Abstract
Dentin sialophosphoprotein (DSPP) consists of dentin sialoprotein (DSP) and dentin phosphoprotein (DPP). DSPP is highly expressed in mineralized tissues. However, recent studies have shown that DSPP is also expressed in several active metabolic ductal epithelial tissues and exists in a variety of sequences. We have investigated DSPP expression in various mouse tissues using RT-PCR, in situ hybridization and immunohistochemical analyses. To identify DSPP gene polymorphisms, we screened a mouse tooth cDNA library as well as isolated and characterized DSPP variations. Our results show that DSPP is predominantly expressed in teeth and moderately in bone tissues. We also have characterized a full-length DSPP cDNA clone with an open-reading frame of 940 codons and this polyadenylation signal. Compared to previously reported mouse DSPP cDNAs, 13 sequence variations were identified, including 8 non-synonymous single nucleotide polymorphisms and an in-frame indel (8 amino acids) at DPP domain of the mouse DSPP. These 8 amino acids are rich in aspartic acid and serine residues. Northern blot assay showed a prominent band at 4.4 kb. RT-PCR demonstrated that this mouse DSPP gene was dominantly expressed in teeth. The predicted secondary structure of DPP domain of this DSPP showed differences from the previously published mouse DPPs, implying that they play different roles during tooth development and formation.
Keywords: dentin sialophosphoprotein (DSPP), dentin sialoprotein (DSP), dentin phosphoprotein (DPP), polymorphism, dentinogenesis
1. Introduction
Dentin is the principal mineralized tissue of teeth and originates from odontoblasts which synthesize and secrete collagenous and non-collagenous proteins (NCPs) to form dentin extracellular matrix (ECM). It is believed that the NCPs are associated at specific sites on the collagen molecule to promote the nucleation and growth of hydroxyapatite crystals (Traub et al., 1992; He et al., 2005). Among the NCPs, dentin sialoprotein (DSP) and dentin phosphoprotein (DPP) are highly expressed in teeth (Veis and Perry, 1967; D’Souza et al., 1997; Ritchie et al., 1997). DSP and DPP have been shown to be cleavage products of a primary transcript precursor encoded by a single gene termed dentin sialophosphoprotein (DSPP) (MacDougall et al., 1997). This gene consists of 5 exons and 4 introns. The DSP sequence is found at the NH2-terminus (exons 1–4 and the 5′ region of exon 5), while the DPP sequence is located at the COOH-region (exon 5) (Gu et al., 2000; Ritchie et al., 2001; Yamakoshi et al., 2003). In the pig, a small polypeptide, dentin glycoprotein (DGP), has been identified (Yamakoshi et al., 2005). DSPP transcript is expressed predominantly in odontoblasts and transiently in preameloblasts and involved in odontoblast differentiation and mineralization (D’Souza et al., 1997; Chen et al., 2005). Defects in human DSPP are associated with various types of hereditary dentin disorders (Xiao et al., 2001; Rajpar et al., 2002; Kim and Simmer, 2007; Song et al., 2008 McKnight et al; 2008). Patients with these diseases are present with discolored teeth, enlarged pulp chambers, a wider predentin zone, decreased dentin width, hypomineralization and the prevalence of pulp exposures. Further study showed that teeth with DSPP knock out mice are similar to human dentinogenesis imperfecta type III (DGI-III) (Sreenath et al., 2003). In addition to teeth, low levels of DSPP expression recently have been detected in bone (Qin et al., 2002) and non-mineralized tissues (Ogbureke and Fisher, 2005; Alvares et al., 2006). Moreover, one has shown that DSPP null mice exhibit abnormal bone metabolism (Verdelis et al., 2008). However, DSPP expression levels in various tissues related to its biological roles remain unknown.
Analyses of expressed sequence tags (ESTs) and alterative splicing (AS) microarray data have shown that more than two-thirds of mammary and 40% of Drosophila genes contain more than one exon variation (Johnson et al., 2003; Stolc et al., 2004). These different arrangements produce structurally and functionally distinct mRNA and protein variations and may be one of the most extensively used mechanisms that account for the greater macromolecular and cellular complexity of higher eukaryotic organisms (Benjamin, 2006). Several variations of NCP genes from different species have been reported including osteocalcin (Laize et al., 2006), dentin matrix protein1 (DMP1) (George et al., 1993; MacDougall et al., 1998) and DSPP (MacDougall et al., 1997; Sfeir et al., 1998; Gu et al., 2000; Ritchie and Wang, 2000; Ritchie et al., 2001; Yamakoshi et al., 2003; Yamakoshi et al., 2008; Song et al., 2008; McKnight et al; 2008). In DSPP genes, variations often occur in DDP domain due to abundant AGT/C (serine) and GAT/C (aspartic acid) repetitive sequences. These variations (polymorphisms) in normal human populations include in-frame indels (trinucleotide insertion/deletion) and single nucleotide polymorphism (SNP).
As the mouse is a favored model for biological study, we investigated tissue-specific DSPP expression in mouse tissues using RT-PCR, in situ hybridization and immunohistochemical analyses as well as characterized the molecular basis for its variability from a mouse tooth cDNA library and predicted the secondary structure of DPP domain by computer software program.
2. Materials and Methods
2.1 Genomic DNA isolation and PCR
All experimental procedures involving the use of animals were reviewed and approved by the Institutional Animal Care at the University of Texas Health Science Center at San Antonio, TX, USA. Genomic DNA from different tissues of 6 month-old mice was isolated by using DNA purification kit (Wizard® Genomic, Promega, Madison, WI, USA). PCR amplification was performed with RED Taq™ DNA polymerase (Sigma, St Louis, MO, USA). The reaction had a cycle at 95°C for 5 min, followed by 30 cycles each with denaturation at 95°C for 30 s, annealing at 58°C for 40 s and extension at 72°C for 60 s. The final cycle was 72°C for 10 min. The PCR products were run on 1.5% agarose gels with ethidium bromide staining and purified by PCR purification kit (Promega). The purified PCR DNA was sequenced by an ABI automatic sequencer model 375 (Applied Biosystems, Foster City, CA, USA).
2.2 RNA isolation from mouse tissues
Tissues including bone, tooth, pancreas, intestine, liver, kidney, muscle, brain and tongue were harvested from 6 month-old mice. Total RNAs were extracted with RNA STAR-60 (TEL-TEST, Inc., Friendswood, TX, USA), then chloroform was added, and the mixture precipitated with isopropanol before treatment with DNase I (Promega). Total RNAs were quantified by UV spectroscopy at 260 nm.
2.3 Reverse transcription polymerase chain reaction (RT-PCR) and quantitative real time PCR
Complementary DNA synthesis and PCR amplification were performed using standard protocols (Sambrok and Russell, 2001). The PCR condition for the detection of mouse DSPP gene was described previously (Chen et al., 2005). DSPP primer sets were used: sense (exon 3), 5′-AACTCTGTGGCTGTGCCTCT-3′, antisense (exon 4), 5′-TATTGACTCGGAGCCATTCC-3′; sense (exon 5), 5′-ACTCACATGCGGGAGAAGAC-3′, antisense (exon 5), 5′-CACTGCTGTTGTCATGGTCA-3′ (GenBank Accession No. : NM_010080). A pair of primers of a house keeping gene, glyceraldehydes 3-phosphate dehydrogenase (GAPDH) (GenBank Accession No.: BC145810.1), was used as an internal positive control as follows: sense, 5′-CCATGGAGAAGGCCGGG-3′, antisense, 5′-CAAAGTCATGGATGACC-3′. The cycling parameters were as follows: 94°C for 4 min, followed by 25 cycles of 94°C for 30s, 58°C for 30s and 72°C for 1 min. The final cycle was 72°C for 10 min. The amplified PCR products were run on 1.5 % agarose gels with ethidium bromide staining, purified and confirmed by DNA sequencing. Gene expression level of DSPP and GAPDH was calculated by using image J software (NIH, USA). Expression level of DSPP gene from each tissue was divided by GDPDH gene from each tissue. Value of kidney group was taken as one fold increase. The individual value was divided by the kidney group value. For quantitative real-time PCR (qRT-PCR), amplification reactions were analyzed on an ABI 7500 (Applied Biosystems) using SYBR Green chemistry, with the threshold values calculated using SDS2 software (Applied Biosystems) as described before (Chen et al., 2005). The primer sequences used for qRT-PCR were as follows: ribosomal protein S18 gene, forward, 5′-GTGTGACCTGCTGGGAGATGA-3′; reverse, 5′-GTGCACAGCCCCTTCAAGA-3′ (GenBank Accession No. : NT039649.7). DSPP primers were described above.
2.4 Northern blot analysis
Radiolabeled cDNA probes were synthesized by Prime-It II Random Primer Labeling Kit (Stratagene, La Jolla, CA, USA) from gel-purified 171 and 1,590 bp fragments containing the mouse DSP and DSP-DPP cDNAs, respectively. Total RNA (50 μg) from mouse teeth was fractionated by agarose-formaldehyde gel electrophoresed, transferred to a nitrocellular membrane and hybridized to the radio-labeled probes using the Hydrosol ® I hybridization solution according to manufacture’s protocol (Chemicon International Inc., Temecula, CA, USA). The hybridization membrane was exposed to X-ray film (Kodak BioMax film) at −80°C for 2 days.
2.5 Isolation of mouse DSPP cDNA from a lambda cDNA library
The mouse tooth cDNA library was generated by using ZAP-cDNA Synthesis Kit (Stratagene) and screened using a mouse DSP-specific cDNA probe as described previously (MacDougall et al., 1997). Positive plaques were performed by a secondary screening using the same probe. The cDNAs from the positive plaques were converted from the phage vector into the pBluescript SK-phagemid vector using M13 helper phage according to the manufacture’s protocol (Stratagene).
2.6 DNA sequencing and data analyses
Both strands of the mouse DSPP cDNA were determined with an ABI automatic sequencer model 375 (Applied Biosystems) at the Human Genome DNA Core Laboratory (University of Texas Health Science Center, San Antonio, TX, USA). A database search was performed at the National Center for Biotechnology Information Website (http://www.ncbi.nlm.nih.gov/blast) using the BLAST program. The mouse DSPP nucleotides and derived from amino acids were aligned with those of the mouse DSPP genes (Genbank Accession No.: AJ002141, AF135799, NM_010080, ENSMUST00000112771, ENSMUST00000065590) using DNAsis® Max v2.5 software and Gene Runner (http://www.generunner). The molecular weight, isoelectric point and the protein secondary structure were also predicted using DNAsis ® Max v2.5 software and Gene Runner programs.
2.7 In situ hybridization
Mouse mandibles were fixed and processed. Serial sections prepared in either the sagittal or frontal plane were mounted on saline-treated slides. Representative sections from each block were stained with hematoxylin. A 479-bp mouse cDNA corresponding to a coding region of the DSP segment of DSPP was generated by PCR (MacDougall et al., 1997). Amplified PCR product was subcloned into pCRII vector containing Sp6 and T7 promoters (Invitrogen, Carslbad, CA, USA). Labeled [α-35S]-UTP antisense and sense DSPP probes were generated using Sp6 or T7 RNA polymerases after linearization with appropriate restriction enzymes. The method of in situ hybridization was performed as described earlier (Chen et al., 2002).
2.8 Immunohistochemical analysis
Immunohistochemistry assay was performed with the use of the ABC Vectastain Kit (Vector Laboratories, Inc., Burlingme, CA, USA) according to the manual’s instruction. Briefly, sections were incubated in a dry oven at 62°C for 1 h and slides were deparaffinized in xylene and rehydrated through a graded ethanol series 100, 95 and 75%. For antigen retrieval, slides were micowaved for 15 min in 10 mM citrate buffer (pH 7.6). Slides were then pretreated with 3% hydrogen peroxide solution for 6 min. After washing with PBS, slides were incubated with normal serum (1:50) for 60 min to block the nonspecific immunoglobulin staining serum-binding sites. Immunohistochemistry was performed using anti-DSP rabbit polyclonal antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA). The samples were incubated for overnight at 4°C and washed with PBS for 3 times. The slides were incubated with the biotinylated goat anti-rabbit antibody, followed by the alkaline phosphatase-conjugated streptavidin complex. All reagents were diluted in Tris-buffered saline and incubated at room temperature for 30 min and washed in PBS. Phosphatase activity was revealed by the new fuchsin substrate-chromogen system (DAKO), and the color reaction was allowed to develop for 20 min. Finally, the slides were washed in tap water, counterstained with Meyer’s hemotoxylin, and mounted with coverslips. A negative control for immunostaining was obtained by substituting the first antibody with normal goat serum.
3. Results
3.1 Analysis of spatial and temporal DSPP expression in mouse tissues
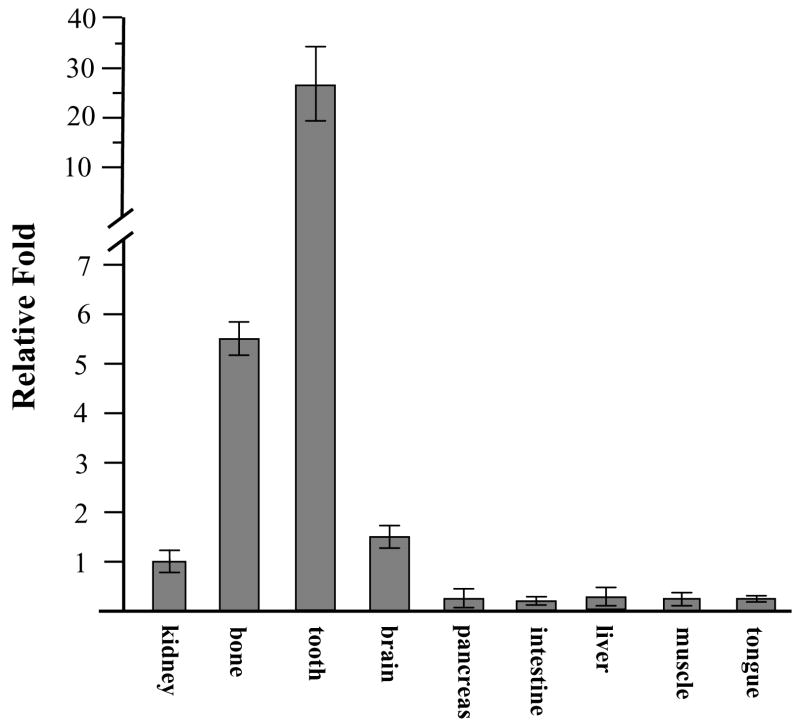
To assess DSPP expression in various tissues, we measured its expression in 6 month-old male and female mouse tissues using qRT-PCR. The results in Figure 1 show that DSPP expression was highly detected in teeth, moderately in bone, but DSPP transcription was also slightly seen in kidney and brain. Similar results were confirmed by semiquantitative RT-PCR (supplemental Fig. 1). We examined the DSPP spatial-temporal expression pattern in tooth and craniofacial tissues using in situ hybridization assay. At day 1 of postnatal development, a DSPP signal was strongly detected in differentiating and differentiated odontoblasts, ameloblasts and moderately in dental pulp cells in the first molar tooth (Fig. 2B). At post-natal days 3 and 10, DSPP signal was weakly detected in cells within alveolar bone in addition to odontoblasts, ameloblasts and dental pulp (Fig. 2D, F). In 2- and 8-week old mice, DSPP remained actively transcribed by odontoblasts and dental pulp (Fig. 2H, J). In addition, weak hybridization for DSPP signal was observed in alveolar bones, periodontal ligament and cellular cementum (Fig. 2H′, J′). These data for in situ hybridization were matched to those of RT-PCR analysis in tooth and bone studies.
Fig. 1. Expression of the mouse DSPP mRNA by qRT-PCR.
Total RNAs from various tissues were isolated and reversely transcribed. Quantitative RT-PCR was used to measure DSPP mRNA expression from different tissues described as above. Value was obtained from ratio between DSPP and S18 expression. The value from kidney group was taken as a 1.0-fold increase. The relative fold value was calculated by dividing the individual value by the kidney group value. The data show the means ± S.E. from 3 separate experiments.
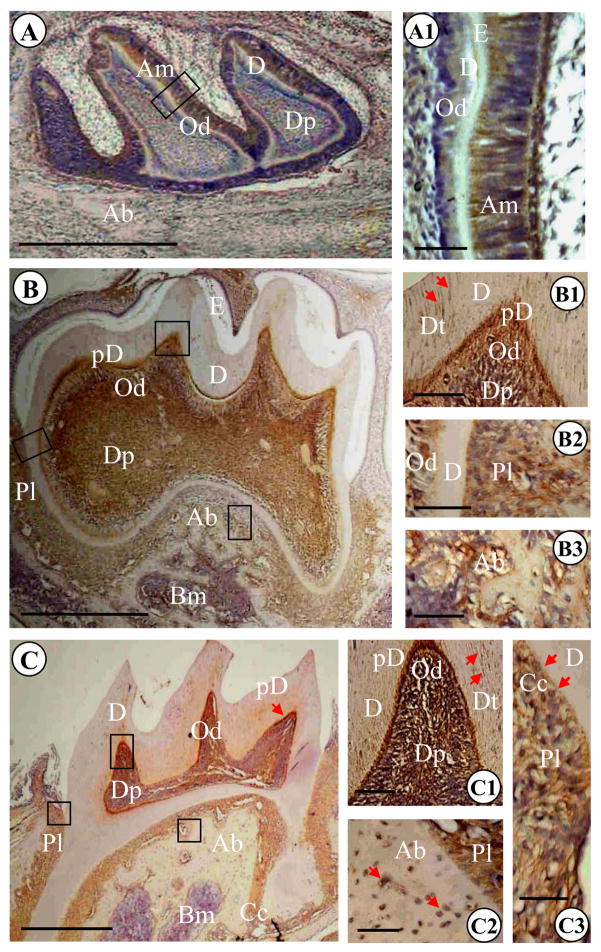
Fig. 2. Expression of DSPP gene in tooth organs during craniofacial development.
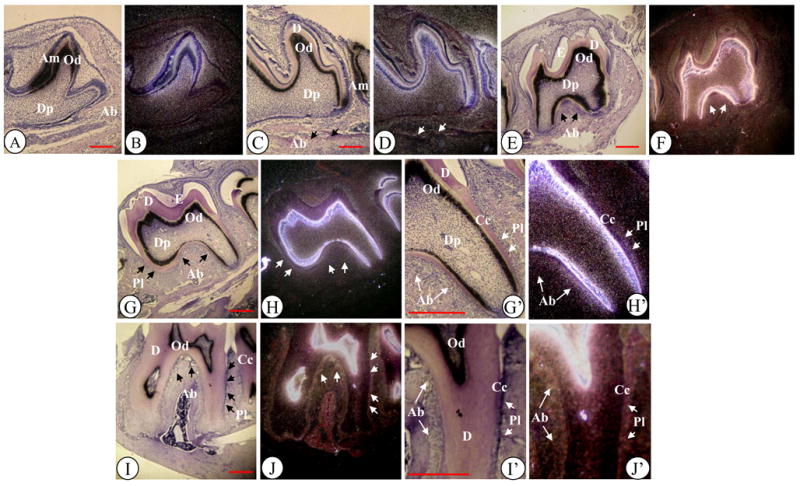
(A–J). Hematoxylin and in situ hybridization of mouse tooth developmental stages from postnatal day (PN) 1 to 8 weeks (8 WK) after birth with DSPP (B, D, F, H and J) antisense probe. Molars at PN1 (A), PN 3 (C), PN 10 (E), 2 WK (G) and 8 WK (I) were stained with hematoxylin. Molars at PN1 (B), PN3 (D), PN 10 (F), 2WK (H) and 8 WK (J) were hybridized with antisense DSPP probe. G′-J′ show high magnifications of G-J. Ab, alveolar bone; Am, ameloblasts; Cc, Cellular cementum; D, dentin; Dp, dental pulp cells; E, enamel; Od, odontoblasts; Pl, periodontal ligament (or its space). Bar, 200 μM
To determine DSP protein expression, we performed immunohistochemistry in developing teeth from 2 postnatal day-old to 13.5 month-old mice. At post-natal day 2, ECM secretion was seen in some of the cusps (Fig. 3A and 3A1). Positive DSP signal was detected in odontoblasts, dentin, dental pulp and differentiating and differentiated ameloblasts. However, DSP expression in differentiated ameloblasts was highest compared to other cells. At this stage, DSP signal was not observed in cells in alveolar bone.
Fig. 3. Light microscopic images of mouse mandibular molars of 2, 17 postnatal day- and 1.4 month-old mice.
(A) Tooth organ at postnatal day 2. Dentin and enamel formation started and thin layer of predentin and dentin were observed. Immunopositive reactions were detected in odontoblasts, ameloblasts and dental pulp. The highest expression of DSP was present in differentiated ameloblast cells. At this stage, DSP signal was not seen in cells and ECM within alveolar bone. A1 is high magnification of A. (B). The first mandibular molar at postnatal day 17. The first molar was not erupted and root dentin formation was ongoing. DSP signaling was observed in dentinal tubules of mantle dentin (arrows in B1), predentin, odontoblasts and dental pulp. Immunopositive reactions were also seen in periodontal ligament and alveolar bone (B2, B3). B1, B2 and B3 are high magnifications of B. (C). The first mandibular molar in a 1.4 month-old mouse. Tooth was erupted and root dentin formation completed. DSP expression was evident in dentinal tubules of mantle dentin (arrow in C1), predentin, odontoblasts and dental pulp. Also, signal for DSP was seen in periodontal ligament, cellular cementum (arrows in C3) as well as cells and ECM within alveolar bone (arrows in C2). C1, C2 and C3 are high magnifications of C. Bar: 500 μM in A, B and C; 50 μM in A1, B1, B2, B3 and C1, C2, C3. Ab, alveolar bone; Am, ameloblasts; Cc, cellular cementum; D, dentin; Dp, dental pulp; Dt, dentinal tubules; E, enamel; Od, odontoblasts; Pd, predentin; Pl, periodontal ligament.
At post-natal day 17, the first mandibular molar was unerupted, root dentin formation was ongoing, and formation of dentin and enamel was essentially complete (Fig. 3B). DSP positive reaction was detected in dental pulp, odontoblasts, predentin, dentinal tubules of mantle dentin (Fig. 3B1). At this stage, DSP expression level in predentin was highest. Moreover, bone tissue surrounding the root of the first molar was not well developed and appeared sponge-like and DSP expression in osteoblasts within alveolar bone, cellular cementum and periodontal tissues was positive. However, DSP expression level in these tissues was lower than predentin (Fig. 3B1, 2 and 3).
In the 1.4-month mouse, the first mandibular molar was erupted and root dentin formation completed. Cellular cementum was well developed at the apical area of the root (Fig. 3C). At this stage, DSP immunopositive signal was observed in dental pulp cells, differentiated odontoblasts, predentin and dentinal tubules (Fig. 3C, C1). The positive reaction in odontoblasts and predentin was more intense than that of dental pulp and dentinal tubules. Furthermore, cellular cementum, periodontal ligament and cells within alveolar bone were also immunopositive (Fig. 3C2, C3).
In 3.5-, 7.5- and 13.5-month mice, DSP expression patterns were quite similar to those from the 1.4 month mouse tissues (Fig. 4A, B, C). In alveolar bone, distinct DSP signal was also apparent in cells and ECM.
Fig. 4. Light microscopic images of mouse mandibular molars of 3.5, 7.5 and 13.5 month-old mice.
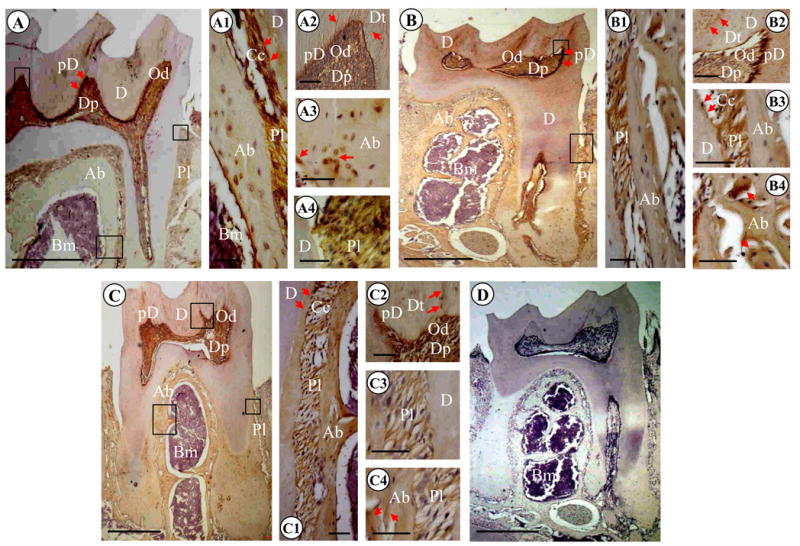
(A) 3.5 month mouse molars. Immunopositive reactions using anti-DSP antibody were observed in dentinal tubules of mantle dentin, predentin (arrows), odontoblasts and dental pulp. DSP expression was also detected in cells and ECM within alveolar bone and periodontal ligament. A1, A2, A3 and A4 are high magnifications of A. (B). 7.5 month first mandibular molar. DSP signal was seen in dentinal tubules of mantle dentin, predentin (arrows), odontoblasts and dental pulp. Positive reactions for DSP were also evident in cells and ECM within alveolar bone and periodontal ligament. B1, B2, B3 and B4 are high magnifications of B. (C). 13.5 month first mandibular molar. Immunostaining reactions for DSP were present in dentinal tubules of mantle dentin, predentin (arrows), odontoblasts and dental pulp. DSP signal was also intense in cells and ECM within alveolar bone and periodontal ligament. C1, C2, C3 and C4 are high magnifications of C. (D). 7.5 month mandibular molars. The section was stained with hematoxylin and eosin with negative control. Bar: 500 μM in A, B, C and D; 50 μM in A1, A2, A3, A4; B1, B2, B3, B4; and C1, C2, C3, C4. Ab, alveolar bone; Am, ameloblasts; Cc, cellular cementum; D, dentin; Dp, dental pulp; Dt, dentinal tubules; E, enamel; Od, odontoblasts; Pd, predentin; Pl, periodontal ligament.
No positive staining was apparent in soft tissues from 1.4 month mouse including pancreas, intestine, liver, kidney, muscle and brain (data not shown).
3.2. Determination of polymorphisms of DSPP gene in mouse tissues
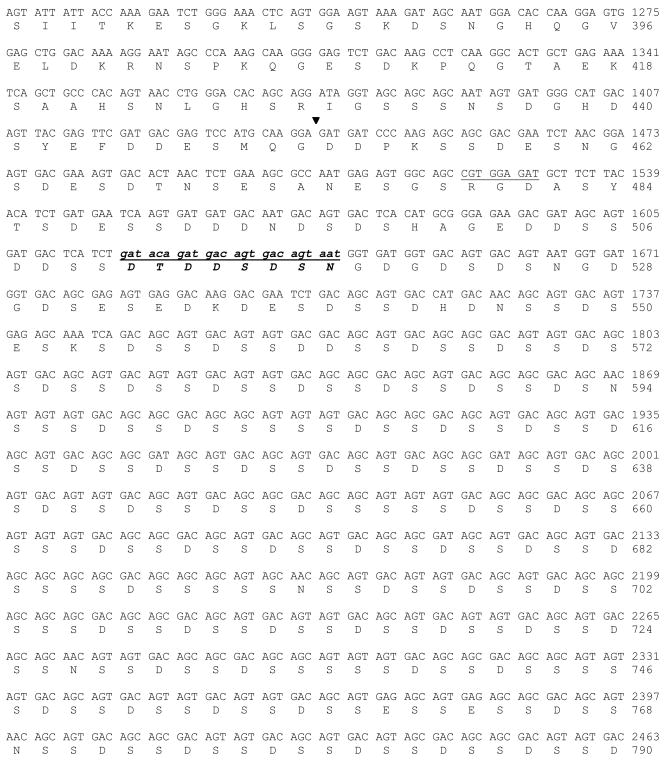
Using a mouse tooth cDNA library, we isolated and characterized a full-length mouse DSPP cDNA gene (Fig. 5). The 4,405 bp DNA sequences contained an open reading frame of 940 amino acids starting with a translation initiation site (ATG) located at nucleotide 88 that has a 3 adenine nucleotide representative of the Kozak initiation consensus sequence. The stop codon at nucleotide 2,913 begins a large untranslated region of 1,482 nucleotides with several polyadenylation signals. Compared between our cloned mouse DSPP and the published mouse DSPP sequences (MacDougall et al., 1997; Sfeir et al., 1998; Genbank Accession No.: AJ002141, AF135799, NM_010080, ENSMUST00000112771, ENSMUST00000065590), 13 polymorphisms were found (Table 1). Two non-synonymous single nucleotide polymorphisms (SNPs) are located at exon 4 within DSP domain, 8 non-synonymous SNPs and one in-frame indel (insertion) were found at exon 5 in DPP portion. Two SNPs were observed in the 3′ untranslated region. A 24-bp (8 amino acids) is located at NH2-part of the DPP domain at amino acids position 60 of the DPP starting site (Fig. 5 and Fig. 6A–B). This 24-bp sequence is present in one mouse DSPP gene (termed mDSPP940/DPP489) (NM_010080, ENSMUST00000112771), but lacks another one (termed mDSPP932/DPP481) (AJ002141, AF135799, ENSMUST00000065590), indicating that there are at least 2 variants of mouse DSPP gene.
Fig. 5. DNA sequence of the mouse dentin sialophosphoprotein and deduced amino acid sequence.
The amino acid and DNA sequence numbering was indicated on the right side. The signal peptide was shown with italic letters. The inserted 24-bp, 8 amino acid sequence, was indicated in small and bold italic font with a single underline. Stop codon was shown by an asterisk. The signal polyadenylation signal was bold with a single underline. Junction between exons was shown by arrows. The beginning of the DPP coding region was indicated by an arrowhead.
Table 1.
Summary of Sequence Differences of Mouse DSPP Genes
| Location | Corresponding Sequences | Positions of this Study | Differences | |
|---|---|---|---|---|
| cDNA | Protein | |||
| Exon 4 | nt 1180, aa 332 | nt 1183, aa 332 | AAG>AAC, | K>N |
| Exon 4 | nt 1181, aa 333 | nt 1184, aa 333 | GAA>CAA, | E>Q |
| Exon 5 | nt 1615. aa 510 | nt 1618, aa 511 | added GATACAGATGACAGTGACAGTAAT, | DTDDSDSN |
| Exon 5 | nt 1871, aa 596 | nt 1897, aa 604 | GGT>AGT, | G>S |
| Exon5 | nt 1902, aa 606 | nt 1928, aa 614 | ACC>AGC, | T>S |
| Exon5 | nt 1904, aa 607 | nt 1930, aa 615 | TGT>AGT, | C>S |
| Exon5 | nt 2045, aa 654 | nt 2071, aa 662 | TGT>AGT, | C>S |
| Exon5 | nt 2229, aa 713 | nt 2255, aa 721 | GGC>GAC, | G>D |
| Exon5 | nt 2246, aa 721 | nt 2272, aa 727 | GAC>AAC, | D>N |
| Exon5 | nt 2256, aa 722 | nt 2282, aa 730 | GCC>GAC, | A>D |
| Exon5 | nt 2347, aa 752 | nt 2373, aa 760 | GAT>GAG, | D>E |
| 3′ Untranslated | nt 2935/2936 | nt 2960/2961 | del G | |
| 3′ Untranslated | nt 4161 | nt 4176 | T>A | |
nt, nucleotide; aa, amino acid.
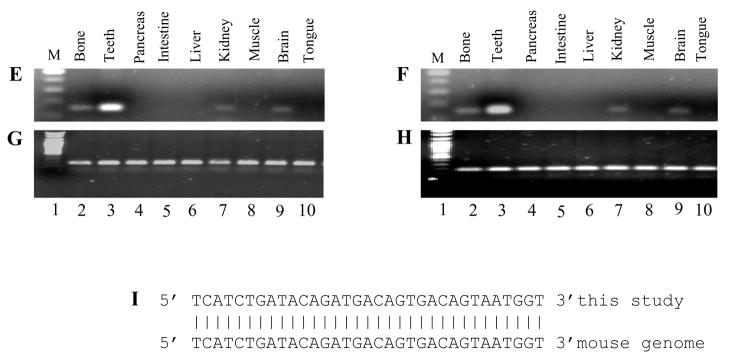
Fig. 6. Twenty-four base pair sequence of mouse DSPP940/DPP489. A.
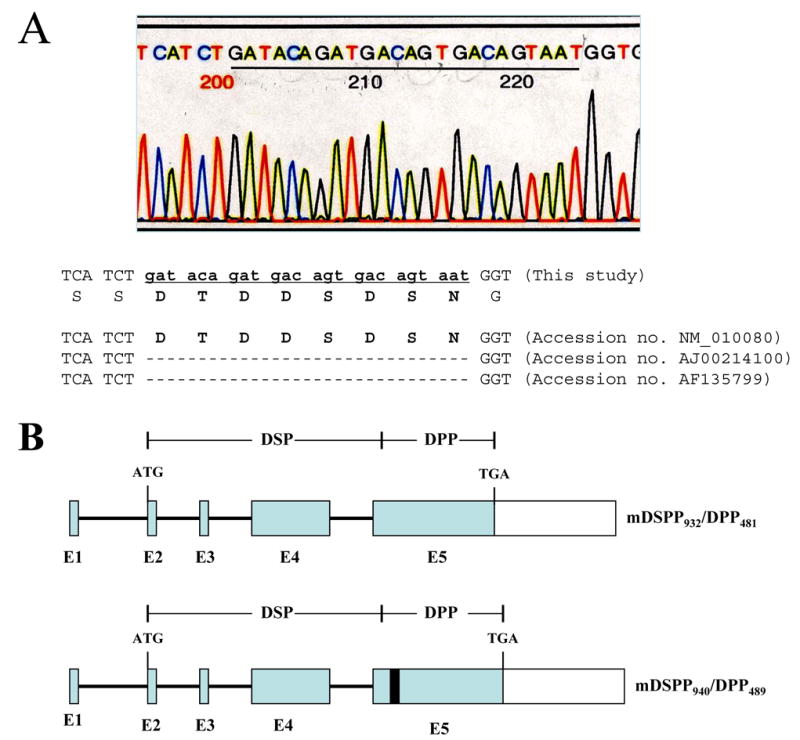
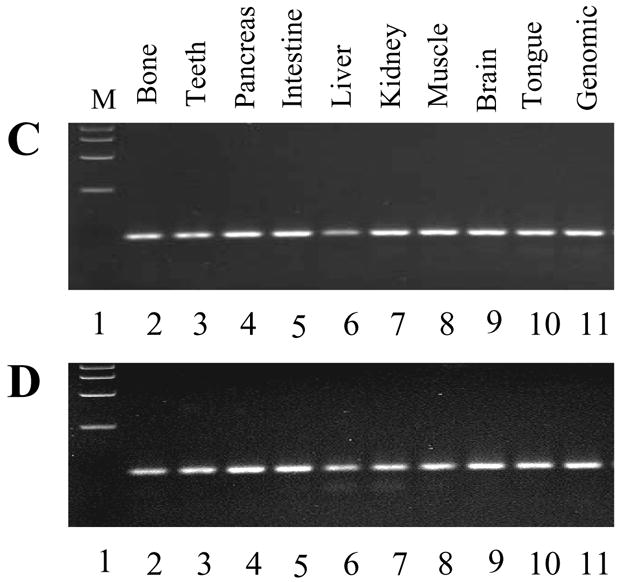
the 24 bp inserted DNA sequence. The amino acid sequence was shown below the 24 nucleotides. Missing nucleotides were indicated by gaps. B. Previous DSPP organization (mDSPP932/DPP481; E1-E5, exons 1–5) and the 24 bp in-frame indel of mDSPP940/DPP489 in exon 5 with a black box. C and D. PCR amplification of the mouse DSPP genomic DNA spanning the region of the unique 24 bp DNA sequence from various tissues of 6 month male (C) and female (D) mice, respectively. E and F. RT-PCR was used to amplify the mouse DSPP cDNA spanning the unique 24-bp region from various tissues of 6 month male (E) and female (F) mice. Specific DSPP primer sets defining this region from nucleotides 1577 to 1732 were used for PCR. G and H. GAPDH was internal positive control from 6 month male (G) and female (H) mouse cDNAs. The PCR products were run on 1.5 % agarose gels with ethidium bromide staining. Lane 1, DNA size marker; lane 2, long bone; lane 3, teeth; lane 4, pancreas; lane 5, intestine; lane 6, liver; lane 7, muscle; lane 8, kidney; lane 9, brain; lane 10, tongue. I. Analysis of the 24 bp sequence with the mouse genome (Genbank accession no. NM_010080).
To investigate which variation of the two mouse DSPP genes predominantly exists in the mouse genome, we performed a PCR analysis of two 6 month male and female mouse tissues using DSPP specific primers which cover the 24 bp elements of the mouse DSPP gene. PCR results show that all of mouse tissues tested contains the 24 bp DNA sequences (Fig. 6C–D), indicating that the mDSPP940/DPP489 different from the mDSPP932/DPP481 is predominantly present in mouse tissues. The results were confirmed by RT-PCR assay (Fig. 6E–F).
Using this 24-bp sequence as a template, we searched for the mouse genome with the BLAST program. This procedure showed that the 24-bp sequence is present in mouse genome (Fig. 6I). Furthermore, studies demonstrated that these 8 amino acid residues (24-bp) also exhibited the same sequence located at amino acid position 58 of the rat DPP starting site (Ritchie et al., 2001), but there is no evidence of this in pig and human DSPP (Gu et al., 2000; Yamakoshi et al., 2003). Northern blot analyses of total RNA extracted from 6 month mouse teeth with DSP and DSP-DPP probes revealed those probes hybridized to a prominent transcript of 4.4 kb (Fig. 7).
Fig. 7. Northern blot analysis of the mouse DSPP gene transcript in mouse teeth.
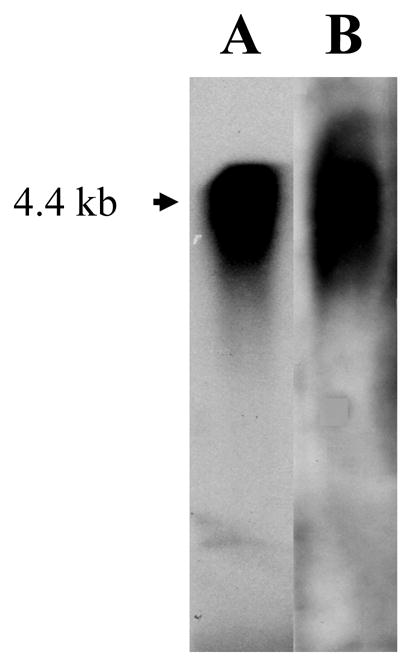
Total RNA was isolated from 6 month-old mouse teeth, separated on a denaturing agarose-formaldehyde gel, and transferred to a membrane. Panel A: 32P-labled mouse DSP as a probe. Panel B: 32P-labled mouse DSP-DPP as a probe. A single transcript of approximately 4.4 kb was recognized by both the probes.
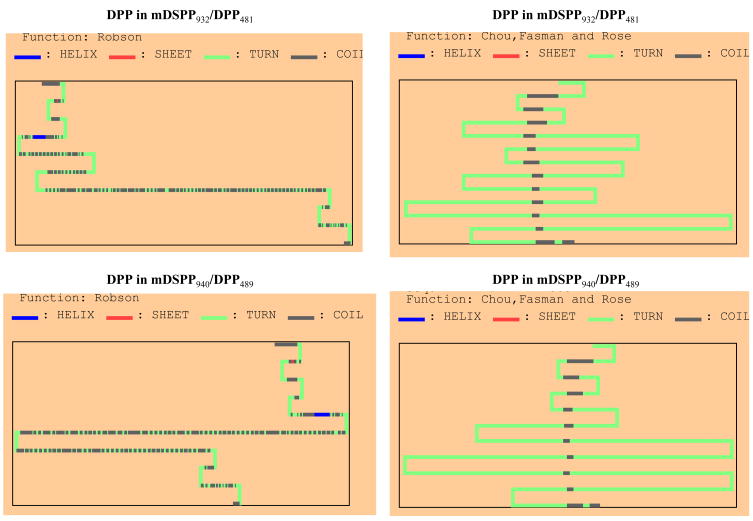
The 8 amino acid sequences (24-bp) within the DPP domain of the mDSPP940/DPP489 contain Asp-Thr-Asp-Asp-Ser-Asp-Ser-Asn. Interspersed arrangement of Asp-Ser or Ser-Asp residues provides an excellent substrate for casein kinase I and II phosphorylation action (Flotow et al.; 1991). Additionally, the threonine residue acts as a substrate for casein kinase I (Flotow et al.; 1990). Furthermore, the phosphorylation of the Ser and Thr residues in the DPP domain of the mDSPP940/DPP489 may lead to the generation of many acidic patches due to Asp-Asp, Ser-Asp residues (Veis and, Perry, 1967; Linde and Goldberg, 1993; He et al., 2005), resulting in the DPP of the mDSPP940/DPP489 to be slightly negatively charged and resulting in changes in its protein structure compared to that of the mDSPP932/DPP481. Therefore, we predicted secondary structures of DPP proteins between mDSPP940/DPP489 and mDSPP932/DPP481 using a computer program. These results demonstrated that the DPP secondary structure of mDSPP940/DPP489 is different from that of mDSPP932/DPP481 (Fig. 8). Also, The DPP of the mDSPP940/DPP489 is slightly negatively charged and has a lower isoelectric point than that of mDSPP932/DPP481 (Table 2). Taken together, this implied that the DPP of the mDSPP940/DPP489 might have different roles during tooth development and biomineralization.
Fig. 8. Predicted secondary structure of mouse DPP proteins between mDSPP940/DPP489 and mDSPP932/DPP481.
The secondary structures of DDP domains of the mDSPP940/DPP489 and mDSPP932/DPP481 were predicted by methods of Robson, Chou, Fasman and Rose. The different DPP secondary structures between the mDSPP940/DPP489 and mDSPP932/DPP481 were observed.
Table 2.
Comparison of Isoelectric Point of DSPP from Different Species
| Species | Isoelectric Point | References | Genbank Accession No. |
|---|---|---|---|
| Human | 3.35 | Gu et al.,2000 | NM_014208 |
| Pig | 4.28 | Yamakoshi et al.,2003 | NM_213777 |
| Rat | 3.35 | Ritchie et al.,2001 | AF251219 |
| Mouse (mDSPP932/DPP481) | 3.43 | Feng et al., 1998 | AF135799 |
| Mouse (mDSPP932/DPP481) | 3.43 | Sfeir et al.,1998 | AJ002141 |
| Mouse (mDSPP940/DPP489) | 3.40 | MacDougall et al.,1997 | NM_010080 |
| Mouse (mDSPP940/DPP489) | 3.40 | This study |
4. Discussion
4.1. Tissue-specific expression of DSPP
DSPP is most abundant protein after collagen type I in dentin ECM. The spatial-temporal expression of DSPP is largely restricted to mineralizing and mineralized tissues, especially in teeth (D’Souza et al., 1997; Chen et al., 2005). DSPP is processed into 2 major dentin matrix proteins, DSP and DPP (MacDougall et al., 1997). DSP and DPP have their unique biological roles during dentin and enamel development and formation (Paine et al., 2005; White et al., 2007). Mutations of either DSP or DPP domain in the human cause various types of inherited dental disorders, which are the most common genetic diseases. These include dentinogenesis impecfecta type II (DGI-II OMIM 125490) and III (DGI-III OMIM 125500) and dentin dysplasia type II (DD-II, OMIM 125420) (Kim and Simmer, 2007; McKnight et al; 2008). DSPP homologous deficient (−/−) mice show teeth similar to human DGI-III (Sreenath et al., 2003). The data indicate that DSPP plays an important role in tooth formation and mineralization. DSPP is also expressed in lower levels in bone and non-mineralized tissues (Xiao et al., 2001; Qin et al., 2003; Ogbureke and Fisher, 2005; Alvares et al., 2006). In DSPP null mice (−/−), abnormal bone microstructures and metabolism were observed (Verdelis et al., 2008), implying that DSPP expression level may be relevant to its biological functions. In this study, we investigated DSPP expression in tissues and cells other than dentin, odontoblasts and ameloblasts at the transcriptional and translational levels using qRT-PCR, in situ hybridization and immunohistochemical analyses. With these approaches, we clearly showed that DSPP was[UE9] expressed highly[UE10] in teeth and moderately in bones of the 6 month-old male and female mice using qRT-PCR method with primer sets specific for DSPP gene (Fig. 1). In situ hybridization and immunostaining assays documented that DSP expression was observed in cells and ECM at postnatal day 2 and the later stages of developing alveolar bones surrounding the forming root (Fig. 2). Immunopositive reactions were also detected in periodontal ligament and cellular cementum (Fig. 3B2, 3C3, 4A4, 4B3, 4C3). These data suggest that DSP is synthesized and secreted by osteoblasts, osteocytes, cementoblasts, cementocytes and fibroblasts that are associated with the initial formation of the periodontium. The expression of DSPP transcripts by osteoblasts, cementoblasts and fibroblasts was verified by an in situ hybridization assay (Fig. 2D, F, H, J). However, the expression levels of DSPP in those cells and ECM were relatively low compared to that of odontoblasts and predentin (Fig. 2, 3, 4). Our results for DSP expression in alveolar bone, cellular cementum and periodontal ligament are consistent with those of Baba et al. (2004). We also observed that the DSP signal was highly apparent in the cytoplasm of differentiating and differentiated ameloblasts at postnatal day 2 (Fig. 3A1). The DSPP transcript in ameloblasts was confirmed by an in situ hybridization assay (Fig. 2B). Our results are consistent with the findings of other Hao et al. (2008). This suggests that DSP is involved in ameloblast differentiation and enamel formation. Paine et al (2005) reported that DSP overexpression in transgenic mice resulted in increased rate of enamel formation and enamel hardness, a result confirmed by White et al. (2007). Based on our systematic research design using sequential stages of mouse molar development, along with well defined transcriptional and translational studies, our data show that DSP expression is mainly restricted to mineralizing/mineralized tissues (D’Souza et al., 1997; Baba et al., 2004) and its functions are involved in tooth formation and biominerlaization and possibly in bone metabolism (Verdelis et al., 2008).
4.2 DSPP polymorphisms
DSPP is the largest member of the SIBLING gene family and the size of DPP domain greatly varies among species. DPPs from mouse, rat, porcine and bovine are smaller than that of human (MacDougall et al., 1997; Gu et al., 2000; Ritchie et al., 2001; Yamakoshi et al., 2003). Recent studies have demonstrated various polymorphisms in DSPP gene from different species, particularly in DPP domain (Song et al., 2008; McKnight et al; 2008). These polymorphisms include synonymous SNP, non-synonymous SNP and in-frame indels (trinucleotide insertion/deletion). These observations imply that DPP function may not be affected by length variations and certain amino acid changes. Therefore, the functional significance or insignificance of DPP sequence changes is important when considering if a particular sequence variation is disease-causing or simply only polymorphism as mutations of DSPP gene are the most common hereditary dental disorders (Kim and Simmer, 2007; McKnight et al; 2008). Since DPP has a unique composition with rich aspartic acid and phsophserine residues comprising >85% of the exceptional extensive triple amino acid repeat motifs of (DSS)n, (DS)n and (NSS)n (Gu et al., 2000), it is rather difficult to amplify of the entire DPP by PCR. We screened a mouse tooth cDNA library and identified an entire mouse DSPP cDNA gene (Fig. 5). Compared to previous publications (MacDougall et al., 1997; Sfeir et al., 1998), the mouse DSPP gene contains 13 polymorphisms including 2 non-synonymous SNPs in exon 4, 8 non-synonymous SNPs, 1 in-frame indel in exon 5 and 2 SNPs in the 3′ untranslated region (Table 1). Notably, a 24 bp DNA sequence in exon 5 exists in one mouse DSPP gene termed mDSPP940/DPP489 (NM_010080, ENSMUST00000112771) and lacks another one, mDSPP932/DPP481 (AJ002141, AF135799, ENSMUST00000065590), suggesting at least two mouse DSPP gene variations. PCR and Northern blot assays demonstrated that mDSPP940/DPP489 dominantly exist in the mouse tissues examined (Fig. 6, 7). This 8 amino acid sequence (24-bp) is present in rat DSPP (Ritchie et al., 2001), but not in pig or human (Gu et al., 2000; Yamakoshi et al., 2003). Since the 8 amino acids contain Asp-Ser or Ser-Asp sequences and are located at the NH2-terminal domain of the DPP, the DPP domain of mDSPP940/DPP489 is relatively larger, acidic (Table 2) and probably highly phosphorylated compared to the DPP of the mDSPP932/DPP481.
The predicted protein secondary structure of the DPP domain of the mDSPP940/DPP489 was different from that of the DPP of mDSPP932/DPP481 (Fig. 8). Addition of Thr, Ser and Asp residues of this peptide within the DPP domain of the mDSPP940/DPP489 provides numerous phosphorylated and acid residues that are able to bind high amounts of calcium or calcium containing crystals. Multiple phosphorylated residues are associated with bone and tooth biomineralization and act as a promoter/nucleator of crystal growth in dentin (Veis and Perry, 1967; Linde and Goldberg, 1993; He et al., 2005). Furthermore, this peptide in the DPP of the mDSPP940/DPP489 contains Asp-Ser and Ser-Asp sequences. Asp-Ser* (*phosphorylation) motif is able to give rise to a ridge of phosphate and carboxyllate while the Ser*-Asp sequence forms a ridge of phosphate and carboxyllate on opposite side of this peptide. Dahlin et al (1998) showed that Asp-Ser* motif has the best fit to crystal structure of hydroxyapatite and octacalcium since the Asp-Ser* oligo is the energetically most favored adaptations. Recent studies have clearly shown that various lengths of in-frame indels (trinucleotide insertion/deletion) within DPP in normal human population contain Asp-Ser*/Ser*-Asp sequences (Song et al., 2008; McKnight et al; 2008). The maximal trinucleotide deletion is up to 135 bp (45 amino acids) (Song et al; 2008). With the 45 amino acid variations, the phosphorylated sites of the Asp-Ser*/Ser*-Asp sequences would be decreased and the subsequent functions associated with phsophoserine residues could be changed. It is known that certain neurological degenerative diseases are associated with trinucleotide repeat mutations (deletions/insertions) (Everett and Wood, 2004; Greenamyre, 2007). These repetitive mutations change normal gene structures and enhance genetic instability (Mooers et al., 2005; Napierala et al., 2005). Therefore, this raises questions as to how the length of DPP can maintain its normal biological function and which variations are disease-causing and normal SNPs. To resolve these questions additional research is required.
Supplementary Material
Acknowledgments
We thank Dr Russel Reiter for critical reading of the manuscript. This work was supported by NIDCR, National Institutes of Health, Grant DE113221 (M.M.), DE014484 (S.C.) and China Scholarship Council Award CSC20073020 (G. Y.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvares K, Kanwar YS, Veis A. Expression and potential role of dentin phosphophoryn (DPP) in mouse embryonic tissues involved in epithelial-mesenchymal interactions and branching morphogenesis. Dev Dyn. 2006;235:2980–90. doi: 10.1002/dvdy.20935. [DOI] [PubMed] [Google Scholar]
- Baba O, Qin C, Brunn JC, Jones JE, Wygant JN, McIntyre BW, Butler WT. Detection of dentin sialoprotein in rat periodontium. Eur J Oral Sci. 2004;112:163–70. doi: 10.1111/j.0909-8836.2004.00110.x. [DOI] [PubMed] [Google Scholar]
- Benjamin JB. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Chen S, Gu TT, Sreenath T, Kulkarni AB, Karsenty G, MacDougall M. Spatial expression of Cbfa1/Runx2 isoforms in teeth and characterization of binding sites in the DSPP gene. Connect Tissue Res. 2002;43:338–44. doi: 10.1080/03008200290000691. [DOI] [PubMed] [Google Scholar]
- Chen S, Rani S, Wu Y, Unterbrink A, Gu TT, Gluhak-Heinrich J, Chuang HH, Macdougall M. Differential regulation of dentin sialophosphoprotein expression by Runx2 during odontoblast cytodifferentiation. J Biol Chem. 2005;280:29717–27. doi: 10.1074/jbc.M502929200. [DOI] [PubMed] [Google Scholar]
- Dahlin S, Angstrom J, Linde A. Dentin phosphoprotein sequence motifs and molecular modeling: conformational adaptations to mineral crystal. Eur J Oral Sci. 1998;106:239–248. doi: 10.1111/j.1600-0722.1998.tb02182.x. [DOI] [PubMed] [Google Scholar]
- D’Souza RN, Cavender A, Sunavala G, Alvarez J, Ohshima T, Kulkarni AB, MacDougall M. Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. J Bone Miner Res. 1997;12:2040–2049. doi: 10.1359/jbmr.1997.12.12.2040. [DOI] [PubMed] [Google Scholar]
- Everett CM, Wood NW. Trinucleotide repeats and neurodegenerative disease. Brain. 2004;127:2385–405. doi: 10.1093/brain/awh278. [DOI] [PubMed] [Google Scholar]
- Flotow H, Graves PR, Wang AQ, Fiol CJ, Roeske RW, Roach PJ. Phosphate groups as substrate determinants for casein kinase I action. J Biol Chem. 1990;265:14264–9. [PubMed] [Google Scholar]
- Flotow H, Roach PJ. Role of acidic residues as substrate determinants for casein kinase I. J Biol Chem. 1991;266:3724–7. [PubMed] [Google Scholar]
- George A, Sabsay B, Simonian IPA, Veis A. Characterization of a novel dentin matrix acidic phosphoprotein. Implications for induction of biomineralization. J Biol Chem. 1993;268:12624–12630. [PubMed] [Google Scholar]
- Greenamyre JT. Huntington’s disease--making connections. New Engl J Med. 2007;356(5):518–20. doi: 10.1056/NEJMcibr067022. [DOI] [PubMed] [Google Scholar]
- Gu K, Chang S, Ritchie HH, Clarkson BH, Rutherford RB. Molecular cloning of a human dentin sialophosphoprotein gene. Eur J Oral Sci. 2000;108:35–42. doi: 10.1034/j.1600-0722.2000.00765.x. [DOI] [PubMed] [Google Scholar]
- Hao J, Ramachandran A, George A. Temporal and Spatial Localization of the Dentin Matrix Proteins During Dentin Biomineralization. J Histochem Cytochem. 2009;57:227–37. doi: 10.1369/jhc.2008.952119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Ramachandran A, Dahl T, George S, Schultz D, Cookson D, Vis A, George A. Phosphorylation of phosphoryn is crucial for its function as a mediator of biomineralizatiom. J Biol Chem. 2005;280:33109–14. doi: 10.1074/jbc.M500159200. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–4. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP. Hereditary dentin defects. J Dent Res. 2007;86:392–9. doi: 10.1177/154405910708600502. [DOI] [PubMed] [Google Scholar]
- Laize V, Viegas CS, Price PA, Cancela ML. Identification of an osteocalcin isoform in fish with a large acidic prodomain. J Biol Chem. 2006;281 :15037–43. doi: 10.1074/jbc.M600373200. [DOI] [PubMed] [Google Scholar]
- Linde A. Goldberg M. Dentinogenesis Crit Rev Oral Biol Med. 1993;4:679–728. doi: 10.1177/10454411930040050301. [DOI] [PubMed] [Google Scholar]
- MacDougall M, Simmons D, Luan X, Nydegger J, Feng J, Gu TT. Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4. Dentin phosphoprotein DNA sequence determination J Biol Chem. 1997;272:835–842. doi: 10.1074/jbc.272.2.835. [DOI] [PubMed] [Google Scholar]
- MacDougall M, Gu TT, Luan X, Simmons D, Chen J. Identification of a novel isoform of mouse dentin matrix protein 1: spatial expression in mineralized tissues. J Bone Miner Res. 1998;13:422–431. doi: 10.1359/jbmr.1998.13.3.422. [DOI] [PubMed] [Google Scholar]
- Mooers BH, Logue JS, Berglund JA. The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc Nat Acad Sci USA. 2005;102:16626–31. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight DA, Suzanne Hart P, Hart TC, Hartsfield JK, Wilson A, Wright JT, Fisher LW. A comprehensive analysis of normal variation and disease-causing mutations in the human DSPP gene. Hum Mutat. 2008;29:1392–404. doi: 10.1002/humu.20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napierala M, Bacolla A, Wells RD. Increased negative superhelical density in vivo enhances the genetic instability of triplet repeat sequences. J Biol Chem. 2005;280:37366–76. doi: 10.1074/jbc.M508065200. [DOI] [PubMed] [Google Scholar]
- Ogbureke KU, Fisher LW. Renal expression of SIBLING proteins and their partner matrix metalloproteinases (MMPs) Kidney Int. 2005;68:155–66. doi: 10.1111/j.1523-1755.2005.00389.x. [DOI] [PubMed] [Google Scholar]
- Paine ML, Luo W, Wang HJ, Bringas P, Jr, Ngan AY, Miklus VG, Zhu DH, MacDougall M, White SN, Snead ML. Dentin sialoprotein and dentin phosphoprotein overexpression during amelogenesis. J Biol Chem. 2005;280:31991–8. doi: 10.1074/jbc.M502991200. [DOI] [PubMed] [Google Scholar]
- Qin C, Brunn JC, Cadena E, Ridall A, Tsujigiwa H, Nagatsuka H, Nagai N, Butler WT. The expression of dentin sialophosphoprotein gene in bone. J Dent Res. 2002;81:392–4. doi: 10.1177/154405910208100607. [DOI] [PubMed] [Google Scholar]
- Rajpar MH, Koch MJ, Davies RM, Mellody KT, Kielty CM, Dixon MJ. Mutation of the signal peptide region of the bicistronic gene DSPP affects translocation to the endoplasmic reticulum and results in defective dentine biomineralization. Hum Mol Genet. 2002;11:2559–2565. doi: 10.1093/hmg/11.21.2559. [DOI] [PubMed] [Google Scholar]
- Ritchie HH, Berry JE, Somerman MJ, Hanks CT, Bronckers AL, Hotton D, Papagerakis P, Berdal A, Butler WT. Dentin sialoprotein (DSP) transcripts: developmentally-sustained expression in odontoblasts and transient expression in pre-ameloblasts. Eur J Oral Sci. 1997;105:405–13. doi: 10.1111/j.1600-0722.1997.tb02137.x. [DOI] [PubMed] [Google Scholar]
- Ritchie H, Wang LH. The presence of multiple rat DSP-PP transcripts. Biochem Biophys Acta. 2000;1493:27–32. doi: 10.1016/s0167-4781(00)00150-0. [DOI] [PubMed] [Google Scholar]
- Ritchie HH, Wang LH, Knudtson K. A novel rat 523 amino acid phosphophoryn: nucleotide sequence and genomic organization. Biochim Biophys Acta. 2001;1520:212–222. doi: 10.1016/s0167-4781(01)00274-3. [DOI] [PubMed] [Google Scholar]
- Sambrok J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor Laboratory press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Sfeir C, Taylor S, Lin E, George A, Veis A. From mouse to zebrafish: dentin matrix proteins genomic characterization. Chemistry and Biology of mineralized tissue; Proceeding of the sixth International Conference; Vittel, France. 1998. pp. 181–184. [Google Scholar]
- Song YL, Wang CN, Fan MW, Su B, Bian Z. Dentin phosphoprotein frameshift mutations in hereditary dentin disorders and their variation patterns in normal human population. J Med Genet. 2008;45 :457–64. doi: 10.1136/jmg.2007.056911. [DOI] [PubMed] [Google Scholar]
- Sreenath T, Thyagarajan T, Hall B, Longenecker G, D’Souza R, Hong S, Wright JT, MacDougall M, Sauk J, Kulkarni AB. Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. J Biol Chem. 2003;278 :24874–80. doi: 10.1074/jbc.M303908200. [DOI] [PubMed] [Google Scholar]
- Stolc V, Gauhar Z, Mason C, Halasz G, van Batenburg MF, Rifkin SA, Hua S, Herreman T, Tongprasit W, Barbano PE, Bussemaker HJ, White KP. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–60. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Traub W, Arad T, Weiner S. Origin of mineral crystal growth in collagen fibrils. Matrix. 1992;12:251–5. doi: 10.1016/s0934-8832(11)80076-4. [DOI] [PubMed] [Google Scholar]
- Veis A, Perry A. The phosphoprotein of the dentin matrix. Biochemistry. 1967;6:2409–2416. doi: 10.1021/bi00860a017. [DOI] [PubMed] [Google Scholar]
- Verdelis K, Ling Y, Sreenath T, Haruyama N, Macdougall M, van der Meulen MC, Lukashova L, Spevak L, Kulkarni AB, Boskey AL. DSPP effects on in vivo bone mineralization. Bone. 2008;43:983–90. doi: 10.1016/j.bone.2008.08.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SN, Paine ML, Ngan AY, Miklus VG, Luo W, Wang H, Snead ML. Ectopic expression of dentin sialoprotein during amelogenesis hardens bulk enamel. J Biol Chem. 2007;282:5340–5. doi: 10.1074/jbc.M604814200. [DOI] [PubMed] [Google Scholar]
- Xiao S, Yu C, Chou X, Yuan W, Wang Y, Bu L, Fu G, Qian M, Yang J, Shi Y, Hu L, Han B, Wang Z, Huang W, Liu J, Chen Z, Zhao G, Kong X. Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat Genet. 2001;27:201–204. doi: 10.1038/84848. [DOI] [PubMed] [Google Scholar]
- Yamakoshi Y, Hu JC, Liu S, Zhang C, Oida S, Fukae M, Simmer JP. Characterization of porcine dentin sialoprotein (DSP) and dentin sialophosphoprotein (DSPP) cDNA clones. Eur J Oral Sci. 2003;111:60–7. doi: 10.1034/j.1600-0722.2003.00009.x. [DOI] [PubMed] [Google Scholar]
- Yamakoshi Y, Hu JC, Fukae M, Zhang H, Simmer JP. Dentin glycoprotein: the protein in the middle of the dentin sialophosphoprotein chimera. J Biol Chem. 2005;280:17472–9. doi: 10.1074/jbc.M413220200. [DOI] [PubMed] [Google Scholar]
- Yamakoshi Y, Lu Y, Hu JC, Kim JW, Iwata T, Kobayashi K, Nagano T, Yamakoshi F, Hu Y, Fukae M, Simmer JP. Porcine dentin sialophosphoprotein: length polymorphisms, glycosylation, phosphorylation, and stability. J Biol Chem. 2008;283:14835–44. doi: 10.1074/jbc.M800633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.