represent a mechanism for dampening phosphatidylcholine-mediated activation of PPARα.
Phosphatidylcholine is ubiquitous in the cell and comprises a substantial proportion of the nuclear volume. It would have a limited capacity to regulate PPARα if high nuclear concentrations ensured constant occupation of the ligand binding site, but the putative PPARα ligand we identified, 16:0/18:1-GPC, is a minor PtdCho species in liver as shown by the current work (Results) and previous work (Hsu et al., 1998), consistent with a signaling role for this molecule. Conversely, 16:0/18:1- GPC is the most abundant PtdCho in brain (Hsu et al., 1998), raising the possibility that a different PtdCho species may activate PPARα in this tissue.
Both PPARα activation and phosphatidylcholine are thought to be anti-inflammatory (Cuzzocrea et al., 2004; Li et al., 2006; Straus and Glass, 2007; Stremmel et al., 2007; Treede et al., 2007). Based on our findings of a discrete PtdCho molecular species serving as a hepatic ligand for PPARα, tissue-specific phospholipid ligands, either induced endogenously or perhaps provided in the diet, could modify inflammatory processes so that off target side effects are minimized.
Experimental Procedures
Animals and Reagents
Animal protocols were approved by the Washington University Animal Studies Committee. Mice were genotyped using previously described primer sets (Bernal-Mizrachi et al., 2003; Chakravarthy et al., 2005) and fed either chow (Purina 5053) or a zero-fat diet (ZFD) (Harlan Teklad, TD03314). Experiments were performed at 16–20 wks of age to allow for maximal effects of albumin-Cre. Assays were performed as described (Chakravarthy et al., 2005). 1-Palmitoyl-2-Oleoyl-sn-Glycero-3-Phosphocholine (PC16:0/18:1) and its regioisomer, 1-Oleoyl-2-Palmitoyl-sn-Glycero-3-Phosphocholine (PC18:1/16:0) were obtained from Avanti Polar Lipids (Alabaster, AL). Stealth™ siRNA oligonucleotides for mouse ChPT1 (GGAGGAGCAACAAUGUGGGACUAUA) and CEPT1 (UGGCAGUGAUUGGAGGACCACCUUU) were from Invitrogen (Carlsbad, CA).
Isolation of Tagged PPARα
Freshly harvested livers (∼100 mg) were gently homogenized in ice-cold non-detergent hypotonic buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 100 mM DTT, protease and phosphatase inhibitor cocktail). After an additional 10 min incubation in the hypotonic buffer, the homogenate was centrifuged at 8000 × g at 4°C for 20 min. The pellet was homogenized in ice-cold extraction buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 0.21 M NaCl, 0.2 mM EDTA, 25% (v/v) glycerol, 100 mM DTT, protease and phosphatase inhibitor cocktail), placed on a rotating shaker at 4°C for 1 h, then centrifuged at 18,000 × g for 10 min. The supernatant (nuclear fraction) was incubated with anti-FLAG M2-Agarose affinity gel (A2220, Sigma) overnight at 4°C on a rotating shaker. Four washes (50 mM Tris HCl, pH 7.4, 100 mM NaCl, protease and phosphatase inhibitor cocktail) were followed by elution using competition with excess 3X FLAG peptide (F4799, Sigma; 150 ng/µl). An aliquot of the complex was processed for immunoblotting; the remainder was transferred to methanol/chloroform and processed for mass-spectrometry.
Electrospray Ionization Mass Spectrometry
Phosphatidylcholine (GPC), lysophosphatidylcholine (LPC), sphingomyelin (SM), and ceramide (CM) were analyzed as Li+ adducts by positive ion ESI/MS on a Finnigan (San Jose, CA) TSQ-7000 triple stage quadrupole mass spectrometer with an ESI source controlled by Finnigan ICIS software. Lipids were dissolved in methanol/chloroform (2/1, v/v) containing LiOH (10 pmol/µl), infused with a Harvard syringe pump, and analyzed as described (Hsu et al., 1998; Hsu and Turk, 2000, 2003; Hsu et al., 2003). For tandem MS, precursor ions selected in the first quadrupole were accelerated into a chamber containing argon to induce collisionally-activated dissociation, and product ions were analyzed in the final quadrupole. Constant neutral loss scanning was performed to monitor GPC [M+Li]+ ions that undergo loss of 183 or 189 (phosphocholine or its Li+ salt) and similar scans were performed to monitor LPC and SM [M+Li]+ ions that undergo loss of 59 (trimethylaine) and to monitor CM [M+Li]+ ions that undergo loss of 48 (water plus formaldehyde). Intensities of ions for internal standards were compared to those of ions for endogenous species followed by interpolation from calibration curves. Glycerophospho-ethanolamine, -glycerol, -serine, and -inositol, were analyzed as [M-H]– ions by negative ion ESI/MS/MS relative to internal standards (Nowatzke et al., 1998; Ramanadham et al., 1998) and their tandem spectra were obtained.
DNA binding activity
Cos-7 cells maintained in DMEM containing 10% fetal bovine serum were transiently transfected with 2 µg of wild type- and DBD-mutant-PPARα plasmids using FuGENE6 (Roche) as described (Lodhi et al., 2007). Nuclear extract DNA binding activity was determined using the PPARα transcription factor assay kit (Cayman Chem, Inc.).
Mouse Hepatocytes
The C57BL/6 mouse hepatoma cell line Hepa 1–6 (ATCC, CRL-1830) was expanded in DMEM with 10% fetal bovine serum. For experiments, cells were cultured to 50–60% confluence, washed twice with PBS, and the medium was changed to serum-free DMEM supplemented with the various types and concentrations of phosphatidylcholine (sonicated to homogeneity in PBS/1% ethanol/4% fatty acid-free BSA), Wy14,643 (dissolved in 80% PBS/20% DMSO), or vehicle-only solutions. 24 h later, cells were washed, RNA prepared, and ACO and CPT-1 expression levels were determined by quantitative RT-PCR. For siRNA experiments, 50–60% confluent cells were treated with siRNAs or their scrambled controls (all diluted in PBS) for 72 h. Following the 72 h siRNA treatment, another set of cells received 16:0/18:1-GPC (using a concentration based on dose-response experiments) for an additional 24 h. For overexpression of CEPT1, cells were transfected with a vector containing human CEPT1 cloned in the pCMV6-XL4 plasmid (Origene, Rockwell, MD) using Lipofectamine 2000 (Invitrogen). After 24 hr, RNA was isolated.
CEPT1 shRNA Adenovirus
pLKO.1 plasmid (TRCN0000103315) encoding mouse CEPT1 shRNA under control of the U6 promoter was obtained from Open Biosystems (Huntsville, AL). The shRNA sequence is: ccggCCCATCCTATAAACTGAATATctcgagATATTCAGTTTATAGGATGGGtttttg To construct the CEPT1 knockdown adenovirus, the shRNA expression cassette from the pLKO.1 plasmid was subcloned into the Dual-Basic adenoviral shuttle vector and recombined with Ad5 (ΔE1/ΔE3) vector (Vector Biolabs, Philadelphia, PA). The adenovirus was packaged in HEK 293 cells and purified using cesium chloride ultracentrifugation. The knockdown virus or a control adenovirus expressing GFP was administered at 8 × 109 PFU in a total volume of 200 µl. On day 5 post-injection, animals were sacrificed and livers were harvested.
PPAR Binding Assays
Binding of 16:0/18:1-GPC and known agonists to the ligand binding domain (LBD) of PPARα in the presence of various peptide motifs was determined by AlphaScreen assay (Pioszak and Xu, 2008). Experiments used 100 nM receptor LBD, purified as 6X His tag fusion proteins (Li et al., 2005), and 20 nM of N-terminal biotinylated CBP1 peptide or other coactivator peptides in the presence of 5 µg/ml donor and acceptor beads in 50 mM MOPS (pH 7.4), 100 mM NaCl, and 0.1 mg/ml BSA for 90 min at 25°C Signals were generated in the absence or the presence of ligand. Identical experiments were performed using the PPARδ and PPARγ LBDs.
Portal Vein Infusion
Portal veins were cannulated as described (Strubbe et al., 1999). The catheter (0.025 mm OD × 0.012 mm ID, Braintree Scientific), prefilled with 55% (w/v) polyvinylpyrrolidone (Sigma) in heparin (100 IU/ml saline) to prevent clotting, was anchored to the abdominal wall, and its free end was inserted under the skin and tunneled to a small midline incision slightly distal to the scapula on the back. Body weight and food intake returned to baseline levels (usually by day 3) before experiments. Starting on post-op day 4, animals were fed a zero fat diet (TD 03314, Harlan Teklad) for 5 days, which induces modest to severe hepatic steatosis in C57BL/6 and PPARα−/− mice, respectively. During this period, animals received three intraportal infusions a day of either 10 mg/kg phosphatidylcholine (16:0/18:1-GPC) sonicated to homogeneity in a 37°C solution of saline/0.5% ethanol/0.5% fatty acid-free BSA, or vehicle alone, based on appropriate time-course and dose-response preliminary experiments (See Supplemental Experimental Procedures). Animals were then fasted for 24 h, and livers were harvested.
Statistical Analyses
Comparisons were performed using an unpaired, two-tailed Student’s t-test or analysis of variance (ANOVA). If the overall F was significant for the latter, comparisons between means were made using appropriate post hoc tests.
Supplementary Material
Acknowledgements
This work was supported by NIH grants DK076729, P50 HL083762, R37 DK34388, P41 RR00954, HL089301, the Clinical Nutrition Research Unit (DK56341), the Diabetes Research and Training Center (DK20579), the Jay and Betty Van Andel Foundation, and by the American Diabetes Association (Junior Faculty Award 1-07-JF-12 to MVC and a Mentor-Based Postdoctoral Fellowship Award).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayers SD, Nedrow KL, Gillilan RE, Noy N. Continuous nucleocytoplasmic shuttling underlies transcriptional activation of PPARgamma by FABP4. Biochemistry. 2007;46:6744–6752. doi: 10.1021/bi700047a. [DOI] [PubMed] [Google Scholar]
- Barter PJ, Rye KA. Is there a role for fibrates in the management of dyslipidemia in the metabolic syndrome? Arterioscler Thromb Vasc Biol. 2008;28:39–46. doi: 10.1161/ATVBAHA.107.148817. [DOI] [PubMed] [Google Scholar]
- Bensinger SJ, Tontonoz P. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature. 2008;454:470–477. doi: 10.1038/nature07202. [DOI] [PubMed] [Google Scholar]
- Bernal-Mizrachi C, Weng S, Feng C, Finck BN, Knutsen RH, Leone TC, Coleman T, Mecham RP, Kelly DP, Semenkovich CF. Dexamethasone induction of hypertension and diabetes is PPARalpha-dependent in LDL receptor-null mice. Nat Med. 2003;9:1069–1075. doi: 10.1038/nm898. [DOI] [PubMed] [Google Scholar]
- Billas IM, Moulinier L, Rochel N, Moras D. Crystal structure of the ligand-binding domain of the ultraspiracle protein USP, the ortholog of retinoid X receptors in insects. J Biol Chem. 2001;276:7465–7474. doi: 10.1074/jbc.M008926200. [DOI] [PubMed] [Google Scholar]
- Chakravarthy MV, Pan Z, Zhu Y, Tordjman K, Schneider JG, Coleman T, Turk J, Semenkovich CF. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005;1:309–322. doi: 10.1016/j.cmet.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Chakravarthy MV, Zhu Y, Lopez M, Yin L, Wozniak DF, Coleman T, Hu Z, Wolfgang M, Vidal-Puig A, Lane MD, et al. Brain fatty acid synthase activates PPARalpha to maintain energy homeostasis. J Clin Invest. 2007;117:2539–2552. doi: 10.1172/JCI31183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton GM, Peak-Chew SY, Evans RM, Schwabe JW. The structure of the ultraspiracle ligand-binding domain reveals a nuclear receptor locked in an inactive conformation. Proc Natl Acad Sci U S A. 2001;98:1549–1554. doi: 10.1073/pnas.041611298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzzocrea S, Di Paola R, Mazzon E, Genovese T, Muia C, Centorrino T, Caputi AP. Role of endogenous and exogenous ligands for the peroxisome proliferators activated receptors alpha (PPARalpha) in the development of inflammatory bowel disease in mice. Lab Invest. 2004;84:1643–1654. doi: 10.1038/labinvest.3700185. [DOI] [PubMed] [Google Scholar]
- Davies SS, Pontsler AV, Marathe GK, Harrison KA, Murphy RC, Hinshaw JC, Prestwich GD, Hilaire AS, Prescott SM, Zimmerman GA, et al. Oxidized alkyl phospholipids are specific, high affinity peroxisome proliferator-activated receptor gamma ligands and agonists. J Biol Chem. 2001;276:16015–16023. doi: 10.1074/jbc.M100878200. [DOI] [PubMed] [Google Scholar]
- Delerive P, Furman C, Teissier E, Fruchart J, Duriez P, Staels B. Oxidized phospholipids activate PPARalpha in a phospholipase A2-dependent manner. FEBS Lett. 2000;471:34–38. doi: 10.1016/s0014-5793(00)01364-8. [DOI] [PubMed] [Google Scholar]
- Forman BM. Are those phospholipids in your pocket? Cell Metab. 2005;1:153–155. doi: 10.1016/j.cmet.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci U S A. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberry AL, Wright MM, McMaster CR. The major sites of cellular phospholipid synthesis and molecular determinants of fatty acid and lipid head group specificity. Mol Biol Cell. 2002;13:3148–3161. doi: 10.1091/mbc.01-11-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu FF, Bohrer A, Turk J. Formation of lithiated adducts of glycerophosphocholine lipids facilitates their identification by electrospray ionization tandem mass spectrometry. J Am Soc Mass Spectrom. 1998;9:516–526. doi: 10.1016/S1044-0305(98)00012-9. [DOI] [PubMed] [Google Scholar]
- Hsu FF, Turk J. Structural determination of sphingomyelin by tandem mass spectrometry with electrospray ionization. J Am Soc Mass Spectrom. 2000;11:437–449. doi: 10.1016/S1044-0305(99)00150-6. [DOI] [PubMed] [Google Scholar]
- Hsu FF, Turk J. Electrospray ionization/tandem quadrupole mass spectrometric studies on phosphatidylcholines: the fragmentation processes. J Am Soc Mass Spectrom. 2003;14:352–363. doi: 10.1016/S1044-0305(03)00064-3. [DOI] [PubMed] [Google Scholar]
- Hsu FF, Turk J, Thukkani AK, Messner MC, Wildsmith KR, Ford DA. Characterization of alkylacyl, alk-1-enylacyl and lyso subclasses of glycerophosphocholine by tandem quadrupole mass spectrometry with electrospray ionization. J Mass Spectrom. 2003;38:752–763. doi: 10.1002/jms.491. [DOI] [PubMed] [Google Scholar]
- Kent C. Regulatory enzymes of phosphatidylcholine biosynthesis: a personal perspective. Biochim Biophys Acta. 2005;1733:53–66. doi: 10.1016/j.bbalip.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, Devchand P, Wahli W, Willson TM, Lenhard JM, et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc Natl Acad Sci U S A. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey G, Braissant O, L’Horset F, Kalkhoven E, Perroud M, Parker MG, Wahli W. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol Endocrinol. 1997;11:779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Lands WEM. Metabolism of glycerolipids. J Biol Chem. 1960;235:2233–2237. [PubMed] [Google Scholar]
- Lee H, Shi W, Tontonoz P, Wang S, Subbanagounder G, Hedrick CC, Hama S, Borromeo C, Evans RM, Berliner JA, et al. Role for peroxisome proliferator-activated receptor alpha in oxidized phospholipid-induced synthesis of monocyte chemotactic protein-1 and interleukin-8 by endothelial cells. Circ Res. 2000;87:516–521. doi: 10.1161/01.res.87.6.516. [DOI] [PubMed] [Google Scholar]
- Li Y, Choi M, Cavey G, Daugherty J, Suino K, Kovach A, Bingham NC, Kliewer SA, Xu HE. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol Cell. 2005;17:491–502. doi: 10.1016/j.molcel.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Li Z, Agellon LB, Allen TM, Umeda M, Jewell L, Mason A, Vance DE. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006;3:321–331. doi: 10.1016/j.cmet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Lodhi IJ, Chiang SH, Chang L, Vollenweider D, Watson RT, Inoue M, Pessin JE, Saltiel AR. Gapex-5, a Rab31 guanine nucleotide exchange factor that regulates Glut4 trafficking in adipocytes. Cell Metab. 2007;5:59–72. doi: 10.1016/j.cmet.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberry EP, Xie Y, Kennedy S, Han X, Buhman KK, Luo J, Gross RW, Davidson NO. Decreased hepatic triglyceride accumulation and altered fatty acid uptake in mice with deletion of the liver fatty acid-binding protein gene. J Biol Chem. 2003;278:51664–51672. doi: 10.1074/jbc.M309377200. [DOI] [PubMed] [Google Scholar]
- Nichols JS, Parks DJ, Consler TG, Blanchard SG. Development of a scintillation proximity assay for peroxisome proliferator-activated receptor gamma ligand binding domain. Anal Biochem. 1998;257:112–119. doi: 10.1006/abio.1997.2557. [DOI] [PubMed] [Google Scholar]
- Nowatzke W, Ramanadham S, Ma Z, Hsu FF, Bohrer A, Turk J. Mass spectrometric evidence that agents that cause loss of Ca2+ from intracellular compartments induce hydrolysis of arachidonic acid from pancreatic islet membrane phospholipids by a mechanism that does not require a rise in cytosolic Ca2+ concentration. Endocrinology. 1998;139:4073–4085. doi: 10.1210/endo.139.10.6225. [DOI] [PubMed] [Google Scholar]
- Ortlund EA, Lee Y, Solomon IH, Hager JM, Safi R, Choi Y, Guan Z, Tripathy A, Raetz CR, McDonnell DP, et al. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat Struct Mol Biol. 2005;12:357–363. doi: 10.1038/nsmb910. [DOI] [PubMed] [Google Scholar]
- Pioszak AA, Xu HE. Molecular recognition of parathyroid hormone by its G protein-coupled receptor. Proc Natl Acad Sci U S A. 2008;105:5034–5039. doi: 10.1073/pnas.0801027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanadham S, Hsu FF, Bohrer A, Nowatzke W, Ma Z, Turk J. Electrospray ionization mass spectrometric analyses of phospholipids from rat and human pancreatic islets and subcellular membranes: comparison to other tissues and implications for membrane fusion in insulin exocytosis. Biochemistry. 1998;37:4553–4567. doi: 10.1021/bi9722507. [DOI] [PubMed] [Google Scholar]
- Reddy JK, Hashimoto T. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu Rev Nutr. 2001;21:193–230. doi: 10.1146/annurev.nutr.21.1.193. [DOI] [PubMed] [Google Scholar]
- Ridgway ND, Lagace TA. Regulation of the CDP-choline pathway by sterol regulatory element binding proteins involves transcriptional and post-transcriptional mechanisms. Biochem J. 2003;372:811–819. doi: 10.1042/BJ20030252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riserus U, Sprecher D, Johnson T, Olson E, Hirschberg S, Liu A, Fang Z, Hegde P, Richards D, Sarov-Blat L, et al. Activation of peroxisome proliferator-activated receptor (PPAR) delta promotes reversal of multiple metabolic abnormalities, reduces oxidative stress, and increases fatty acid oxidation in moderately obese men. Diabetes. 2008;57:332–339. doi: 10.2337/db07-1318. [DOI] [PubMed] [Google Scholar]
- Semenkovich CF. Regulation of fatty acid synthase (FAS) Prog Lipid Res. 1997;36:43–53. doi: 10.1016/s0163-7827(97)00003-9. [DOI] [PubMed] [Google Scholar]
- Shi Y, Hon M, Evans RM. The peroxisome proliferator-activated receptor delta, an integrator of transcriptional repression and nuclear receptor signaling. Proc Natl Acad Sci U S A. 2002;99:2613–2618. doi: 10.1073/pnas.052707099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus DS, Glass CK. Anti-inflammatory actions of PPAR ligands: new insights on cellular and molecular mechanisms. Trends Immunol. 2007;28:551–558. doi: 10.1016/j.it.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Stremmel W, Ehehalt R, Autschbach F, Karner M. Phosphatidylcholine for steroid-refractory chronic ulcerative colitis: a randomized trial. Ann Intern Med. 2007;147:603–610. doi: 10.7326/0003-4819-147-9-200711060-00004. [DOI] [PubMed] [Google Scholar]
- Strubbe JH, Bruggink JE, Steffens AB. Hepatic portal vein cannulation for infusion and blood sampling in freely moving rats. Physiol Behav. 1999;65:885–887. doi: 10.1016/s0031-9384(98)00248-0. [DOI] [PubMed] [Google Scholar]
- Treede I, Braun A, Sparla R, Kuhnel M, Giese T, Turner JR, Anes E, Kulaksiz H, Fullekrug J, Stremmel W, et al. Anti-inflammatory effects of phosphatidylcholine. J Biol Chem. 2007;282:27155–27164. doi: 10.1074/jbc.M704408200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yki-Jarvinen H. Thiazolidinediones. N Engl J Med. 2004;351:1106–1118. doi: 10.1056/NEJMra041001. [DOI] [PubMed] [Google Scholar]
- Yu K, Bayona W, Kallen CB, Harding HP, Ravera CP, McMahon G, Brown M, Lazar MA. Differential activation of peroxisome proliferator-activated receptors by eicosanoids. J Biol Chem. 1995;270:23975–23983. doi: 10.1074/jbc.270.41.23975. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chen YQ, Bonacci TM, Bredt DS, Li S, Bensch WR, Moller DE, Kowala M, Konrad RJ, Cao G. Identification and characterization of a major liver lysophosphatidylcholine acyltransferase. J Biol Chem. 2008;283:8258–8265. doi: 10.1074/jbc.M710422200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.