Abstract
The DNA damage and replication checkpoint kinase Mec1/ATR is a member of the PI3-kinase related kinases that function in response to various genotoxic stresses. The checkpoint clamp 9-1-1 (Rad9-Rad1-Hus1 in S. pombe and mammals; Ddc1-Rad17-Mec3 in S. cerevisiae) executes two distinct checkpoint functions. In S. cerevisiae, DNA-bound 9-1-1 directly activates Mec1 kinase activity, a function that has not been demonstrated in other organisms. A second, conserved activity of 9-1-1 is that of TopBP1/Cut5/Dpb11 recruitment to stalled replication sites; subsequent activation of Mec1/ATR is carried out by TopBP1/Cut5/Dpb11. Biochemical studies indicate that the mode of Mec1/ATR activation by S. cerevisiae 9-1-1 is analogous to activation by S. cerevisiae Dpb11 or by vertebrate TopBP1: activation is mediated by the intrinsically disordered C-terminal tail of each activator. The relative contributions made by multiple activators of Mec1/ATR are discussed.
Introduction
The DNA damage checkpoint machinery is a vital cellular process that coordinates cell cycle progression with DNA repair in response to DNA damage. The checkpoint machinery is highly conserved in eukaryotes and defects in this machinery lead to damage-sensitivity in yeast and to cancer-susceptibility in humans. The ATM (S. cerevisiae and S. pombe Tel1) and ATR (S. cerevisiae Mec1 and S. pombe Rad3) Pi3-kinase-like protein kinases play important roles as the initial sensor kinases in these signal transduction pathways that eventually result in the transcriptional induction of repair genes and the slowing of cell cycle progression [1–3]. These kinases are tightly regulated such that they are only activated when there is a threat to genomic integrity. Checkpoint signal transduction pathways begin with sensor proteins that either recognize DNA damage or DNA structures that have been formed as a result of the initial processing of DNA damage (Table 1). Mec1/ATR and Tel1/ATM also localize to the DNA, and have been variously designated as sensor kinases or transducer kinases, the latter because they are the initial protein kinases in their signal transduction pathways. Mediator proteins often function as scaffolds that bring other factors in the pathway into juxtaposition. The effector kinases Chk1 and Rad53/Chk2 phosphorylate critical targets that promote cell cycle arrest, transcriptional activation, and apoptosis (reviewed in [4,5]).
Table 1.
Nomenclature and abbreviations of checkpoint proteins in the Mec1/ATR pathway1
| S. cerev. | S. pombe | Human Xenopus | Step | Function |
|---|---|---|---|---|
| Rpa1 | Rpa1 | RPA1 | Initiation | RPA large subunit |
| Rpa2 | Rpa2 | RPA2 | Initiation | RPA middle subunit, redirects RPA to DNA repair foci. |
| Rad24 | Rad17 | Rad17 | Sensor | Rfc1 homolog; large subunit of Rad24/Rad17-RFC |
| Rad17 | Rad1 | Rad1 | Sensor | Checkpoint clamp subunit |
| Mec3 | Hus1 | Hus1 | Sensor | Checkpoint clamp subunit |
| Ddc1 | Rad9 | Rad9 | Sensor | Checkpoint clamp subunit; activates Mec1; phosphorylated form binds Dpb11/Cut5/TopBP1 |
| Dpb11 | Cut5/Rad4 | TopBP1 | Sensor | Replication initiation protein; activates ATR; binds phosphorylated clamp; associates with Pol ε. |
| Pol2 | Cdc20 | Pol2 | Sensor | Catalytic subunit of Pol ε |
| Mec1 | Rad3 | ATR | Transducer | PIKK catalytic subunit; phosphorylates Rad53/Chk1/Chk2, and factors upstream of it |
| Ddc2/ | Rad26 | ATRIP | Transducer | Mec1/Rad3/ATR regulatory subunit; binds RPA |
| Tel1 | Tel1 | ATM | Transducer | PIKK; primarily in response to dsDNA breaks |
| Rad9 | Crb2 | 53BP1 MDC1 |
Mediator | Scaffold for Rad53 or Chk1; facilitates transphosphorylation |
| Mrc1 | Mrc1 | Claspin | Mediator | Replication fork-associated scaffold |
| Rad53 | Cds1 | Chk2 | Effector | FHA kinase; phosphorylates factors in effector pathways |
| Chk1 | Chk1 | Chk1 | Effector | kinase; phosphorylates factors in effector pathways |
This is the common set of checkpoint proteins, and other factors contributing to checkpoints relating to dsDNA breaks (the MRN complex, etc.), fork pausing (Tof1, Csm3, etc.), and mitosis (cohesin, etc.) are not listed.
The appropriate DNA substrates for checkpoint initiation can be generated by several pathways. DNA double strand breaks are processed by several nucleases and helicases to form single-stranded DNA regions with 3′-single-stranded DNA overhangs and 5′-single- to double-stranded DNA junctions (5′-ss/ds-junctions) [6,7]. Damage produced by UV irradiation or by other DNA damaging agents that elicit nucleotide excision repair (NER) is processed by the NER machinery to form single stranded DNA gaps [8,9]. Single-stranded DNA also accumulates at stalled replication forks [10]. These various types of ssDNA regions bind the single-strand binding protein RPA. Both RPA-coated ssDNA and the ss/ds DNA junctions that have been generated during DNA processing are instrumental in the recruitment of checkpoint complexes.
In this review, we will focus primarily on the initial steps in checkpoint activation involving the recognition of DNA intermediates by checkpoint factors, and on the activation of Mec1/ATR by two specific activators. The first activator is the checkpoint clamp 9-1-1 that is loaded onto 5′-ss/ds DNA junctions (5′-junctions). The second activator is TopBP1/Dpb11 that may be recruited to the DNA by at least two distinct mechanisms, through interaction with the 9-1-1 camp, or through interaction with the leading strand DNA polymerase ε (Pol ε). The juxtaposition of Mec1/ATR and an activator initiates the checkpoint by stimulating the kinase activity of Mec1/ATR. Unfortunately, the nomenclature for these checkpoint factors is extremely confusing (Table I). We will restrict our discussion wherever possible to the human and budding yeast nomenclature while recognizing that seminal insights in checkpoint function have also been derived from studies in fission yeast. Potential other functions of 9-1-1 in damage-induced mutagenesis [11,12] and in base excision repair [13–16] fall outside the scope of this review.
PCNA and RFC as a model for 9-1-1 and its loader
Proliferating cell nuclear antigen (PCNA) is the homotrimeric circular clamp that functions as a processivity factor during DNA replication and repair. RFC is a heteropentameric complex consisting of the large Rfc1 subunit and four small subunits, Rfc2–5. These clamp loading complexes are conserved in bacteriophages such as T4, bacteria, archaea, and eukaryotes [17]. Each Rfc subunit belongs to the AAA+ protein family with typical ATP-binding motifs that convert the energy from ATP binding and hydrolysis to the mechanical force that mediates the process of clamp loading. The only exception is the Rfc5 subunit, which is structurally conserved with other AAA+ proteins but does not bind ATP. As the prototypical eukaryotic clamp loader, yeast RFC has been most extensively studied (reviewed in [18]), and a crystal structure of the yeast PCNA-RFC complex is available (Figure 1). Since PCNA and RFC represent a structural model for the checkpoint clamp and clamp loader, we will first discuss its mechanism, and then discuss similarities and differences with the checkpoint factors.
Figure 1. Structure of PCNA-RFC as a model for the checkpoint clamp and loader.
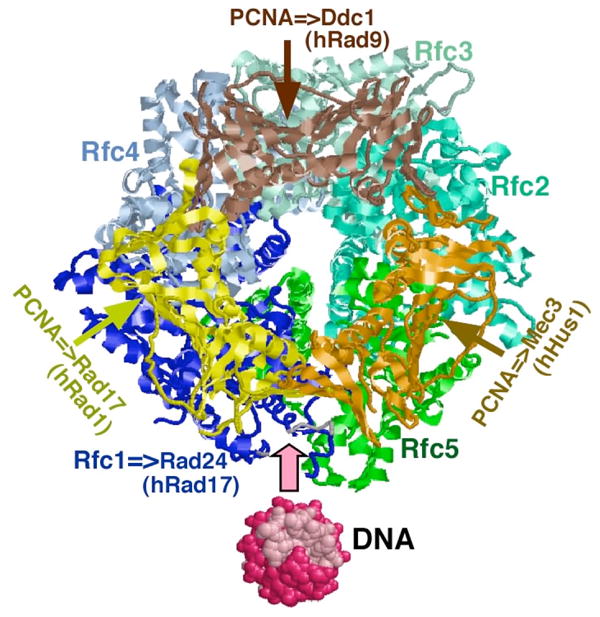
A Rasmol representation of the complex is shown [89], looking down along the dsDNA axis. In the S. cerevisiae PCNA-RFC complex, ATP binding to the Rfc2, Rfc3 and Rfc4 subunits opens the RFC ring between Rfc1 and Rfc5 with associated PCNA opening between two of the monomers [20]. In the analogous PCNA-Rad24-RFC complex, opening between the Rad24 and Rfc5 subunits is proposed to be associated with clamp opening between the Rad17 and Mec3 subunits [31]. Note that S. cerevisiae Rad17 is a clamp subunit, whereas human Rad17 is the loader subunit (Table 1).
PCNA loading by RFC proceeds via an ordered mechanism requiring binding of the clamp to the loader prior to binding of this complex to primer/template DNA. Each step in the loading process is associated with the binding of ATP. Thus, RFC alone binds two ATPs whereas binding of PCNA to RFC induces a conformational change that allows binding of a third ATP [19]. At this stage, the PCNA ring can be observed as a ring-opened complex ready to engage and encircle effector DNA [20]. The association of this ring-opened complex with template primer DNA with a 3′-ss/ds-junction induces another conformational change that allows binding of a fourth ATP molecule. Hydrolysis of bound ATP with concomitant closure of the PCNA ring and release of RFC completes the process of clamp loading. With the exception of the final step involving PCNA closure around the DNA, all steps in this sequential pathway can be carried out with ATPγS, a non-hydrolysable analog of ATP, indicating that those steps are solely driven by the energy derived from ATP binding. Remarkably, such an exquisite regulation by step-wise ATP binding does not exist in prokaryotic and phage clamp loading systems, and is therefore not a priori required. However, this stepwise mechanism should in principle not only permit editing of the process at multiple steps, but also allow for divergence of the default process into more specialized modes. Indeed, multiple clamp loading systems have evolved in eukaryotes, each system consisting of a core containing the Rfc2, Rfc3, Rfc4, and Rfc5 subunits together with a separate large subunit that replaces Rfc1.
Currently, three alternative RFC-like clamp loaders have been identified (reviewed in [18]). The checkpoint clamp loader Rad24-RFC will be the focus of our discussion below. The Ctf18-RFC complex links the biochemical activities of PCNA loading and unloading to the establishment of sister chromatid cohesion [21–24]. The Elg1-RFC clamp loader is more enigmatic because of the pleiotropic nature of the ELG1 deletion [25–27]. So far no PCNA loading or unloading function has been ascribed to this complex. Neither RFC nor Ctf18-RFC or Elg1-RFC show demonstrable activity with the 9-1-1 clamp, which so far is solely targeted by Rad24-RFC [28]. Additional Rfc1-like AAA+ proteins exist in eukaryotes that function in DNA metabolism, e.g. yeast Mgs1 and Rvb2, and might be potential candidates for forming alternative clamp loaders.
The heterotrimeric 9-1-1 clamp is structurally related to PCNA. Its S. cerevisiae subunits are Ddc1, Mec3 and Rad17; they are the respective homologs of S. pombe and human Rad9, Rad1, and Hus1, hence the designation 9-1-1 [29] (Table 1). Protein threading analyses coupled with biochemical studies and structural modeling have provided a good working model for the PCNA-like domains of the three subunits [29–31]. In contrast, each of the C-terminal tails of the three subunits shows high evolutionary divergence, and can be poorly fitted to secondary structure assignments. However, despite this apparent lack of conservation, in particular the C-terminal tail of Ddc1/Rad9 is important for the transmission of the regulatory functions of 9-1-1 (see below). The checkpoint clamp loader is a heteropentamer of the Rfc2-5 core assembly together with Rad24/Rad17. The Rad24 subunit of S. cerevisiae Rad24-RFC has an ATP binding site that is essential for loading 9-1-1 in vitro and for checkpoint function in vivo [32,33].
The strong structural parallels between the RFC+PCNA and the 9-1-1+Rad24-RFC systems suggest an analogous loading and operating mechanism for the checkpoint clamp. Key characteristics of the RFC+PCNA system are the requirement for ATP for loading, the requirement for a 3′-ss/ds-junction as an entry site for the RFC-PCNA complex, and the ability of PCNA to slide across dsDNA after loading. Indeed, proper loading of the 9-1-1 clamp also requires the energy of ATP hydrolysis [34–37]. Like PCNA, the yeast 9-1-1 ring has the ability to slide across dsDNA [35]. However, sliding by 9-1-1 was only observed when the clamp was loaded onto naked DNA, and it was strongly inhibited when the ssDNA adjoining the ss/ds junction was coated with RPA. Presumably, the protein-protein interactions between RPA and the loader, between RPA and 9-1-1, and between the loader and 9-1-1 serve to stabilize this complex of three factors at the site of loading [36,38]. These properties suggest that 9-1-1 may act primarily at the very site where it is loaded by the loader.
Some disagreement existed in the literature regarding the exact DNA substrate for loading of the checkpoint clamp, that with a 3′-ss/ds-junction, a 5′-ss/ds-junction, or either junction. In both the yeast and human systems, indiscriminate 9-1-1 loading was observed onto either the 3′- or 5′-junction of naked DNA [35,36]. However, another study with the human factors concluded that a marked preference existed for a RPA-coated DNA substrate with a 5′-junction [37]. From a study with the yeast factors, it was concluded that these differential loading results could be reconciled if the role of RPA was taken into account [38]. Under conditions of complete coating of the ssDNA by RPA, 9-1-1 loading onto 3′-junctions was inhibited, thereby endowing a unique specificity of the checkpoint loader for 5′-junctions (Figure 2).
Figure 2. RPA-coated DNA restricts loading of the checkpoint clamp to 5.
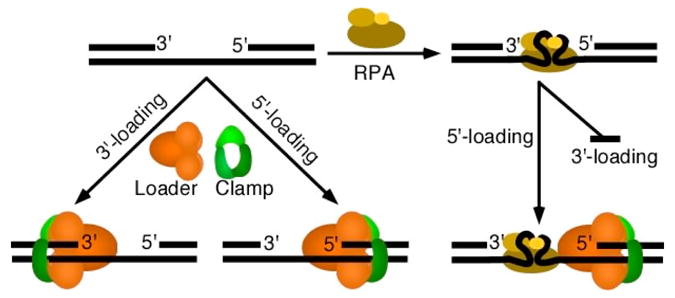
′-junctions. The loader is Rad24/Rad17-RFC and the clamp is 9-1-1. For a full discussion of the role of RPA in loading specificity, see [38].
Recruitment of 9-1-1 to sites of damage
Genetic and cell biological studies in mammals and in both yeast models, S. cerevisiae and S. pombe, have been instrumental in identifying pathways in which 9-1-1 functions, and in suggesting biochemical functions that these complexes should carry out in order to fulfill their proposed pathway functions. The DNA damage checkpoint in both organisms is abrogated when any of the three subunit genes of 9-1-1 is deleted, or the Rad24/Rad17 subunit of the clamp loader [39–42]. Since the clamp loader is required for the known functions of 9-1-1, this indicates that the clamp only shows checkpoint activity when loaded onto DNA. The loaded 9-1-1 clamp is involved in the recruitment of the replication initiator protein Dpb11/Cut5/TopBP1, which activates Mec1/ATR kinase activity. Since both the recruitment and deployment of Dpb11/Cut5/TopBP1 varies between organisms, this factor will be discussed in a separate section below.
During the G1 phase of the cell cycle, the processing of DNA damage by the nucleotide excision repair (NER) machinery is a required step for checkpoint activation in S. cerevisiae and human, suggesting that the gaps generated during NER may form loading sites for 9-1-1 [8,9]. Checkpoint activation in human G0 and G2/M fibroblasts, but not in S phase cells, depends on several NER components. The processing of lesions by the global genome NER repair machinery rather than the transcriptionally coupled machinery is required for triggering the DNA damage checkpoint after UV radiation [9]. Furthermore, the S. cerevisiae Rad14 protein (human XPA) was shown to bind 9-1-1, suggesting a model for NER-facilitated recruitment of 9-1-1 to sites of active repair [8]. When replication forks stall because of DNA damage or nucleotide precursor depletion, for instance in hydroxyurea treated cells, the uncoupling of helicase and polymerase activities generates long stretches of ssDNA. Given the loading specificity of Rad24/Rad17-RFC, we hypothesize that the 5′-ends of Okazaki fragments are the preferred loading sites for 9-1-1.
Mec1/ATR is the transducer kinase required for checkpoint initiation in response to some forms of DNA damage and in response to stalling of the replication fork. The Mec1/ATR kinase is recruited by its subunit Ddc2/ATRIP to damage sites through interactions between RPA and Ddc2 [43,44]. Although earlier studies in S. cerevisiae reported that 9-1-1 and Mec1-Ddc2 are recruited to double strand DNA breaks independently of each other [45,46], recent studies suggest that 9-1-1 may also participate in recruiting Mec1-Ddc2 to double strand breaks [47,48]. This could partly be due to one proposed function of 9-1-1, its ability to recruit a 5′-exonuclease [49]. Multiple nucleases such as Exo1, Dna2, Sae2 and Mre11-Rad50-Xrs2 seem to play a role in the generation of 5′-junctions at double strand breaks, and it remains to be determined which of these nucleases, or perhaps an additional nuclease, are regulated by the 9-1-1 clamp [6,7]. The enhanced generation of ssDNA that is coated with RPA would provide increased loading sites for Mec1/ATR [50]. Genetic studies of the checkpoint at dysfunctional telomeres in S. cerevisiae indeed shows that the regulated 5′-exonucleolytic resection of DNA is in part dependent on the presence of 9-1-1 [49].
The S. cerevisiae 9-1-1 clamp activates Mec1
Mec1 has a low basal kinase activity that phosphorylates its partner Ddc2 during G2 [51]. Phosphorylation of the other known targets of Mec1 requires its activation. A biochemical analysis of the S. cerevisiae checkpoint machinery shows that loading of the 9-1-1 clamp onto DNA is both required and sufficient for the activation of Mec1 [28]. Mec1 activation by the 9-1-1 clamp requires that it is loaded onto ss/ds junction DNA by the Rad24-RFC clamp loader. Alternative clamp loaders like Elg1-RFC and Ctf18-RFC do not substitute for Rad24-RFC in loading 9-1-1 onto DNA and hence do not mediate activation of Mec1. The polarity of the ss/ds junction plays a decisive role in clamp loading and subsequent Mec1 activation. As discussed above, in the absence of RPA, Rad24-RFC loads 9-1-1 on either 3′- or 5′-junctions and either complex can activate Mec1, suggesting that the mere encircling of DNA by 9-1-1 suffices for activation. However, when the effector DNA is coated with RPA, Mec1 activation occurs only with 5′-junction substrates and not with 3′-junction substrates that are inactive for loading 9-1-1 (Figure 2). In addition, RPA enhances Mec1 activation by the 9-1-1 clamp, likely by bringing Mec1 into juxtaposition with the clamp through RPA-Ddc2 interactions [28,44]. Studies in Xenopus egg extracts are consistent with this specificity for 9-1-1 loading as the initial step in ATR activation [52]. Addition of primed ssDNA to Xenopus extracts suffices for ATR activation. By blocking either the 5′-junction or the 3′-junction, it was determined that accessibility to the 5′-junction is essential for checkpoint activation, consistent with a model of checkpoint initiation through loading of 9-1-1 at 5′-junctions.
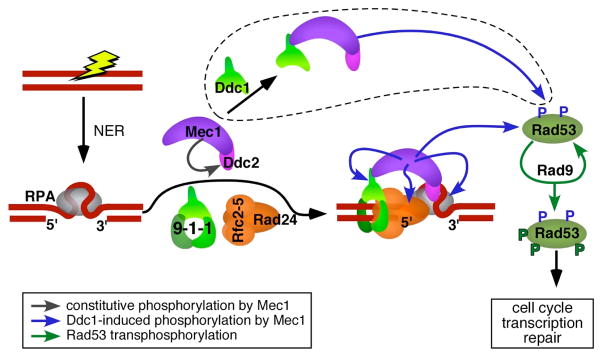
The activation of Mec1 appears to be global in nature in which all targets examined are phosphorylated at a greatly increased rate (Figure 3). Among these are the Rad24 subunit of the loader and the Mec3 and Ddc1 subunits of the clamp, the Rpa1 and Rpa2 subunits of RPA, and the effector kinase Rad53 [28]. All of these proteins fulfill essential checkpoint functions and their phosphorylation is important in mediating protein-protein interactions, in the DNA repair process itself, and in propagating the checkpoint signal transduction pathway. Rpa2 phosphorylation by ATR in human cells is critical in slowing down the DNA replication and in the redirection of RPA to DNA repair [53,54]. Phosphorylation of the human and S. pombe checkpoint clamp subunit Rad9 and the homologous S. cerevisiae Ddc1 subunit by Mec1/ATR is instrumental in further checkpoint activation by linking 9-1-1 to TopBP1/Cut5/Dpb11, discussed below. Rad53 phosphorylation by Mec1 is crucial in mediating cell cycle arrest and transcriptional induction of repair genes in response to genotoxic stress [55–57](Figure 3). In addition, activated Rad53 and Mec1 function in the stabilization of stalled replication forks [58,59].
Figure 3. Model for checkpoint activation during the DNA damage response in S. cerevisiae.
The loading of 9-1-1 at RPA-coated gaps, generated during NER of DNA damage, activates Mec1. The various types of phosphorylations by Mec1 are indicated with arrows, but not all targets are shown. The Ddc1-only bypass pathway of Mec1 activation, surrounded by a dotted line, is only operative at low salt in vitro and appears not to operate in the cell, unless Mec1 and Ddc1 are artificially colocalized by protein fusions [60]. Multiple Mec1-phosphorylated Rad53 proteins bind the mediator Rad9, and undergo transphosphorylation to complete full activation of Rad53. The S. cerevisiae factors are shown. Note that S. cerevisiae Rad9 is a mediator protein, and human Rad9 a clamp protein (Table 1).
The biochemical analysis of these checkpoint factors has allowed the utilization of assay conditions that permit additional insights into checkpoint activation, but that may not be of relevance in the cell. Thus, at very low NaCl concentrations, a modest activation of Mec1 by 9-1-1 was observed in the absence of Rad24-RFC or DNA [28]. This activation potential was shown to reside in the Ddc1 subunit of 9-1-1. The Rad17 and Mec3 subunits of 9-1-1 were neither required for low-salt activation nor could they activate Mec1 under these conditions. Consistent with these results, the Ddc1 subunit binds Mec1-Ddc2. The Mec1 activation domain maps to the unstructured C-terminal tail of Ddc1 (Navadgi-Patil and Burgers, unpublished results).
In a remarkable validation of the biochemical approach to checkpoint studies, all essential conclusions derived from these biochemical studies were confirmed by an in vivo fusion approach that short-circuits the checkpoint pathway. In this approach, the Ddc1 subunit of the clamp and the Ddc2 subunit of the Mec1-Ddc2 complex were brought into juxtaposition by fusing the genes to the LacI protein in a strain harboring a large array of Lac operator sequences [60]. This juxtaposition sufficed to cause some activation of the checkpoint in the absence of DNA damage. This activation relied neither on Rad24-RFC nor on the other clamp subunits. Substituting LacI-Ddc1 for another 9-1-1 fusion subunit, LacI-Rad17 or LacI-Mec3, failed to cause gratuitous activation in a ddc1Δ deletion mutant indicating a requirement for Ddc1. Therefore, both the biochemical and genetic studies agree that the critical step in Mec1 activation is accomplished by bringing Ddc1 and Mec1 into close proximity.
TopBP1/Cut5/Dpb11 is an essential replication and checkpoint protein
Vertebrate TopBP1, S. pombe Cut5/Rad4, and S. cerevisiae Dpb11 are multiple-BRCT domain (breast cancer 1 C-terminal domain) containing proteins that are essential for replisome assembly and for the DNA replication checkpoint (reviewed in [61]). The mechanistic details of the essential replication role of this protein have recently been elucidated in budding yeast. The BRCT repeats in Dpb11 interact with the Sld2 and Sld3 initiator proteins that have previously been phosphorylated by the S-phase CDK/cyclin kinase complex, and the formation of a stable Sld2-Dpb11-Sld3 complex is a crucial step in the initiation of DNA replication [62,63]. S. cerevisiae Dpb11 also interacts with Pol ε and loading of Pol ε onto the pre-replication complex is dependent on Dpb11 [64,65]. A C-terminal truncation mutant of DPB11 shows replication defects at elevated temperatures and is defective for the replication checkpoint in hydroxyurea-treated cells [64]. Similar to Dpb11, S. pombe Cut5/Rad4 and human and Xenopus TopBP1 are also essential proteins that function in DNA replication and in the replication checkpoint [66–69].
Once the replisome has been assembled and replication initiated, TopBP1/Cut5/Dpb11 does not appear to remain with the replication fork machinery during elongation. In S. cerevisiae, Dpb11 is not found as a component of the Replication Progression Complex that contains many other replication factors associated with the MCM helicase, as well as the replication checkpoint mediator protein Mrc1, and several chromatin- and cohesion-associated proteins [70,71], nor could Dpb11 be detected at replication forks by chromatin immunoprecipitation analysis [65]. Similarly, in human cells, TopBP1 does not localize to PCNA-containing replication foci [67]. However, when replication forks are stalled by hydroxyurea treatment, TopBP1 foci co-localize with PCNA foci. If this mode of localization prevails in other organisms, TopBP1/Cut5/Dpb11 needs to be recruited back to stalled replication forks. TopBP1/Cut5/Dpb11 interacts with the Rad9/Ddc1 subunit of the 9-1-1 clamp in all model organisms in which this has been studied. One function of 9-1-1 is to recruit Cut5/TopBP1/Dpb11 to stalled DNA replication forks.
Role of the 9-1-1 clamp in recruiting TopBP1/Dpb11/Cut5 to stalled replication forks
Although S. pombe Cut5 is required for phosphorylation of the downstream effector kinases Chk1 and Cds1 after DNA damage, it is not required for phosphorylation of the 9-1-1 clamp subunits and Rad26/ATRIP, suggesting that Cut5/TopBP1 does not function in the very early steps of checkpoint activation [72]. In order for S. pombe Cut5 to form a complex with Rad3/ATR and activate it, Cut5 has to bind to phosphorylated serines/threonines in the C-terminal tail of Rad9 (Figure 4). Substitution of these critical residues with alanine leads to checkpoint defects and reduced cell survival after hydroxyurea treatment [73]. This type of phosphorylation-mediated Rad9-TopBP1 interaction is conserved in the other model organisms, S. cerevisiae, X. laevis, and human [74–78].
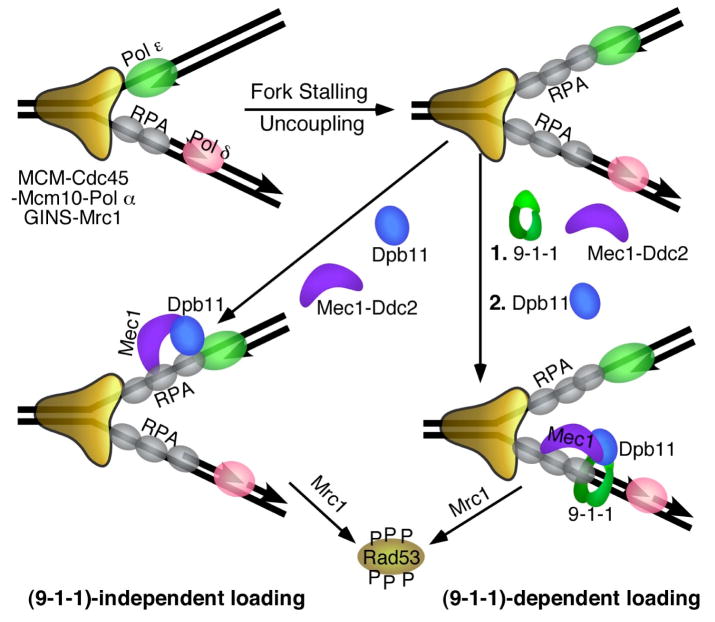
Figure 4. The DNA replication checkpoint.
S. cerevisiae proteins that are involved in checkpoint function are shown. See Table 1 for the homologous human proteins. Stalling of the fork results in uncoupling of the leading and lagging strands, and in the formation of extended RPA-coated ssDNA regions that recruit Mec1. The 9-1-1 clamp is shown loaded onto the 5′-junction of an Okazaki fragment because of its demonstrated loading specificity, but might also load onto the leading strand after fork restart by Pol α-primase [90]. Recruitment of Dpb11/TopBP1/Cut5 to the fork is facilitated through interactions with 9-1-1 ((9-1-1)-dependent loading, right). In S. cerevisiae, (9-1-1)-independent recruitment of Dpb11 may also occur, possibly through interactions with Pol ε. Dpb11/TopBP1/Cut5 activates Mec1/ATR to phosphorylate Rad53/Chk1 and additional factors. Mrc1/Claspin is the proposed downstream chromatin-associated mediator that activates transphosphorylation of Rad53/Chk1 [91].
BRCT repeats I and II of Xenopus and human TopBP1 are involved in the interaction with Rad9 [77], while in S. pombe and S. cerevisiae BRCT repeats III and IV perform this function [73,74]. In addition to its phosphorylation after DNA damage, Ddc1/Rad9 is also phosphorylated during S phase [73,79], suggesting that 9-1-1 may form a complex with Dpb11/Cut5 throughout S phase. This would facilitate the loading of Dpb11/Cut5 at stalled replication forks through stable interactions with 9-1-1.
In S. pombe and metazoan cells, the 9-1-1-mediated recruitment of Cut5/TopBP1 appears to be the dominant mechanism. No significant S phase damage checkpoint activity remains in a Rad9 mutant in S. pombe and in avian DT40 cells [40,80], nor in a Rad9-depleted Xenopus extract [77]. The checkpoint defect of avian Rad9−/− cells could be suppressed by fusion of the ATR-activation domain of TopBP1 to either PCNA or to histone H2A, thereby bypassing the requirement for 9-1-1 in localizing TopBP1 to stalled replication forks [76]. In contrast, in S. cerevisiae, a functional replication checkpoint remains in ddc1Δ cells defective for 9-1-1, although the presence of 9-1-1 does mediate a more robust checkpoint [74]. In ddc1Δ cells, Dpb11 may be recruited to stalled replication forks by an alternative mechanism (Figure 4). One likely candidate for recruitment is Pol ε that is known to bind Dpb11 [64,65]. Apparently, in other eukaryotes this mode of recruitment is either lacking or inefficient. Interestingly, mutations in the catalytic POL2 gene for S. cerevisiae Pol ε are defective for the replication checkpoint, whereas mutations in the S. pombe Pol2 gene fail to show a checkpoint defect [81,82].
Mec1/ATR activation by TopBP1/Dpb11
So far, there are no reports that the 9-1-1 clamp from higher eukaryotes can activate the ATR kinase though it is required for checkpoint activation through its TopBP1 recruitment function. Progress in this field came from the discovery that Xenopus TopBP1 activates ATR kinase [83]. In these in vitro studies, purified Xenopus TopBP1 was incubated with ATR/ATRIP that had been immunoprecipitated from Xenopus egg extracts and the activation of ATR kinase activity observed. Activated ATR phosphorylated several targets at a greatly increase rate including Mcm2, Chk1, and human PHAS-1. The latter is a general substrate for PI3-kinase-like kinases, and is in fact also phosphorylated by 9-1-1-activated yeast Mec1 [28,84]. Remarkably, Xenopus TopBP1 can directly activate ATR without the necessity of either DNA or RPA [83]. The 1513 a.a. long Xenopus TopBP1 has eight BRCT domains. The ATR activating domain (AAD) lies between BRCT6 and BRCT7, and this single, bacterially expressed 215 a.a. domain suffices for activation. Consistent with the strong conservation between Xenopus and human TopBP1, human TopBP1 was also found to activate human ATR [85]. However, in this study it was reported that the presence of DNA further stimulated the activation of ATR by TopBP1. Protein mapping studies and mutational studies showed that the activation of ATR by TopBP1 is mediated by interactions with both ATR and ATRIP, the regulatory subunit of ATR [86].
The architecture of the C-terminal half of TopBP1 including the AAD domain is only conserved in vertebrate cells and cannot be identified in either yeast. Considering that S. cerevisiae Dpb11 and S. pombe Cut5 do not possess this conserved activation domain, it was questionable whether these proteins would serve as Mec1/ATR activators. However, in analogy to Xenopus TopBP1, the simple addition of S. cerevisiae Dpb11 to Mec1 suffices for kinase activation towards all of its downstream targets [87.92]. Neither DNA nor RPA is required for robust activation of Mec1. Furthermore, in agreement with the demonstrated checkpoint defect of the dpb11-1 mutant lacking its C-terminal tail [64], this truncated form of Dpb11 also fails to activate Mec1, suggesting that the determinants for binding and activating Mec1 reside in the C-terminal tail of Dpb11, beyond its BRCT domains. In this biochemical assay, substantial synergism in activation is observed when both the Dpb11 and 9-1-1 activators are present. Dpb11-activated Mec1 and 9-1-1-activated Mec1 show very similar kinase activities; the phosphorylation of targets such as Rad53, RPA, and PHAS-1 is dramatically enhanced by either activator [87].
ATR activation mechanisms in different organisms
One interesting feature of Mec1/ATR checkpoint activation is that the relevant activation domains of both activators reside in flexible C-terminal tails. In contrast to the structured PCNA-like domains of Rad9/Ddc1 that constitute the major section of this 9-1-1 subunit, the C-terminal tail lacks a well-defined structure and is poorly conserved. Yet, the phosphorylation sites on Rad9/Ddc1 that mediate the interaction with TopBP1/Cut5/Dpb11 reside in this tail, and the Mec1-activation region of S. cerevisiae Ddc1 has also been mapped to the unstructured tail of this protein (Navadgi-Patil and Burgers, unpublished results). Dpb11 consists mainly of well-defined BRCT motifs, yet its C-terminal tail required for activating Mec1 is also poorly conserved. It cannot be reliably aligned with the AAD region of vertebrate TopBP1 that activates ATR. Both the activation region of TopBP1 and that of Dpb11 lack a well-defined structure. The flexible nature that these protein domains likely possess may be important in mediating the protein-protein interactions that are important for functionality.
Considering the strong conservation of checkpoint proteins in eukaryotic organisms it is puzzling to note that both S. cerevisiae 9-1-1 and Dpb11 can activate Mec1, whereas in vertebrates only TopBP1 has been shown to activate ATR, with 9-1-1 serving a recruitment function. Because of the essential function of TopBP1/Cut5/Dpb11 in DNA replication, it is difficult to approach this problem by genetic means because mutations in TopBP1/Cut5/Dpb11 that affect checkpoint function may also affect its replication function. An extensive mutational study of S. cerevisiae DPB11 illustrates this problem. All four dpb11 alleles isolated based upon hydroxyurea sensitivity were temperature-sensitive for growth and showed replication defects while all seven dpb11 alleles isolated on the basis of temperature-sensitivity were sensitive to hydroxyurea and showed checkpoint defects [74]. This study and similar studies in S. pombe with Cut5/Rad4 suggest that the replication initiation and checkpoint functions of TopBP1 are more closely linked than is generally assumed [61].
The question whether 9-1-1 can activate ATR in organisms other than S. cerevisiae remains. Studies in Xenopus and S. pombe have shown that the Rad1 and Rad9 subunits of the 9-1-1 clamp are phosphorylated by ATR in response to DNA damage or to inhibition of DNA replication, and this phosphorylation is independent of TopBP1/Cut5 [72,88]. The question raised by these studies is the following: is this phosphorylation of 9-1-1 carried out by the basal activity of ATR, or does 9-1-1 activate ATR just to mediate phosphorylation of its own subunits? If so, the further activation of ATR by TopBP1/Cut5 would be required for the phosphorylation of all other targets of ATR. Biochemical studies of checkpoint factors from these organisms should help in resolving this uncertainty.
Acknowledgments
The authors thank Jurek Majka for help and critical discussions during the course of this work. Checkpoint studies in the authors’ laboratory are supported in part by Grant GM32431 from the National Institutes of Health.
Abbreviations
- ATR
ataxia telangiectasia (ATM) related protein
- BRCT
Brca1 C-terminus
- RPA
replication protein A
- ssDNA
single stranded DNA
- PCNA
proliferating cell nuclear antigen
- RFC
complex of Rfc1-5 subunits
- 9-1-1
Rad9-Hus1-Rad1 checkpoint clamp in S. pombe and higher eukaryotes and Ddc1-Rad17-Mec3 in S. cerevisiae
- Rad24/Rad17-RFC
complex of Rfc2-5 with S. cerevisiae Rad24 (or human Rad17)
- NER
nucleotide excision repair
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 2.Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Lambert S, Carr AM. Checkpoint responses to replication fork barriers. Biochimie. 2005;87:591–602. doi: 10.1016/j.biochi.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 4.Cobb JA, Shimada K, Gasser SM. Redundancy, insult-specific sensors and thresholds: unlocking the S-phase checkpoint response. Curr Opin Genet Dev. 2004;14:292–300. doi: 10.1016/j.gde.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 6.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giannattasio M, Lazzaro F, Longhese MP, Plevani P, Muzi-Falconi M. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J. 2004;23:429–438. doi: 10.1038/sj.emboj.7600051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marini F, Nardo T, Giannattasio M, Minuzzo M, Stefanini M, Plevani P, Muzi Falconi M. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc Natl Acad Sci U S A. 2006;103:17325–17330. doi: 10.1073/pnas.0605446103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulovich AG, Armour CD, Hartwell LH. The Saccharomyces cerevisiae RAD9, RAD17, RAD24 and MEC3 genes are required for tolerating irreparable, ultraviolet-induced DNA damage. Genetics. 1998;150:75–93. doi: 10.1093/genetics/150.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabbioneda S, Minesinger BK, Giannattasio M, Plevani P, Muzi-Falconi M, Jinks-Robertson S. The 9-1-1 checkpoint clamp physically interacts with polzeta and is partially required for spontaneous polzeta-dependent mutagenesis in Saccharomyces cerevisiae. J Biol Chem. 2005;280:38657–38665. doi: 10.1074/jbc.M507638200. [DOI] [PubMed] [Google Scholar]
- 13.Wang W, Brandt P, Rossi ML, Lindsey-Boltz L, Podust V, Fanning E, Sancar A, Bambara RA. The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc Natl Acad Sci U S A. 2004;101:16762–16767. doi: 10.1073/pnas.0407686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi G, Chang DY, Cheng CC, Guan X, Venclovas C, Lu AL. Physical and functional interactions between MutY glycosylase homologue (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem J. 2006;400:53–62. doi: 10.1042/BJ20060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gembka A, Toueille M, Smirnova E, Poltz R, Ferrari E, Villani G, Hubscher U. The checkpoint clamp, Rad9-Rad1-Hus1 complex, preferentially stimulates the activity of apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta in long patch base excision repair. Nucleic Acids Res. 2007;35:2596–2608. doi: 10.1093/nar/gkl1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan X, Madabushi A, Chang DY, Fitzgerald ME, Shi G, Drohat AC, Lu AL. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates DNA repair enzyme TDG glycosylase. Nucleic Acids Res. 2007;35:6207–6218. doi: 10.1093/nar/gkm678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cullmann G, Fien K, Kobayashi R, Stillman B. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol. 2004;78:227–260. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- 19.Gomes XV, Schmidt SL, Burgers PM. ATP utilization by yeast replication factor C. II. Multiple stepwise ATP binding events are required to load proliferating cell nuclear antigen onto primed DNA. J Biol Chem. 2001;276:34776–34783. doi: 10.1074/jbc.M011743200. [DOI] [PubMed] [Google Scholar]
- 20.Zhuang Z, Yoder BL, Burgers PM, Benkovic SJ. The structure of a ring-opened proliferating cell nuclear antigen-replication factor C complex revealed by fluorescence energy transfer. Proc Natl Acad Sci U S A. 2006;103:2546–2551. doi: 10.1073/pnas.0511263103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell. 2001;7:959–970. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- 22.Bermudez VP, Maniwa Y, Tappin I, Ozato K, Yokomori K, Hurwitz J. The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc Natl Acad Sci U S A. 2003;100:10237–10242. doi: 10.1073/pnas.1434308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiomi Y, Shinozaki A, Sugimoto K, Usukura J, Obuse C, Tsurimoto T. The reconstituted human Chl12-RFC complex functions as a second PCNA loader. Genes Cells. 2004;9:279–290. doi: 10.1111/j.1356-9597.2004.00724.x. [DOI] [PubMed] [Google Scholar]
- 24.Bylund GO, Burgers PM. Replication Protein A-Directed Unloading of PCNA by the Ctf18 Cohesion Establishment Complex. Mol Cell Biol. 2005;25:5445–5455. doi: 10.1128/MCB.25.13.5445-5455.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bellaoui M, Chang M, Ou J, Xu H, Boone C, Brown GW. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. Embo J. 2003;22:4304–4313. doi: 10.1093/emboj/cdg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-Aroya S, Koren A, Liefshitz B, Steinlauf R, Kupiec M. ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc Natl Acad Sci U S A. 2003;100:9906–9911. doi: 10.1073/pnas.1633757100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanellis P, Agyei R, Durocher D. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr Biol. 2003;13:1583–1595. doi: 10.1016/s0960-9822(03)00578-5. [DOI] [PubMed] [Google Scholar]
- 28.Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burtelow MA, Roos-Mattjus PM, Rauen M, Babendure JR, Karnitz LM. Reconstitution and molecular analysis of the hRad9-hHus1-hRad1 (9-1-1) DNA damage responsive checkpoint complex. J Biol Chem. 2001;276:25903–25909. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- 30.Thelen MP, Venclovas C, Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins [letter] Cell. 1999;96:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- 31.Venclovas C, Colvin ME, Thelen MP. Molecular modeling-based analysis of interactions in the RFC-dependent clamp-loading process. Protein Sci. 2002;11:2403–2416. doi: 10.1110/ps.0214302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majka J, Chung BY, Burgers PM. Requirement for ATP by the DNA damage checkpoint clamp loader. J Biol Chem. 2004;279:20921–20926. doi: 10.1074/jbc.M400898200. [DOI] [PubMed] [Google Scholar]
- 33.Naiki T, Shimomura T, Kondo T, Matsumoto K, Sugimoto K. Rfc5, in cooperation with rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5888–5896. doi: 10.1128/mcb.20.16.5888-5896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci U S A. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci USA. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellison V, Stillman B. Biochemical Characterization of DNA Damage Checkpoint Complexes: Clamp Loader and Clamp Complexes with Specificity for 5′ Recessed DNA. PLoS Biol. 2003;1:231–243. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Majka J, Binz SK, Wold MS, Burgers PM. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem. 2006;281:27855–27861. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- 39.al-Khodairy F, Carr AM. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. Embo J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchetti MA, Kumar S, Hartsuiker E, Maftahi M, Carr AM, Freyer GA, Burhans WC, Huberman JA. A single unbranched S-phase DNA damage and replication fork blockage checkpoint pathway. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7472–7477. doi: 10.1073/pnas.112702399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paciotti V, Lucchini G, Plevani P, Longhese MP. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. Embo J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondo T, Matsumoto K, Sugimoto K. Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol Cell Biol. 1999;19:1136–1143. doi: 10.1128/mcb.19.2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rouse J, Jackson SP. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Molecular Cell. 2002;9:857–869. doi: 10.1016/s1097-2765(02)00507-5. [DOI] [PubMed] [Google Scholar]
- 44.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes.[see comment] Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 45.Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes & Development. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kondo T, Wakayama T, Naiki T, Matsumoto K, Sugimoto K. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science. 2001;294:867–870. doi: 10.1126/science.1063827. [DOI] [PubMed] [Google Scholar]
- 47.Dubrana K, van Attikum H, Hediger F, Gasser SM. The processing of double-strand breaks and binding of single-strand-binding proteins RPA and Rad51 modulate the formation of ATR-kinase foci in yeast. J Cell Sci. 2007;120:4209–4220. doi: 10.1242/jcs.018366. [DOI] [PubMed] [Google Scholar]
- 48.Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell. 2008;30:73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zubko MK, Guillard S, Lydall D. Exo1 and Rad24 Differentially Regulate Generation of ssDNA at Telomeres of Saccharomyces cerevisiae cdc13-1 Mutants. Genetics. 2004;168:103–115. doi: 10.1534/genetics.104.027904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aylon Y, Kupiec M. The checkpoint protein Rad24 of Saccharomyces cerevisiae is involved in processing double-strand break ends and in recombination partner choice. Mol Cell Biol. 2003;23:6585–6596. doi: 10.1128/MCB.23.18.6585-6596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paciotti V, Clerici M, Lucchini G, Longhese MP. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- 52.MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olson E, Nievera CJ, Klimovich V, Fanning E, Wu X. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J Biol Chem. 2006;281:39517–39533. doi: 10.1074/jbc.M605121200. [DOI] [PubMed] [Google Scholar]
- 54.Vassin VM, Wold MS, Borowiec JA. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol Cell Biol. 2004;24:1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 57.Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO Journal. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 59.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects.[see comment] Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 60.Bonilla CY, Melo JA, Toczyski DP. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol Cell. 2008;30:267–276. doi: 10.1016/j.molcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst) 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 62.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 63.Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 64.Araki H, Leem SH, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci USA. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Masumoto H, Sugino A, Araki H. Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol Cell Biol. 2000;20:2809–2817. doi: 10.1128/mcb.20.8.2809-2817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saka Y, Fantes P, Sutani T, McInerny C, Creanor J, Yanagida M. Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. Embo J. 1994;13:5319–5329. doi: 10.1002/j.1460-2075.1994.tb06866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Makiniemi M, Hillukkala T, Tuusa J, Reini K, Vaara M, Huang D, Pospiech H, Majuri I, Westerling T, Makela TP, Syvaoja JE. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J Biol Chem. 2001;276:30399–30406. doi: 10.1074/jbc.M102245200. [DOI] [PubMed] [Google Scholar]
- 68.Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol. 2002;22:555–566. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parrilla-Castellar ER, Karnitz LM. Cut5 is required for the binding of Atr and DNA polymerase alpha to genotoxin-damaged chromatin. J Biol Chem. 2003;278:45507–45511. doi: 10.1074/jbc.C300418200. [DOI] [PubMed] [Google Scholar]
- 70.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 71.Hodgson B, Calzada A, Labib K. Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol Biol Cell. 2007;18:3894–3902. doi: 10.1091/mbc.E07-05-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harris S, Kemplen C, Caspari T, Chan C, Lindsay HD, Poitelea M, Carr AM, Price C. Delineating the position of rad4+/cut5+ within the DNA-structure checkpoint pathways in Schizosaccharomyces pombe. J Cell Sci. 2003;116:3519–3529. doi: 10.1242/jcs.00677. [DOI] [PubMed] [Google Scholar]
- 73.Furuya K, Poitelea M, Guo L, Caspari T, Carr AM. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang H, Elledge SJ. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics. 2002;160:1295–1304. doi: 10.1093/genetics/160.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.St Onge RP, Besley BD, Pelley JL, Davey S. A role for the phosphorylation of hRad9 in checkpoint signaling. J Biol Chem. 2003;278:26620–26628. doi: 10.1074/jbc.M303134200. [DOI] [PubMed] [Google Scholar]
- 76.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 78.Puddu F, Granata M, Di Nola L, Balestrini A, Piergiovanni G, Lazzaro F, Giannattasio M, Plevani P, Muzi-Falconi M. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol. 2008;28:4782–4793. doi: 10.1128/MCB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Longhese MP, Paciotti V, Fraschini R, Zaccarini R, Plevani P, Lucchini G. The novel DNA damage checkpoint protein ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO Journal. 1997;16:5216–5226. doi: 10.1093/emboj/16.17.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kobayashi M, Hirano A, Kumano T, Xiang SL, Mihara K, Haseda Y, Matsui O, Shimizu H, Yamamoto K. Critical role for chicken Rad17 and Rad9 in the cellular response to DNA damage and stalled DNA replication. Genes Cells. 2004;9:291–303. doi: 10.1111/j.1356-9597.2004.00728.x. [DOI] [PubMed] [Google Scholar]
- 81.Navas TA, Zhou Z, Elledge SJ. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 82.Durso G, Nurse P. Schizosaccharomyces pombe cdc20(+) encodes dna polymerase epsilon and is required for chromosomal replication but not for the s phase checkpoint. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:12491–12496. doi: 10.1073/pnas.94.23.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 84.Lawrence JC, Lin TA, McMahon LP, Choi KM. Modulation of the protein kinase activity of mTOR. Curr Top Microbiol Immunol. 2004;279:199–213. doi: 10.1007/978-3-642-18930-2_12. [DOI] [PubMed] [Google Scholar]
- 85.Choi JH, Lindsey-Boltz LA, Sancar A. Reconstitution of a human ATR-mediated checkpoint response to damaged DNA. Proc Natl Acad Sci U S A. 2007;104:13301–13306. doi: 10.1073/pnas.0706013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem. 2008;283:35853–35859. doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lupardus PJ, Cimprich KA. Phosphorylation of Xenopus Rad1 and Hus1 defines a readout for ATR activation that is independent of Claspin and the Rad9 carboxy terminus. Mol Biol Cell. 2006;17:1559–1569. doi: 10.1091/mbc.E05-09-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 90.You Z, Kong L, Newport J. The role of single-stranded DNA and polymerase alpha in establishing the ATR, Hus1 DNA replication checkpoint. J Biol Chem. 2002;277:27088–27093. doi: 10.1074/jbc.M204120200. [DOI] [PubMed] [Google Scholar]
- 91.Zhao H, Russell P. DNA binding domain in the replication checkpoint protein Mrc1 of Schizosaccharomyces pombe. J Biol Chem. 2004;279:53023–53027. doi: 10.1074/jbc.M410449200. [DOI] [PubMed] [Google Scholar]
- 92.Mordes DA, Nam EA, Cortez D. Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci U S A. 2008;105:18730–18734. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]