Abstract
Kinases in the phosphoinositide 3-kinase related kinase (PIKK) family include ATM (ataxia-telangiectasia mutated), ATR (ATM and Rad3-related), DNA-PKcs (DNA-dependent protein kinase catalytic subunit, mTOR (mammalian target of rapamycin), and SMG1 (suppressor with morphological effect on genitalia family member). These atypical protein kinases regulate DNA damage responses, nutrient-dependent signaling, and nonsense-mediated mRNA decay. This review focuses on the mechanisms regulating the PIKK family with a strong emphasis on the DNA damage regulated kinases. We outline common regulatory themes and suggest how discoveries about the regulation of one PIKK can be informative for the other family members.
Keywords: ATR, ATM, DNA-PK, mTOR, SMG1, PIKK
1. INTRODUCTION
Genome maintenance is critical to prevent disease. Challenges to genome integrity come from environmental mutagens, byproducts of cellular respiration, and errors during nucleic acid metabolism including DNA replication. Cells have DNA damage response (DDR) activities that continually monitor the integrity of the DNA and function to prevent the occurrence of deleterious mutations and rearrangements. The DDR is regulated by the phosphoinositide-3-kinase-related protein kinases (PIKKs). The PIKKs primarily responsible for signaling the presence of DNA damage include ATM, ATR and DNA-PKcs. These PIKKs phosphorylate hundreds of proteins that maintain genome integrity through regulation of cell cycle progression, DNA repair, apoptosis, and cellular senescence.
Human cells also contain three additional PIKKs (SMG1, mTOR, and TRRAP) with activities in other biological pathways. SMG1 primarily controls nonsense-mediated mRNA decay, mTOR regulates nutrient-dependent signaling, and TRRAP regulates transcription but lacks kinase activity. Insights into mTOR and SMG1 regulation are pertinent to the DNA-damage regulated PIKKs.
2. PIKK COMPLEXES
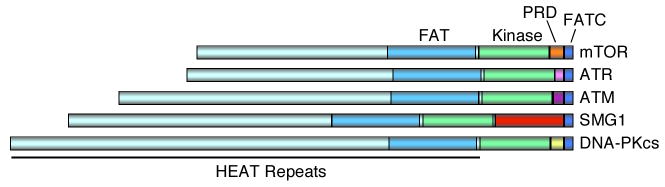
The PIKK enzymes are large proteins (between 2547-4128 amino acids) that share a common domain structure (Figure 1). The kinase domain is located near the C-terminus and is flanked by two regions of sequence similarity among all the PIKKs called the FAT (FRAP, ATM, TRRAP) and FATC (FAT C-terminus) domains. The FAT domain consists of HEAT (Huntingtin, Elongation factor 3, A subunit of protein phosphatase 2A and TOR1) repeats. The FATC domain is small (32 amino acids) and is required for PIKK kinase activity. However, the precise functions of these domains remain unclear. Since the FAT and FATC domains flank the PIKK kinase domain it has been suggested that they may interact and participate in kinase regulation [1]. The remainder of each protein consists of more HEAT repeats with little sequence similarity between the kinases and may serve as a protein-protein interaction surface [2].
Figure 1.
Diagram of PIKK domain structures showing the locations of the kinase, FAT, and FATC domains.
The signal that activates ATM and DNA-PKcs is a double strand break (DSB), while ATR responds to single stranded DNA (ssDNA) gaps. All three kinases are recruited to the DNA lesion site, which promotes kinase activation (although it is possible that soluble ATM can be activated indirectly as discussed below). While the PIKKs are at the apex of their signaling pathways and all three kinases have been reported to have some affinity for nucleic acids, they depend on associated proteins for DNA lesion recognition. ATM is recruited to double strand breaks (DSBs) indirectly through binding Mre11-Rad50-Nbs1 (MRN), a complex that binds DNA ends [3]. DNA-PKcs is recruited to DSBs by interacting with the end binding heterodimer Ku70/80 [4]. ATR is recruited to ssDNA through its binding partner ATRIP, which indirectly recognizes ssDNA through an interaction with the ssDNA binding protein replication protein A (RPA) [5]. Where it has been mapped, the primary binding site for these accessory proteins is within the N-terminal HEAT repeat region of each PIKK, and is mediated through a common motif near the C-terminus of Nbs1, Ku70, and ATRIP [6].
The MRN complex and Ku70/80 have functions in double strand break repair independent of their ATM and DNA-PKcs regulatory activity. ATRIP, however, has only been shown to function as part of a complex with ATR. All known functions of ATR require ATRIP and vice-versa and each protein is dependent on the other for stability [7], thus ATR-ATRIP is thought to be a holo-enzyme complex. mTOR also forms one of two holo-enzyme complexes with either Raptor or Rictor (mTORC1 and mTORC2 respectively) [8, 9]. Similar to ATR, the stability of mTOR also depends on it forming a holo-enzyme complex.
3. LOCALIZATION
3.1 ATR and ssDNA gaps
ATR appears to be the most versatile of the PIKK family of DNA damage-responsive kinases, being activated by a variety of DNA lesions including base adducts, crosslinks, DSBs, and compounds that directly promote replication stress such as hydroxyurea and aphidicolin. In addition to responding to environmental mutagens that induce replication stress, ATR is essential for the viability of replicating cells [7, 10]. ATR is activated during every cell cycle to regulate replication origin firing and repair damaged replication forks. Disruption of ATR results in an accumulation of DSBs during S phase, cell cycle arrest or apoptosis, and early embryonic lethality in mice [7, 11].
ATR-activating DNA lesions have in common the ability to expose single-stranded DNA (ssDNA), often as a consequence of stalling the replicative polymerases. The relative insensitivity of the replicative helicase to these lesions causes an uncoupling of polymerase and helicase activities resulting in ssDNA gaps [12]. The ssDNA is a common ATR-activating signal [13, 14].
ssDNA is rapidly coated by the heterotrimeric ssDNA binding protein RPA [15]. Several observations have highlighted the importance of RPA-coated ssDNA in ATR activation. The extent of ssDNA generated by a lesion influences the amount of DDR activation observed, with larger regions producing greater ATR activation [12]. Additionally, S. cerevisiae ATR (Mec1) activation is compromised by mutations in RPA, and knockdown of RPA in mammalian cells impairs ATR activation [14, 16, 17]. These defects may be partially reflective of the important role that RPA has in the recruitment of ATR to DNA lesions in eukaryotic organisms. ATR-ATRIP recruitment to ssDNA gaps is dependent on both ATR and ATRIP [7, 14]. An evolutionarily conserved RPA binding surface called the checkpoint protein recruitment domain (CRD) binds an N-terminal domain of RPA70 [18]. Deletion of the ATRIP CRD severely compromises the localization of ATR-ATRIP to DNA lesions, although it has only mild effects on ATR signaling [18, 19]. The lack of strong phenotype may be explained by additional RPA interactions, alternative mechanisms of ATR-ATRIP recruitment, or possibly ATR activation with only transient localization to ssDNA gaps.
The interaction with RPA-coated ssDNA may be sufficient for the recruitment of ATR-ATRIP to DNA lesions, but it is not sufficient to activate ATR. Co-localization of the RAD9-RAD1-HUS1 (9-1-1) complex to ssDNA gaps is also required. The 9-1-1 heterotrimeric ring is similar to the replicative sliding clamp PCNA, and is loaded onto ssDNA-dsDNA junctions by the RAD17-RFC2-5 clamp loader [20–23]. In vitro, the 9-1-1 complex is preferentially loaded at ssDNA gaps at the free 5′ DNA end [20], and a 5′ primer end appears to be the relevant checkpoint-activating structure [24]. A ssDNA gap with a 5′ primer end can be found at resected DSBs, unprotected telomeres, during nucleotide excision repair, and at stalled replication forks. Thus, all of these structures can activate ATR. RPA directs the recruitment of the 9-1-1 complex to the 5′ primer ends, effectively placing the components of the checkpoint activating structure in close proximity [20, 21]. A checkpoint recruitment domain on RAD9 with sequence similarity to the ATRIP CRD binds the same RPA70N binding surface as ATRIP and may help direct 9-1-1 loading or retain it at the 5′ junction [25]. Thus, multiple RPA-checkpoint protein interactions promote assembly of two checkpoint complexes (ATR-ATRIP and 9-1-1) at ssDNA gaps formed as a consequence of many types of DNA lesions.
3.2 ATM, DNA-PK, and DNA double strand breaks
ATM and DNA-PKcs are also recruited to DNA lesions sites through their accessory binding proteins NBS1 and Ku70/80. In the case of DNA-PKcs, it is clear that its function and activation is dependent on Ku70/80-dependent localization [26, 27]. Ku70/80 form a ring shaped molecule with high affinity for DNA ends [28]. Binding of Ku70/80 to DNA ends provides a scaffold for the association of DNA-PKcs and other proteins involved in non-homologous end joining. DNA-PKcs activation requires both the Ku proteins and DNA.
Whether ATM must be recruited to a double strand break to be activated is less clear. ATM autophosphorylates (a measure of activation) when cells are treated with agents like chloroquine or placed in hypotonic conditions, which reportedly do not cause DNA strand breaks [29]. In these conditions, ATM activation may depend on a protein, ATMIN, that is dispensable for DSB-induced ATM activation [30]. It is unclear what physiological stimulus these treatments mimic.
Considerable data indicate that recruitment to the damage site is important for ATM activation in response to DNA damage. Disruption of MRN function through mutation or viral infection causes defects in ATM localization and activation [31–33]. The MRN complex directly binds DNA through DNA binding domains in Mre11, and Nbs1 contains a region with sequence similarity to both ATRIP and Ku70/80 that binds ATM and is important for ATM signaling [6, 34]. Thus, the MRN complex bridges ATM with the DNA end.
Controversy remains, however, about where the initial activation of ATM occurs in response to a DSB. The chloroquine/hypotonic ATM activation data and chromatin immunoprecipitation experiments indicating that kinase activity and ATM autophosphorylation are important for ATM localization [35] have been interpreted as evidence that chromatin structure changes initiate ATM activation. In this model, the recruitment of an already active ATM to DSBs by Nbs1 amplifies ATM activity and allows it to phosphorylate additional proteins on chromatin. The alternative model is that the initial ATM activation happens at the DSB site. Support for this idea includes experiments in Xenopus egg extracts that demonstrate both the DSB end and the flanking chromatin provide a platform for ATM autophosphorylation and activation [36]. Both ATM autophosphorylation and substrate phosphorylation depended on the DNA end in this system. While further experiments will be needed to distinguish between these ideas, both models agree that amplification of ATM signaling and phosphorylation of most ATM substrates requires the concentration/localization of ATM at the DSB site.
3.3. mTOR, endomembranes, SMG1, and mRNA
Other PIKKs are also regulated by dynamic changes in cellular localization. Amino acid stimulation causes mTOR to localize to distinct perinuclear, endomembrane compartments using a Raptor- and Rag GTPase-dependent mechanism [37]. This localization is important for mTOR activation because it co-localizes mTOR and its protein activator, Rheb (see below). SMG1 binds to protein complexes at premature termination codons and exon-junction complexes on mRNA. This localization regulates nonsense-mediated mRNA decay. Thus, localization is a common theme for PIKK regulation.
4. PIKK ACTIVATORS
The localization of PIKKs to subcellular compartments promotes kinase activation by concentrating each kinase with an activating protein. ATM, ATR, and DNA-PK are activated through protein-protein interactions with Mre11, TopBP1, and Ku70/80 respectively.
4.1 ATR activation by TopBP1
Topoisomerase binding protein 1 (TopBP1) directly stimulates ATR kinase activity [38]. TopBP1 is not required for the localization of ATR to sites of damage or for the basal kinase activity of ATR [39, 40]. However, TopBP1-mediated activation of ATR is required for the damage-induced phosphorylation of ATR substrates, and is essential for cell viability in the absence of exogenous DNA damage [39]. TopBP1 is recruited to lesions independently of ATR-ATRIP by binding the phosphorylated C-terminal tail of RAD9 [41–45]. The concentration of these proteins at sites of DNA damage is hypothesized to facilitate an interaction between TopBP1 and the ATR-ATRIP holo-enzyme complex, which allows TopBP1 to promote activation of ATR kinase activity.
TopBP1 functions in both replication initiation and DDR signaling [46]. However, its activity in ATR activation is largely separable from its replication function. An “ATR-activation domain” (AAD) within TopBP1 binds directly to ATR-ATRIP [38] primarily through a binding surface on ATRIP [39]. TopBP1 also makes contact with a region on ATR called the PIKK regulatory domain (PRD) that lies between the kinase and FATC domains [39]. These interactions are essential for ATR activation and checkpoint signaling.
4.2 Activation of ATM, DNA-PK and mTOR by protein activators
In vitro, ATM is activated by Mre11/Rad50, particularly in the presence of DNA [47]. Ku70/80 activates DNA-PK in the presence of DNA ends [27]. A small GTPase, Rheb, activates mTOR [48]. The exact binding surfaces for these protein activators on their respective PIKK are unknown.
The ATM PRD is a site for acetylation by the Tip60 acetylase [49]. Mutations in the ATM PRD that prevent acetylation also prevent ATM activation. It will be important to test whether the ATM PRD acetylation alters its interaction with Mre11/Rad50, or perhaps changes its interaction with another ATM regulator such as Aven. Aven is an apoptotic inhibitor that also binds ATM and stimulate its activity [50].
Ku70/80 combined with DNA ends activates DNA-PKcs kinase activity. Mutations in the DNA-PKcs PRD block autophosphorylation [39], and low resolution structures suggest that Ku70/80 may contact the C-terminal region of DNA-PKcs containing the PRD. Further experiments will be needed to determine whether the DNA-PKcs PRD does make contact with either DNA or Ku70/80.
mTORC1 kinase activity is stimulated by mLST8 and Rheb-GTP. Rheb binds to a C-terminal portion of mTOR that includes the PRD [48]. Binding of an antibody to or deletion of the mTOR PRD creates a constitutively active mTOR kinase [51] consistent with the model that protein interactions with the PRD regulate mTOR activation.
Thus, specific protein activators regulate ATR, ATM, DNA-PKcs and mTOR. Where it has been investigated, the binding of the activator to the kinase complex causes a conformational change that allows increased substrate access to the kinase domain (see below). The PRD of each kinase has sequence conservation among orthologs but lacks significant similarity between paralogs. In each case, with the exception of SMG-1, this region is small and is unlikely to fold into an independent protein domain. As a linker region between the kinase and FATC domains with divergent amino acid sequence, the PRD provides an opportunity for unique regulatory inputs (post-translational modifications and protein-protein interactions) for each of the PIKKs.
5. POST-TRANSLATIONAL MODIFICATIONS AND PIKK STRUCTURES
How does the interaction of TopBP1 with ATR or MRN with ATM activate these kinases? Most likely, these interactions cause a conformational change that increases the ability of these kinases access substrates [52]. In response to DNA damage, ATM transitions from an inactive dimer to an active monomer [29]. This transition may be facilitated by the MRN complex and function to release ATM autoinhibition. ATM autophosphorylation correlates with the transition. Mutations that remove these autophosphorylation sites impair ATM activation in human cell culture models [29, 53]. However, in mouse models, ATM autophosphorylation mutants are completely functional [54]. Furthermore, in vitro activation of ATM by the MRN complex does not appear to require autophosphorylation [47]. It is possible that autophosphorylation stabilizes the active ATM conformation or amplifies ATM activation, but may not be essential in all biological contexts.
ATR also forms a higher order oligomeric complex in cells; however, it is unclear whether it is regulated in response to DNA damage or TopBP1 binding [55]. Both ATR-ATR and ATRIP-ATRIP contacts participate in the oligomerizations. It is possible that the ATR-ATR contacts could be disrupted upon activation but the ATRIP-ATRIP contacts remain intact. ATR is also autophosphorylated, but again there is no data indicating whether any ATR post-translational modification regulates its activity. ATRIP is phosphorylated, and this modification modulates the checkpoint function of ATR [56, 57]. Specifically, serines 224 and 239 are phosphorylated in cells. S224 is a cyclin-dependent kinase site, and kinase for S239 has not been identified. Non-phosphorylateable ATRIP mutants do not support G2 checkpoint responses as well as wild type ATRIP; however, the mechanism(s) underlying this regulatory effect remain undetermined.
DNA-PKcs also forms a dimeric complex at the double-strand break. Single particle electron microscopy images suggest that dimeric DNA-PKcs/Ku structures may help to tether two DNA ends together [58]. Tethering the ends may be essential for end-joining. The presence of DNA-PKcs at the DNA end prevents premature end processing. Current models suggest that ligation requires DNA-PKcs displacement that is triggered by the trans-phosphorylation of the DNA-PKcs dimer [59]. While the details of this process are unclear, many of the DNA-PKcs autophosphorylation sites have been mapped and non-phosphorylateable mutants cause radiosensitivity and defects in NHEJ [60, 61].
Understanding the conformational changes in the PIKKs that result in activation will require structural studies of these kinases. Unfortunately, no high resolution structures have been described. The low-resolution data indicates that DNA-PKcs is globular. The Ku70/80 heterodimer appears to bridge the N-terminal HEAT repeats and a C-terminal region distal to the kinase domain [58]. It is possible that the Ku proteins in combination with DNA may alter the position of the C-terminal region freeing the kinase domain so it can phosphorylate substrates. High resolution structures of DNA-PK and the other PIKKs are required to test these hypotheses.
6. SUMMARY AND PERSPECTIVE
The PIKKs are unusual protein kinases in that their sequence suggests common evolution from lipid kinases. The common evolution is apparent in the similarity of their regulation. While responding to different input signals, common themes of regulation include dynamic localization dependent on partner proteins and a protein or protein/nucleic acid activator (Figure 2).
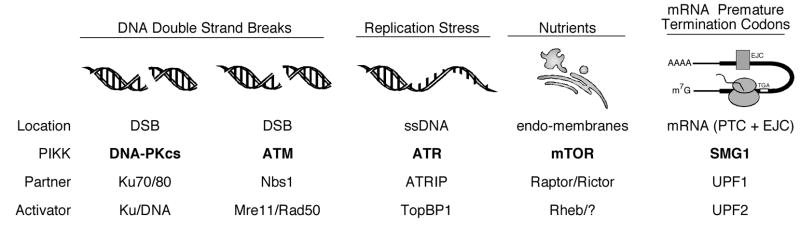
Figure 2.
Common themes of PIKK regulation. Each PIKK is regulated through localization (to a nucleic acid or intracellular compartment), a partner protein that is required for both the recruitment and activation of the kinase, and a protein or protein/nucleic acid activator.
PIKK relocalization concentrates the kinases to where their activator is found. This concentration may be primarily responsible for activation since regulation of both ATR and ATM can by bypassed in experimental systems that increase the kinase/activator local concentrations. For example, overexpression of the TopBP1 ATR-activating domain induces ATR activation in the absence of DNA damage, producing pan-nuclear γH2AX staining and cellular senescence [18, 38, 62]. Furthermore, concentrating the Mec1ATR-DDC2ATRIP and 9-1-1 complexes together on chromatin is sufficient to activate ATR without a DNA lesion in S. cerevisiae [63]. Therefore, RPA-coated ssDNA primarily serves as a scaffold for the assembly of proteins required for ATR activation, with the independent recruitment of checkpoint activating complexes providing a mechanism of regulation to ensure that ATR is not inappropriately activated. Similarly, immobilizing and concentrating ATM or key ATM regulators on chromatin also activates ATM signaling [64].
Kinase signaling is further regulated by post-translational modifications on the PIKKs, accessory proteins, and activator proteins. These modifications may provide opportunities for fine-tuning or amplifying the signal. Finally, conformational changes underlie PIKK activation. We do not yet understand why the dimer to monomer ATM transition is important or how TopBP1 interaction with ATR-ATRIP allows greater substrate access. Further mutagenesis and domain mapping approaches will be useful, but ultimately high-resolution structures will be critical.
While the amount of information about how the PIKKs are activated has exploded in recent years, there remain many unanswered questions about this interesting family of kinases. Experimental results such as the lack of phenotype of the ATM S1981A mutant in mice, and relatively minor defects associated with mutations in the ATRIP-RPA binding surfaces, suggest that the simple activation models need further revision. New proteins like Aven must be incorporated into the models. Of course, the signaling downstream of these kinases has not been addressed in this review, but is equally complex.
Future research into the PIKKs will surely help us to understand not only how cells maintain genome integrity to prevent diseases such as cancer, but also may point to opportunities for drug development. mTOR has been targeted for decades with the natural product rapamycin – an antiproliferative antibiotic useful to cause immunosuppression. ATM and ATR pathway inhibitors are being developed as potential radio- and chemo-sensitizing agents for cancer therapy. Since cancer cells have defects in genome maintenance activities and have considerable genome integrity challenges due to their microenvironment and oncogene-induced replicative stress, targeting the DDR pathways may be useful as a monotherapy in cancer. However, successful targeting of these pathways requires that we understand their genome maintenance activities, how they are regulated, what genome maintenance activities are defective in cancer cells, which proteins might provide the best targets, and which patients might benefit most from this type of therapeutic approach. Furthermore, biomarkers for patient selection and drug evaluation will be needed. Thus, filling in the details of the PIKK signaling pathways is critical. Fortunately, our task is made easier by the significant similarities between the mechanisms regulating these kinases.
Acknowledgments
The authors would like to thank Daniel Mordes and other members of the Cortez laboratory for their insightful contributions. Studies on ATR in the authors’ laboratory are supported by R01 CA102729.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bosotti R, Isacchi A, Sonnhammer EL. FAT: a novel domain in PIK-related kinases. Trends Biochem Sci. 2000;25:225–227. doi: 10.1016/s0968-0004(00)01563-2. [DOI] [PubMed] [Google Scholar]
- 2.Perry J, Kleckner N. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell. 2003;112:151–155. doi: 10.1016/s0092-8674(03)00033-3. [DOI] [PubMed] [Google Scholar]
- 3.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 4.Smith GC, Jackson SP. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 5.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 7.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 8.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 9.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 10.Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 12.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costanzo V, Gautier J. Single-strand DNA gaps trigger an ATR- and Cdc7-dependent checkpoint. Cell Cycle. 2003;2:17. doi: 10.4161/cc.2.1.290. [DOI] [PubMed] [Google Scholar]
- 14.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 15.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Longhese MP, Neecke H, Paciotti V, Lucchini G, Plevani P. The 70 kDa subunit of replication protein A is required for the G1/S and intra-S DNA damage checkpoints in budding yeast. Nucleic Acids Res. 1996;24:3533–3537. doi: 10.1093/nar/24.18.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umezu K, Sugawara N, Chen C, Haber JE, Kolodner RD. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ball HL, Ehrhardt MR, Mordes DA, Glick GG, Chazin WJ, Cortez D. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol Cell Biol. 2007;27:3367–3377. doi: 10.1128/MCB.02238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ball HL, Myers JS, Cortez D. ATRIP Binding to RPA-ssDNA Promotes ATR-ATRIP Localization but Is Dispensable for Chk1 Phosphorylation. Mol Biol Cell. 2005;16:2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majka J, Binz SK, Wold MS, Burgers PM. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem. 2006;281:27855–27861. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- 22.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9–1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci U S A. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci U S A. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X, Vaithiyalingam S, Glick GG, Mordes DA, Chazin WJ, Cortez D. The basic cleft of RPA70N binds multiple checkpoint proteins including RAD9 to regulate ATR signaling. Mol Cell Biol. 2008 doi: 10.1128/MCB.01079-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dvir A, Peterson SR, Knuth MW, Lu H, Dynan WS. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci U S A. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 28.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 29.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 30.Kanu N, Behrens A. ATMIN defines an NBS1-independent pathway of ATM signalling. Embo J. 2007;26:2933–2941. doi: 10.1038/sj.emboj.7601733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. Embo J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. The Mre11 complex is required for ATM activation and the G2/M checkpoint. Embo J. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horejsi Z, Falck J, Bakkenist CJ, Kastan MB, Lukas J, Bartek J. Distinct functional domains of Nbs1 modulate the timing and magnitude of ATM activation after low doses of ionizing radiation. Oncogene. 2004;23:3122–3127. doi: 10.1038/sj.onc.1207447. [DOI] [PubMed] [Google Scholar]
- 34.Stracker TH, Morales M, Couto SS, Hussein H, Petrini JH. The carboxy terminus of NBS1 is required for induction of apoptosis by the MRE11 complex. Nature. 2007;447:218–221. doi: 10.1038/nature05740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 36.You Z, Bailis JM, Johnson SA, Dilworth SM, Hunter T. Rapid activation of ATM on DNA flanking double-strand breaks. Nat Cell Biol. 2007;9:1311–1318. doi: 10.1038/ncb1651. [DOI] [PubMed] [Google Scholar]
- 37.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 39.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol. 2006;26:6056–6064. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.St Onge RP, Besley BD, Pelley JL, Davey S. A role for the phosphorylation of hRad9 in checkpoint signaling. J Biol Chem. 2003;278:26620–26628. doi: 10.1074/jbc.M303134200. [DOI] [PubMed] [Google Scholar]
- 42.Greer DA, Besley BD, Kennedy KB, Davey S. hRad9 rapidly binds DNA containing double-strand breaks and is required for damage-dependent topoisomerase II beta binding protein 1 focus formation. Cancer Res. 2003;63:4829–4835. [PubMed] [Google Scholar]
- 43.Furuya K, Poitelea M, Guo L, Caspari T, Carr AM. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 45.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9–1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst) 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 47.Lee JH, Paull TT. ATM Activation by DNA Double-Strand Breaks Through the Mre11-Rad50-Nbs1 Complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 48.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 49.Sun Y, Xu Y, Roy K, Price BD. DNA damage induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol. 2007;24:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo JY, Yamada A, Kajino T, Wu JQ, Tang W, Freel CD, Feng J, Chau BN, Wang MZ, Margolis SS, Yoo HY, Wang XF, Dunphy WG, Irusta PM, Hardwick JM, Kornbluth S. Aven-dependent activation of ATM following DNA damage. Curr Biol. 2008;18:933–942. doi: 10.1016/j.cub.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekulic A, Hudson CC, Homme JL, Yin P, Otterness DM, Karnitz LM, Abraham RT. A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer Res. 2000;60:3504–3513. [PubMed] [Google Scholar]
- 52.Mordes DA, Cortez D. Activation of ATR and related PIKKs. Cell Cycle. 2008;7:2809–2812. doi: 10.4161/cc.7.18.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozlov SV, Graham ME, Peng C, Chen P, Robinson PJ, Lavin MF. Involvement of novel autophosphorylation sites in ATM activation. Embo J. 2006;25:3504–3514. doi: 10.1038/sj.emboj.7601231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pellegrini M, Celeste A, Difilippantonio S, Guo R, Wang W, Feigenbaum L, Nussenzweig A. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature. 2006;443:222–225. doi: 10.1038/nature05112. [DOI] [PubMed] [Google Scholar]
- 55.Ball HL, Cortez D. ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J Biol Chem. 2005;280:31390–31396. doi: 10.1074/jbc.M504961200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myers JS, Zhao R, Xu X, Ham AJ, Cortez D. Cyclin-dependent kinase 2 dependent phosphorylation of ATRIP regulates the G2-M checkpoint response to DNA damage. Cancer Res. 2007;67:6685–6690. doi: 10.1158/0008-5472.CAN-07-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venere M, Snyder A, Zgheib O, Halazonetis TD. Phosphorylation of ATR-interacting protein on Ser239 mediates an interaction with breast-ovarian cancer susceptibility 1 and checkpoint function. Cancer Res. 2007;67:6100–6105. doi: 10.1158/0008-5472.CAN-07-0369. [DOI] [PubMed] [Google Scholar]
- 58.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 59.Weterings E, Chen DJ. DNA-dependent protein kinase in nonhomologous end joining: a lock with multiple keys? J Cell Biol. 2007;179:183–186. doi: 10.1083/jcb.200705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding Q, Reddy YV, Wang W, Woods T, Douglas P, Ramsden DA, Lees-Miller SP, Meek K. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toledo LI, Murga M, Gutierrez-Martinez P, Soria R, Fernandez-Capetillo O. ATR signaling can drive cells into senescence in the absence of DNA breaks. Genes Dev. 2008;22:297–302. doi: 10.1101/gad.452308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonilla CY, Melo JA, Toczyski DP. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol Cell. 2008;30:267–276. doi: 10.1016/j.molcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soutoglou E, Misteli T. Activation of the Cellular DNA Damage Response in the Absence of DNA Lesions. Science. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]