SUMMARY
Centromeres of higher eukaryotes are epigenetically maintained, however, the mechanism that underlies centromere inheritance is unknown. Centromere identity and inheritance require the assembly of nucleosomes containing the CenH3 histone variant in place of canonical H3. Whereas H3 nucleosomes wrap DNA in a left-handed manner and induce negative supercoils, we show here that CenH3 nucleosomes that are reconstituted from Drosophila histones induce positive supercoils. Furthermore, we show that CenH3 likewise induces positive supercoils in functional centromeres in vivo, using a budding yeast minichromosome system and temperature-sensitive mutations in kinetochore proteins. The right-handed wrapping of DNA around the histone core implied by positive supercoiling indicates that centromere nucleosomes are unlikely to be octameric. Rather, the surfaces that hold the nucleosome together would be available for kinetochore protein recruitment. The mutual incompatibility of nucleosomes with opposite topologies can potentially explain how centromeres are efficiently maintained as a unique loci on chromosomes.
INTRODUCTION
Genomic DNA of eukaryotes is wrapped around octameric histone core particles consisting of two molecules each of histones H2A, H2B, H3 and H4. Each histone core particle contains an (H3/H4)2 tetramer flanked by H2A/H2B dimers, and displays two-fold symmetry around a dyad axis that passes through the H3-H3 4-helix bundle dimerization interface (Luger et al., 1997). Nucleosomal DNA tightly wraps around this core 1.7 times in a left-handed configuration, and in vitro reconstitution studies demonstrate that left-handedness is an inherent feature of nucleosome structure. The direction of DNA wrapping is thought to have important implications for active processes that take place on chromosomes, including replication, transcription and DNA repair. For example, torsional stress (positive supercoiling) created by polymerases moving along DNA would tend to counteract the left-handed wrap of nucleosomes, possibly facilitating their displacement (van Holde et al., 1992).
The presence of a variant histone within a nucleosome has the potential to profoundly alter chromatin structural properties and impact chromosomal processes. At centromeres, the CenH3 variant replaces canonical H3 in centromeric nucleosomes (Buchwitz et al., 1999; Henikoff et al., 2000; Meluh et al., 1998; Palmer et al., 1991; Takahashi et al., 2000) and is essential for the recruitment of other kinetochore components (Heun et al., 2006). Relative to canonical H3, which is one of the most highly conserved proteins known, CenH3s from different organisms are surprisingly diverged, even within the histone core, with N-terminal tails that can be of very different lengths (Malik and Henikoff, 2003). Despite these differences, the function of CenH3 nucleosomes in organizing the kinetochore appears to be invariant. For example, CenH3 from budding yeast (Cse4p) can substitute for human CenH3 (CENP-A) (Wieland et al., 2004), even though only a single Cse4p nucleosome occupies each budding yeast centromere (Furuyama and Biggins, 2007), whereas human centromeres comprise long arrays consisting of thousands of CENP-A nucleosomes (Lam et al., 2006; Schueler et al., 2001). These observations suggest that there are general structural features of CenH3 nucleosomes responsible for their conserved role in forming the foundation of the kinetochore and for their faithful assembly at centromeres every cell cycle (Bloom and Carbon, 1982; Dalal et al., 2007b; Polizzi and Clarke, 1991; Takahashi et al., 1992) . Indeed, CenH3 nucleosomes have been found to differ profoundly from their canonical counterparts. Whereas micrococcal nuclease (MNase) cleaves between canonical nucleosomes to yield familiar nucleosomal ladders with periodicities reflecting internucleosomal distances, no such ladders were observed for fission yeast CenH3 (Cnp1) (Bloom and Carbon, 1982; Polizzi and Clarke, 1991). MNase sensitivity was also seen for native Drosophila melanogaster CenH3 (CID) nucleosomes (Dalal et al., 2007b), in which centromeric DNA was deduced to be wrapped around tetramers of CenH3, H4, H2A and H2B. The tetrameric organization of Drosophila CenH3 nucleosomes observed in chromatin extracts was confirmed by direct measurement of purified native particle heights using atomic force microscopy, and suggested that interphase CenH3 nucleosomes are stable heterotypic tetramers for which a “hemisome” model has been proposed (Dalal et al., 2007b).
Reconstituted (H3/H4)2 tetramers can be wrapped in either direction (Hamiche et al., 1996), and only the addition of H2A/H2B dimers locks them in the left-handed configuration (Alilat et al., 1999). Many archaea package DNA into nucleosomes, which are tetrameric and appear to wrap DNA in a left- or right-handed manner in vitro depending on the salt conditions used (Marc et al., 2002; Musgrave et al., 2000; Musgrave et al., 1991). Intriguingly, topological analysis of a yeast minichromosome suggested that deletion of the centromere resulted in more negatively supercoiled DNA, an observation made prior to the discovery of CenH3s and not interpreted as unusual by the authors of the study (Bloom et al., 1984).
Here we examine the topological state of centromeric nucleosomes in vitro and in vivo to determine the direction of supercoiling induced by substitution of CenH3 for H3 within nucleosomes. We show that Drosophila CID induces positive supercoils when reconstituted into nucleosomes with partner histones in vitro. We confirm this observation in vivo, using wildtype and mutant budding yeast minichromosomes maintained in the presence of temperature-sensitive mutations in kinetochore components. Our findings suggest that positive supercoiling is a general feature of centromeric nucleosomes that has important implications for maintaining centromeres as uniquely defined loci that organize kinetochores.
RESULTS
Reconstituted Drosophila CenH3 nucleosomes induce positive supercoils
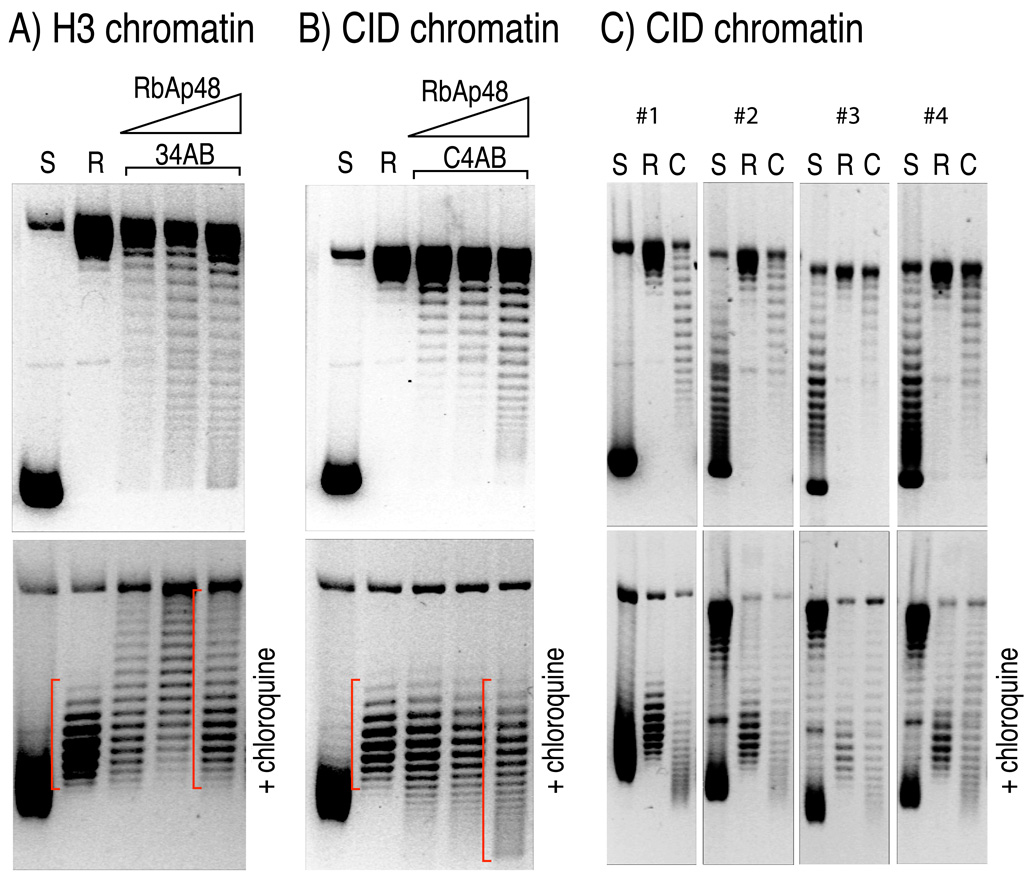
In previous work, we identified the abundant histone chaperone, RbAp48, as the single non-histone stoichiometric component of native CID complexes isolated from soluble Drosophila extracts (Furuyama et al., 2006). Although the the RbAp48-CID/H4 complex by itself was unable to assemble nucleosomes on DNA, addition of H2A/H2B dimers led to the assembly of chromatin particles in vitro as evidenced by electron microscopy, DNaseI digestion and plasmid supercoiling (Furuyama et al., 2006). In the standard plasmid supercoiling assay, reconstitution of assembled particles onto a closed circular plasmid DNA is performed in the presence of topoisomerase I, which relaxes the compensatory torsional stress on DNA during nucleosome assembly. Subsequent removal of proteins yields a closed circular DNA, in which additional “turns”, each originally induced by the wrapping of DNA around one histone core particle, are now irreversibly trapped (Prunell, 1998). When these plasmids are electrophoretically separated, each additional full turn of nucleosome-wrapped DNA contributes to compaction relative to relaxed “open” circles, yielding a ladder of topoisomers. This assay is indicative of the number of nucleosomes assembled on the plasmid, but not the direction of induced writhe, because both positive and negative supercoils cause compaction relative to relaxed circles.
To ascertain the direction of supercoiling induced by CenH3 core particles assembled by RbAp48, we electrophoresed the deproteinized plasmids in the presence of the intercalating drug chloroquine, which reduces the twist of DNA (Figure S1). Because the linking number (Lk) is fixed in a covalently closed plasmid, the reduction in twist (Tw) must be compensated for by an increase in writhe (Wr), (ΔLk = ΔTw+ΔWr) (Prunell, 1998). At a chloroquine concentration of 1 µg/ml, plasmids isolated from Escherichia coli migrate more slowly (Figure 1 bottom panel and Figure S2), because they are negatively supercoiled and so are relaxed by the additional positive writhe. Similarly, the additional positive writhe induced by chloroquine causes plasmids that are relaxed by pre-treatment with topoisomerase to migrate faster than nicked circular DNA (Figure 1A–B bottom panel). The relative position of topoisomers induced with assembled chromatin relative to that of relaxed DNA indicates their topological states (Figure S1). Topoisomers induced by H3-containing nucleosomes are known to be negatively supercoiled; therefore, topoisomers obtained upon chromatin assembly using RbAp48, H3 and its histone partners migrate more slowly in chloroquine-containing gels than initially relaxed plasmids (Figure 1A bottom panel). In striking contrast, chloroquine intercalation causes topoisomers induced by RbAp48-assembled CID chromatin to migrate faster than initially relaxed plasmids (Figure 1B bottom panel); therefore, these topoisomers must have had net positive supercoils compared to relaxed plasmids, just the opposite of supercoiling induced by H3 chromatin.
Figure 1. Drosophila CID chromatin assembled in vitro induces positive supercoils in closed circular plasmids.
(A) An ~7 kb plasmid containing an ~3 kb Drosophila satellite DNA insert (Furuyama et al., 2006) electrophoreses as negatively supercoiled species in an agarose gel after isolation from E.coli (S). The plasmid DNA was relaxed by topoisomerase (R), and H3 chromatin was assembled in vitro (34AB) in the presence of the Drosophila RbAp48 chaperone, as previously described. The top panel shows the topoisomer separation on an agarose gel without chloroquine, whereas the bottom panel shows their migration in the presence of 1µg/ml chloroquine. Gels were stained with ethidium bromide after separation to visualize DNA. The slower migration of topoisomers induced by H3-containing nucleosome relative to the migration of relaxed DNA (R) indicates that H3-nucleosomes induce negative supercoils (red bracket). (B) same as in (A), except that CID (Drosophila CenH3) was used in place of H3 (C4AB). The faster migration of topoisomers induced by CID-containing nucleosome relative to the migration of relaxed DNA (R) in the presence of chloroquine indicates that CID-nucleosomes induce positive supercoils in vitro. (C) Identical chromatin assembly reactions were performed as in (B) using four independent plasmids containing randomly cloned 3-kb Drosophila genomic DNA (Clone #1 through #4; See also Figure S1). CID nucleosomes induce positive supercoils regardless of DNA sequence, as evident from migration patterns that are similar to those in (B). S: negatively supercoiled; R, relaxed; C: CID chromatin.
The DNA that we used in supercoiling assays contains a 3-kb segment of a 359-bp Drosophila melanogaster satellite repeat array inserted into a plasmid vector (Furuyama et al., 2006). To ascertain if DNA sequence might influence supercoiling behavior when CID nucleosomes are assembled, we cloned random 3-kb Drosophila DNA into the same plasmid vector and chose four of the resulting plasmids for supercoiling analysis. In all four cases, we observed positive supercoils induced by RbAp48-assembled CID nucleosomes (Figure 1C), and additional random plasmids yielded the same result (Figure S1). In contrast, reconstituted H3 nucleosomes induced only negative supercoils on a wide variety of DNAs (Bates and Maxwell, 2005). Therefore, we conclude that the direction of supercoiling depends on the presence of CID-containing nucleosomes rather than structural properties of specific DNA sequences.
Loss of CenH3 reduces positive supercoiling in budding yeast in vivo
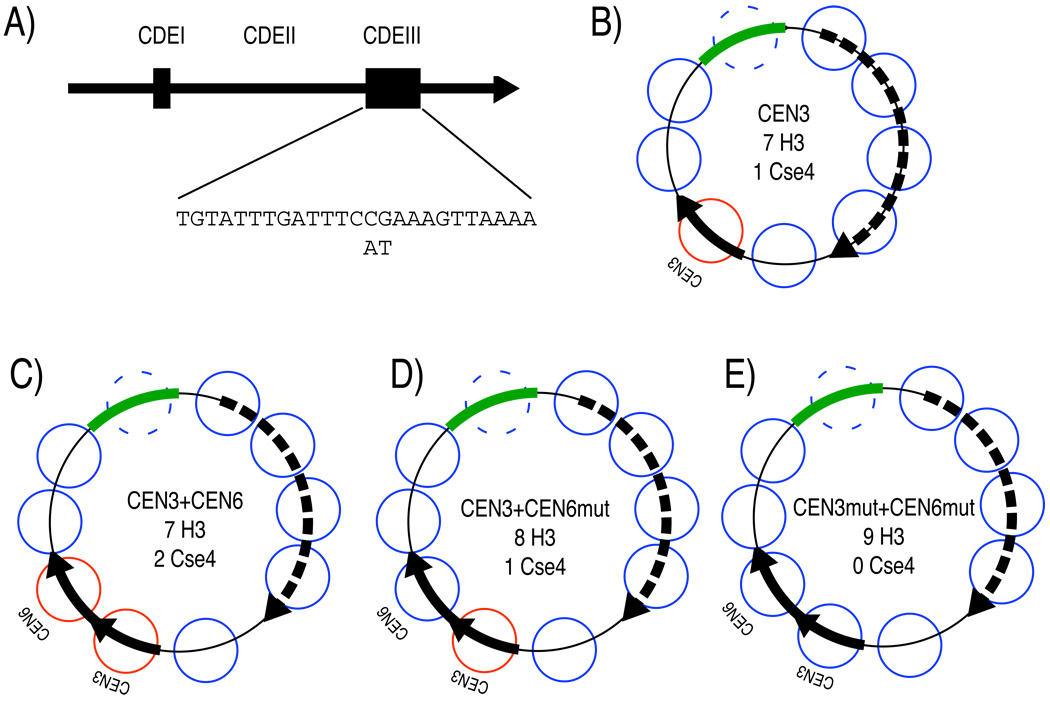
Topology assays in vivo require small covalently closed DNA circles of a defined sequence that can be distinguished from endogenous sequences in the genome. Higher eukaryotes lack plasmid systems that can be manipulated to yield small closed DNA circles with active centromeres. Furthermore, centromeres of most higher eukaryotes are embedded in long tandem arrays of satellite sequences that contain interspersed blocks of both CenH3 and H3 nucleosomes (Blower and Karpen, 2001; Lam et al., 2006), and sequences known to support centromere function are often hundreds of kilobases in length. This situation is even more challenging in the case of Drosophila, where no single centromeric DNA satellite is common to all chromosomes. Because of these considerations, topological assays cannot be practically performed in Drosophila, and probably other complex eukaryotes, using available technologies. In contrast, each Saccharomyces cerevisiae centromere is specified by a ~125-bp centromere-determining element (CDE, Figure 2A) that contains a single Cse4p nucleosome (Furuyama and Biggins, 2007) and supports regular segregation of a plasmid that carries it (Clarke and Carbon, 1980).
Figure 2. Schematic diagrams of minichromosomes used in the study.
(A) A diagram of a 190-bp CEN region. It consists of CDEI (small box), CDEIII (large box), and CDEII between the boxes. The conserved CDEIII sequence with two single base pair substitutions that abolish centromere functions are shown. (B) The CEN3 minichromosome construct used in Figure 3 (a gift from S. Biggins and T. Tsukiyama). This ~2-kb minichromosome contains a TRP selectable marker (Dotted arrow indicates the location of ORF), CEN3 (solid arrow) and a short stretch of bacterial DNA (green bar), which was used as the Southern blot probe target, because it does not cross-hybridize with yeast genomic DNA (data not shown). The diagram is approximately to scale. The known locations of H3 nucleosomes that were previously mapped (Thoma et al., 1984) are shown as blue circles. Red circles indicate the presumed locations of Cse4p nucleosomes. We do not know whether a nucleosome is present over the bacterial sequence used for Southern blotting (dashed circle). Expected numbers of H3 and Cse4p nucleosomes (assuming that there are no nucleosomes corresponding to the dashed circle) are indicated. The relatively low nucleosome density of 2000 bp/8 nucleosomes = 250 bp/nucleosome (yeast average: 165 bp/nucleosome (Nelson and Fangman, 1979)) results from the presence of several well-positioned nucleosome and several nucleosome-free regions on the minichromosome. (C) The CEN3+CEN6 double centromere minichromosome construct used in Figure 4 (See Table S1). (D) same as (C) except that CEN6 carries the 2-bp substitution shown in (A). (E) same as (C) except that both CEN3 and CEN6 carry the 2-bp substitution shown in (A).
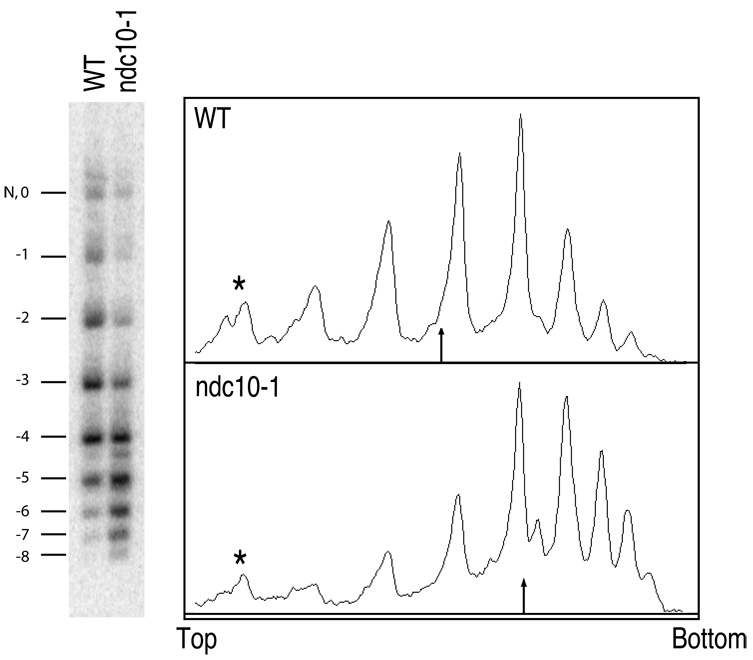
We used a ~2-kb minichromosome derived from the well-characterized TRP-ARS1 construct (Thoma et al., 1984), into which both the Chromosome 3 centromere (CEN3) and a short stretch of bacterial sequence (for Southern blot probing) had been inserted (Figure 2B). The resulting minichromosome contains ~8 total nucleosomes: 1 CenH3 nucleosome at CEN3 plus 7 previously mapped well-positioned H3 nucleosomes (Thoma et al., 1984) (although in what follows, the absolute number of nucleosomes on the minichromosome is not important). To observe this minichromosome in a configuration that lacks a centromere, we maintained it in an ndc10-1 background. ndc10-1 is a temperature-sensitive mutation in a component of the CBF3 complex, which binds to the CDEIII cis-acting element within the 125-bp CDE, and is required for the localization of Cse4p to CEN (Ortiz et al., 1999; Pearson et al., 2003). Both ndc10-1 and wildtype strains carrying the minichromosome were arrested at the G1/S phase boundary with α-factor, then released into S phase at the restrictive temperature (37°C). As expected, loss of Cse4p in ndc10-1 results in 8 left-handed H3 nucleosomes, each of which induces a negative supercoil. However, if Cse4p induces positive supercoiling as we observed in vitro, 7 left-handed H3 nucleosomes and 1 right-handed CenH3 nucleosome in wildtype would contribute 6 net negative supercoils, for a difference of -2 supercoils between ndc10-1 and wildtype (For an explanation of the in vivo DNA topology assay, see Figure S3). We find that the topoisomer distribution of the minichromosome isolated from wildtype is shifted up (becomes less negative) by an average of 1.33 supercoils (−2.86) compared to the identical minichromosome isolated from the ndc10-1 mutant strain (−4.19) (Figure 3). The average number of negative supercoils in wildtype might have been overestimated, because the band corresponding to a topoisomer with no net writhe in the presence of chloroquine co-migrates with a nicked circle (Figure S4), and so was not included in the calculation. Therefore, the difference between wildtype and the ndc10-1 mutant might be closer to 2 than 1.33, which would imply that loss of the centromere results in loss of a positive supercoil induced by the CenH3 nucleosome and gain of a negative supercoil induced by the H3 nucleosome that replaces it. The observed shift in topoisomer distribution between wildtype and ndc10-1 is similar to the shift seen when the centromere was deleted from a minichromosome (Bloom et al., 1984).
Figure 3. A yeast minichromosome loses negative supercoils with a functioning centromere.
Total DNA was isolated from either a wildtype (WT) or a mutant ndc10-1 strain carrying the same CEN3 minichromosome (Figure 2B), released from α-factor arrest at the restrictive temperature (37°C), and purified DNA was resolved on an agarose gel containing 0.3µg/ml chloroquine. A Southern blot to detect the minichromosome is shown on the left. A densitometry trace of each lane is shown on the right. The most slowly migrating band contains both a nicked circle (N) and a topoisomer with no net writhe in the presence of chloroquine (0). The numbers correspond to the value of net writhe in this chloroquine gel. The minichromosome has fewer negative supercoils in the wildtype strain with Cse4p present at the centromere, compared to the ndc10-1 mutant strain, which loses Cse4p at the restrictive temperature. The arrow in the trace indicates the location of the mean topoisomer distribution. The asterisks mark the bands containing nicked and “0” topoisomer, which was omitted from calculating the mean distribution. Nicked and ”0” topoisomer were resolved by two-dimensional electrophoresis, where similar results were obtained (Figure S4).
The number of positive supercoils corresponds to the number of functional centromeres
A complication of comparing yeast plasmids with and without a functional centromere is that the proteinaceous yeast kinetochore is present throughout the cell cycle (McAinsh et al., 2003), and this evidently protects a region of ~200 bp from MNase digestion, compared to ~160-bp protection afforded by the presence of a typical H3 nucleosome (Bloom and Carbon, 1982). Consequently, if Cse4p induces a negative supercoil, then the 1.33 negative supercoil gain with centromere loss might be attributable to the gain of two H3 nucleosomes that would replace the lost Cse4p nucleosome. Therefore, the observed topoisomer shifts do not rule out the possibility that Cse4p induces a negative supercoil.
To address this alternative interpretation, we constructed minichromosomes of identical size with zero, one or two centromeres (Figure 2; Table S1), which effectively increases the resolution of the plasmid supercoiling assay (Figure S3). Yeast sequences taken from CEN3 and CEN6 were inserted as close as possible next to one another (CEN3+CEN6) (Figure 2) to reduce the dicentric chromosome instability, which is known to increase with distance between the two centromeres (Koshland et al., 1987). To eliminate centromere function without changing the size of the minichromosome, two base-pair substitutions were made in the critical CDEIII region, which causes loss of CBF3 complex binding and abolition of centromere function (Jehn et al., 1991). Minichromosomes that contain these base pair substitutions in one or both centromeres were also constructed (CEN3+CEN6mut and CEN3mut+CEN6mut, respectively). The resulting minichromosomes of identical size with 0, 1 or 2 functional centromeres were used to transform wildtype and ndc10-1 strains.
To confirm the dicentricity of our CEN3+CEN6 construct, we measured characteristics previously documented for dicentric minichromosomes (Koshland et al., 1987). Strains carrying a dicentric minichromosome grow more slowly than strains carrying a monocentric minichromosome in selective media. Similar growth delays were consistently observed for our CEN3+CEN6 construct (Table 1). In addition, CEN3+CEN6 displayed a lower mitotic stability phenotype with a value very similar to that reported previously for a dicentric minichromosome (Koshland et al., 1987) (Table 1).
Table 1.
Doubling time, copy number, and mitotic stability of di-, mono-, and a-centric minichromosomes.
| Genetic background |
# of CEN |
Doubling time (min) a |
Mitotic Stability per generation b |
Copy number per cell c |
|---|---|---|---|---|
| WT | 2 | 325.1±8.7 | 0.740±0.029 | 1.53±0.06 |
| 1 | 219.7±16.7 | 0.958±0.057 | 4.01±0.71 | |
| 0 | 219.1±7.8 | 0.952±0.026 | 9.96±2.62 | |
| ndc10 | 2 | 321.0±23.2 | 0.793±0.028 | 1.50±0.17 |
| 1 | 266.6±0 | 0.942±0.034 | 3.36±0.31 | |
| 0 | 217.0±11.4 | 0.938±0.031 | 11.52±1.20 | |
| ncd80 | 2 | 360.4±67.0 | 0.456±0.396 | 1.13±0.10 |
| 1 | 281.1±6.7 | 0.850±0.001 | 1.63±0.12 | |
| 0 | 230.1±6.7 | 0.948±0.044 | 8.39±2.79 |
The average doubling time (± standard deviation) at 25°C of early to mid- log culture for three independent transformants of the indicated genotype.
The mitotic stability (± standard deviation) of each minichromosome was determined by first culturing in non-selective media to allow minichromosome loss for approximately 10 generation, then plating the culture on non-selective or selective plates. Values represent averages from three independent transformants. The tenth root of the ratio of the number of resulting colonies on selective versus non-selective plates approximates the probability of minichromosome loss per generation.
The average copy number (± standard deviation) of minichromosomes per cell in three independent transformants, obtained from a Southern blot detecting chromosomal and minichromosomal copy of TRP loci (Figure S4).
We also tested our CEN3mut+CEN6mut constructs for loss of centromere function. When placed under selection for tryptophan, minichromosomes without a functional centromere are known to be maintained at elevated copy numbers relative to CEN plasmids (Hill and Bloom, 1987). As expected for a minichromosome without centromere function, the CEN3mut+CEN6mut minichromosome is maintained at a higher copy number (~10 copies per cell), as determined by probing Southern blots for both the endogenous TRP locus and the TRP locus on the minichromosome (Table 1; Figure S5 and Figure S6A). A minichromosome with a single functional centromere is known to be maintained at ~2–3 copies per cell (Resnick et al., 1990), and our results are consistent with that observation. The CEN3+CEN6 minichromosome is maintained at lower copy number than a minichromosome with one functional centromere, which probably reflects dicentric chromosome instability. Taken together, the genetic characterization of strains carrying 0, 1, or 2 centromeres show that each wildtype centromere is functional and each mutated centromere is not.
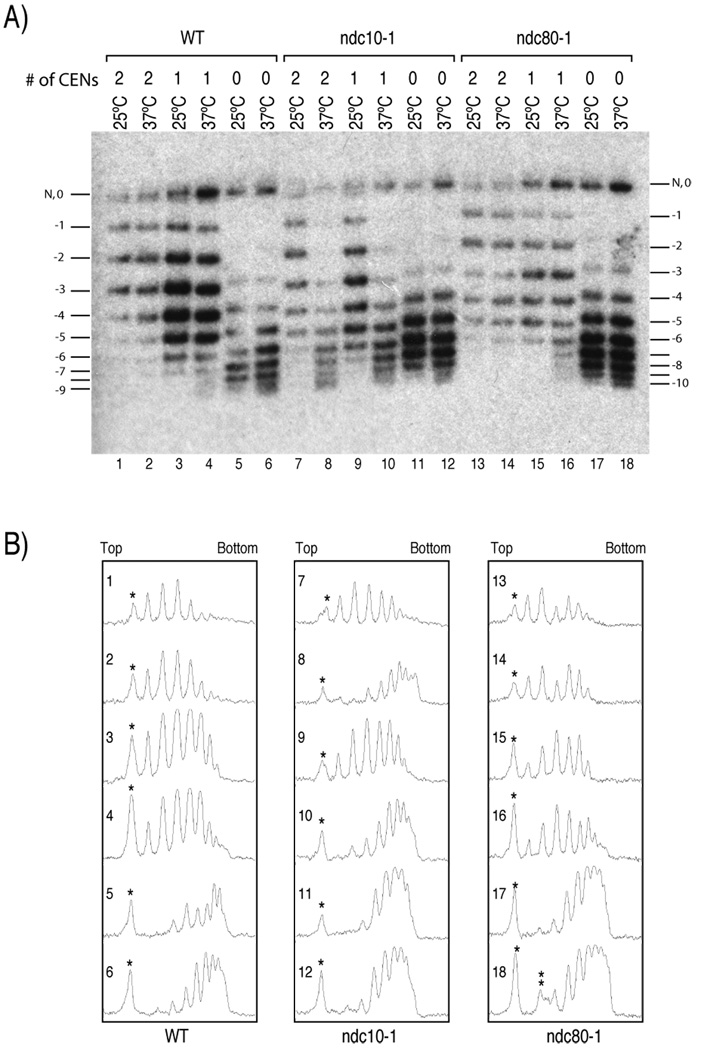
These minichromosome constructs display the topological differences that are expected if each CenH3 nucleosome induces one positive supercoil. The CEN3mut+CEN6mut minichromosome, which contains no functional centromeres, is the most negatively supercoiled construct in wildtype cells, whereas the addition of one or two functional centromeres results in progressively fewer supercoils (Figure 4A, lanes 1–6; Figure S5; Table 2). Small topological differences were also observed for minichromosomes obtained from wildtype cells grown at 25°C and 37°C, as expected from the known partial unwinding of DNA at higher temperature; this produces a compensatory increase in positive writhe that is removed by cellular topoisomerases in vivo, and results in net negative supercoils (Saavedra and Huberman, 1986) (Figure 4: lanes 1 versus 2 and 3 versus 4; Table 2).
Figure 4. Cse4p nucleosomes induce positive supercoils in vivo.
(A) Minichromosomes containing 2, 1 or no functional centromeres (# of CENs) that were derived from wildtype (WT), ndc10-1, or ndc80-1 strains were allowed to go through S phase at either 25°C or 37°C. Total DNA were isolated and electrophoresed on an agarose gel containing 0.3µg/ml chloroquine to resolve topoisomers. Southern blot analysis was performed to detect the minichromosomes. The most slowly migrating band contains both a nicked circle (N) and a topoisomer with no net writhe (0). The numbers correspond to the value of net writhe in this chloroquine gel. When a relaxed circle was run in an identical chloroquine gel, its peak distribution was +2 and +3, which co-migrates with −2 and −3 topoisomers (data not shown). Note that the first detectable band in lanes 5, 6, 11, 12, 17, and 18 corresponds to −3. The Southern blot shown here is a representative of independent experiments. (B) Densitometric trace of (A) with lane numbers as indicated. The asterisks mark the nicked and “0” topoisomer in (A). The double asterisks in the trace of lane 18 indicate a smudge in Figure 4A, which was not included in the analysis provided in Table 2.
Table 2.
Mean supercoiling densities of minichromosomes
| Genetic background |
# of CEN |
Temperature (°C) |
Mean supercoiling Density a |
|---|---|---|---|
| WT | 2 | 25 | −2.07 |
| 2 | 37 | −2.25 | |
| 1 | 25 | −3.09 | |
| 1 | 37 | −3.27 | |
| 0 | 25 | −6.16 | |
| 0 | 37 | −6.16 | |
| ndc10-1 | 2 | 25 | −2.48 |
| 2 | 37 | −5.87 | |
| 1 | 25 | −3.45 | |
| 1 | 37 | −5.48 | |
| 0 | 25 | −5.51 | |
| 0 | 37 | −5.69 | |
| ndc80-1 | 2 | 25 | −1.99 |
| 2 | 37 | −3.20 | |
| 1 | 25 | −3.03 | |
| 1 | 37 | −3.40 | |
| 0 | 25 | −5.97 | |
| 0 | 37 | −6.08 |
The mean supercoiling density was calculated from the densitometry trace in Figure 4B. The area under each peak except for the peak corresponding to nicked and relaxed circle was determined using ImageJ. The supercoiling value corresponding to reaching 50% of the total signal density in each lane was considered to be the mean supercoiling density.
We can quantify the net supercoil change attributable to centromere mutations. The most slowly migrating band in each lane corresponds to nicked circles and topoisomers with no net writhe (0). The second most slowly migrating band corresponds to one with a net writhe value of −1, and so on. In the CEN3mut+CEN6mut construct, which is expected to have ~9 total nucleosomes (Figure 2E), we observed at least 8 topological isomers. However, in the CEN3+CEN6 construct, which has two functional centromeres, we can reliably count only 5 topoisomers, ~4 less than observed for the same construct with mutated centromeres (Figure 4, Figure S3). It is not plausible that two functional centromeres remove 4 H3 nucleosomes from other regions of the minichromosome. It is also not plausible that mutating both centromeres results in a gain of 4 additional H3 nucleosomes, because a gain from 9 to 13 nucleosomes would exceed the maximum capacity of this 2-kb minichromosome for octamers. If two out of nine total nucleosomes are positively supercoiled, we expect net −5 supercoils rather than −9 supercoils predicted from 9 nucleosomes that are negatively supercoiled (Figure S3). Analysis of the densitomery trace of the gel reveals that the mean distribution of topoisomers in lane 1 and lane 5 is shifted by approximately 3 to 4 supercoils (Figure 4B WT panel; Table 2). The CEN3+CEN6mut minichromosome with one functional centromere is intermediate between the two as we expected, showing ~7 topoisomers, and the mean distribution is shifted by 2 to 3 supercoils from one with no functional centromeres. Therefore, we conclude that each functional centromeric nucleosome containing Cse4p induces one positive supercoil into the minichromosome, canceling one negative supercoil induced by a canonical H3-containing nucleosome.
A defective centromeric nucleosome, but not a defective kinetochore causes progressive loss of positive supercoils
In order to show that the differences in topological states of these minichromosome constructs depend on the presence of Cse4p nucleosomes, minichromosomes were isolated from ndc10-1 mutant cells at either the permissive (25°C) or restrictive (37°C) temperature. As discussed above, Cse4p fails to localize to centromeres when the ndc10-1 mutant is allowed to pass through S-phase at the restrictive temperature (Ortiz et al., 1999; Pearson et al., 2003). As expected, all three constructs behave similarly to their respective constructs in the wildtype background at the permissive temperature; however, all three constructs become virtually indistinguishable at the restrictive temperature, reaching the most negatively supercoiled states of the CEN3mut+CEN6mut minichromosome with no functional centromere (Figure 4 lanes 7–12; Table 2). This indicates that in the ndc10-1 mutant at the restrictive temperature, Cse4p nucleosomes that induce positive supercoils are replaced by H3 nucleosomes, which in turn induces negative supercoils into minichromosomes.
As ndc10-1 mutants at restrictive temperature disrupt both Cse4p nucleosomes and kinetochore functions in general, it is formally possible that kinetochore function, which is to attach the centromere to the mitotic spindle, can directly or indirectly alter the level of supercoiling by physically pulling on DNA (Gore et al., 2006). In this case, changes in linking number would occur with kinetochore loss, complicating the interpretation of our experiments. To rule out this possibility, we determined the topological states of minichromosomes isolated from cells that carry a temperature-sensitive mutation in the ndc80 gene, which encodes a component of the central kinetochore (Welburn and Cheeseman, 2008). This ndc80-1 mutation does not affect the localization of Cse4p; however, it will cause failure of the mitotic spindle to attach to centromeres. Topological analysis shows that the ndc80-1 mutation has a much smaller effect on the topological states of minichromosomes relative to ndc10-1 (Figure 4), indicating that microtubule attachment does not substantially change topological states of minichromosomes (see also Table 2). It has been reported that the ndc80-1 mutation genetically interacts with ndc10-1 (Wigge and Kilmartin, 2001); therefore, the small changes in the supercoiling states in a more negative direction in the ndc80-1 mutant strain relative to wildtype at the restrictive temperature might be attributed to a marginally increased loss of Cse4p from centromeres. Importantly, ndc80-1 mutant cells released from α-factor to the restrictive temperature arrest at mitosis, so that the observed shift in the topoisomer distribution occurs during mitosis, when kinetochores function. Therefore, positive supercoiling induced by Cse4p nucleosomes is a feature of functional kinetochores.
Only kinetochore proteins that come into contact with centromeric DNA are candidates for affecting DNA topology. Condensin from Xenopus has been shown to induce positive supercoils in vitro (Kimura and Hirano, 1997), although budding yeast condensin lacks this in vitro activity (Stray et al., 2005). We observed no changes in the distribution of topoisomers on CenH3 minichromosomes in a condensin ycg1 mutant (Figure S7), which indicates that condensin plays no role in inducing positive supercoils on these minichromosomes. Rather, our data indicate that either the Cse4p nucleosome or the CBF3 complex, which contains Ndc10, is responsible for the observed positive supercoils. Because of the small footprint of CBF3 on DNA, it is highly unlikely that this complex alone can change topology by one linking number. Therefore, we conclude that changes in the topological states of minichromosomes result from the presence or absence of Cse4p nucleosomes.
Discussion
We have shown that CenH3 nucleosomes induce positive supercoils, both when D. melanogaster CID is reconstituted into nucleosomes in vitro, and when S. cerevisiae Cse4p is assembled at functional minichromosome centromeres in vivo. This behavior is in stark contrast to canonical nucleosomes, in which the left-handed wrapping leads to induction of negative supercoils in topological assays. Our observations of positive supercoiling induced by CenH3 from eukaryotic taxa as different as animals and fungi can be explained by either of two general models: overtwisting with left-handed wrapping or right-handed wrapping.
In a covalently closed circle, overtwisting of DNA (positive ΔTw) causes compensatory negative writhe that is removed by topoisomerase, resulting in a net positive ΔLk after deproteination (Malcolm and Snounou, 1983). If CenH3 nucleosomes are left-handed octamers (Wr = −1), ΔTw would need to be +2 in order to result in a ΔLk of +1 (ΔLk = ΔTw + ΔW). Although the reported value of ΔWr for left-handed octamers varies (Bancaud et al., 2006; Prunell, 1998), we use the most conservative cited value of −1 to calculate the degree of overtwisting consistent with left-handed wrapping. The change required in the helical periodicity of DNA (Δh) to gain ΔTw = +2 and cancel one negative writhe induced by a left-handed nucleosome can be calculated as Δh = −h2xΔTw/N, where h = N/Tw, where N is the number of base pairs wrapped around the nucleosome. If we assume an octameric CenH3 nucleosome (N=150 bp), Δh equals −1.47 for ΔTw = +2. This corresponds to a helical periodicity of 9.03 bp/turn (whereas h = 10.5 bp/turn for B-DNA free in solution). The situation is even more extreme for CenH3 hemisomes, which wrap 80–120 bp of DNA, because the same amount of twist must be taken up by the shorter span of DNA (helical periodicity of 7.74–8.66 bp/turn). These estimated values for helical twist are conservative in that they assume that the extra twist is distributed over the whole nucleosome, including the DNA that wraps H2A/H2B dimers, whereas in the crystal structure of the H3 nucleosome core particle the twist of DNA wrapping H2A/H2B is similar to that in free solution (Luger et al., 1997). In addition, DNaseI digestion of CID chromatin assembled in vitro resulted in a normal helical periodicity estimate of ~10 bp/turn (Furuyama et al., 2006), and electron microscopy of CID chromatin revealed a beads-on-a-string appearance (Dalal et al., 2007b; Furuyama et al., 2006), suggesting entry/exit crossing. Thus, existing data are inconsistent with positive ΔTw being the reason for the observed positive supercoiling.
The implausibility of such strongly overtwisted DNA wrapping around a left-handed nucleosome leads us to conclude that positive supercoiling instead indicates a right-handed wrap. A right-handed nucleosome would satisfy the observed positive supercoiling of approximately one supercoil per CenH3 nucleosome without a significant change in B-DNA periodicity. Tetrameric archaeal nucleosomes also wrap DNA in a right-handed configuration, with a helical periodicity of 10–11bp/turn (Musgrave et al., 1991). Also, in the absence of H2A/H2B dimers, (H3/H4)2 tetramers are capable of spontaneously shifting between both left- and right-handed configurations (Hamiche et al., 1996), presumably without significant changes in helical twist.
Histone octamers capable of wrapping DNA into a right-handed configuration have never been observed. Because H3/H4 tetramers can wrap DNA in either direction, it is the creation of a left-handed ramp by addition of two H2A/H2B dimers that is incompatible with the right-handed structure (See Figure 5A–B). The crystal structure of the H3 nucleosome (H2A’-H2B’-H4’-H3’-H3-H4-H2B-H2A plus DNA) reveals that the N-terminal helix of H3, as well as the C-terminus of H4, contact the C-terminal docking domain of H2A’, which are essential interactions that hold the octamer together (Luger et al., 1997). In addition, the interaction between H2A and H2A’ within the octamer through their Loop 1 regions hold together the two gyres of the DNA superhelix (Luger et al., 1997). These interactions that hold the octamer together are expected to be disrupted in a right-handed nucleosome because they would face away from each other in the right-handed structure (Figure 5C and D); therefore, there is a strong structural basis for the absence of right-handed octameric nucleosomes in eukaryotes. Without altering the twist of DNA significantly, the only structures that yield ΔLk = +1 other than a right-handed octamer are right-handed hemisomes with right entry/exit crossing, and left-handed hemisomes with right entry/exit crossing. A single superhelical turn of DNA around a hemisome results in a closer physical distance between the entry/exit DNA compared to that in an octameric structure, which has an additional turn between the two entry/exit sites (compare Figure 5B to 5F). Therefore, it is structurally very difficult to make a left-handed hemisome with a right-handed crossing.
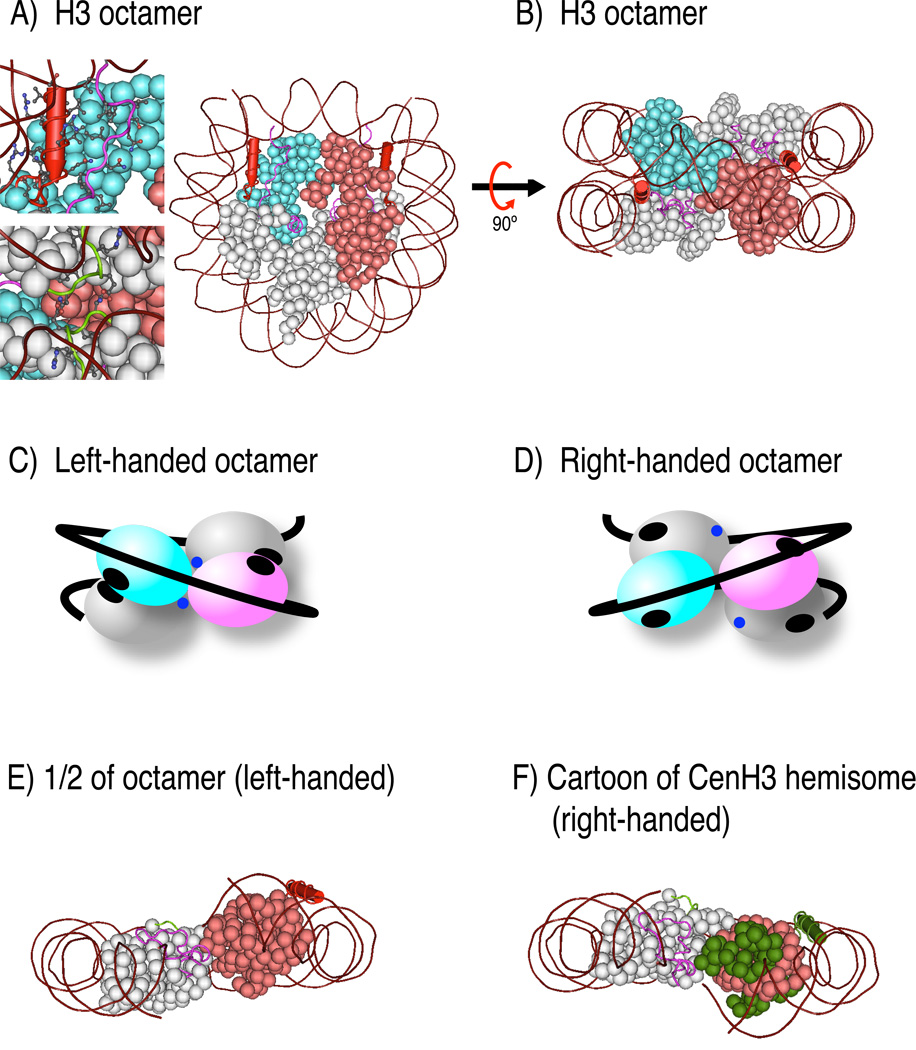
Figure 5. Structures and model of a right-handed hemisome.
A) Crystal structure of a nucleosome (Luger et al., 1997). One H3/H4 dimer is colored in light blue, the second H3/H4 dimer is colored in light red, both H2A/H2B dimers are shown in gray, and DNA is shown in brown wrapping around the histone octamer. A close-up view of the N-terminal helix of H3 (red stick) and H2A docking domain (magenta) with their side chains shown in the top inset. The interaction between two H2A molecules at the bottom of the octameric structure through their Loop 1 (green) is evident in the bottom inset. B) The structure in A) is rotated 90° to emphasize the spiral of 4 histone dimers wrapping DNA in the left-hand orientation. C) and D) Cartoons of left- and putative right-handed octamers. The two H3/H4 dimers were differentially colored in light blue and magenta, while the two H2A/H2B dimers are shown in gray. The DNA double helix is shown as a black line. Black ovals in C) depict the interaction between the N-terminal helix of H3 and the docking domain of H2A. Blue dots indicate H2A Loop1. In the right-handed octamer, which does not exist, the indicated interaction surfaces would face away from each other. E) One half of the left-handed structure shown in B), illustrating exposed surfaces in the absence of the other half. F) a cartoon of what a right-handed structure might look like. F) was produced using Abobe Photoshop, maintaining the approximate orientation of H4/H2B 4-helix bundle.
In budding yeast, various model of Cse4p nucleosomes have been suggested, including octamers (H2A/H2B/H4/Cse4p/Cse4p/H4/H2B/H2A) (Meluh et al., 1998), hemisomes (Cse4p/H4/H2B/H2A) (Dalal et al., 2007a) and nucleosomes containing the non-histone Scm3 protein substituting for H2A/H2B dimers (H4/Cse4p/Cse4p/H4)(Scm3)1–2 (Mizuguchi et al., 2007). Given our finding that Cse4p nucleosomes induce positive supercoils, it is unlikely that they can exist as octamers. Furthermore, the observation that Scm3 binds to the region of Cse4p required for the 4-helix bundle homodimerization interface of the octameric particle (Stoler et al., 2007) would a priori argue against a stable octameric particle. That leaves either Cse4p/H4/H2A/H2B hemisomes or Cse4p/H4/Scm3 particles as candidate yeast CenH3 nucleosomes. Both of these models are consistent with the localization of Cse4p to a small ~80bp CDEII region of CDE. It is attractive to suggest that right-handed hemisomes are conserved in all eukaryotes, because Cse4p can functionally replace human CENP-A (Wieland et al., 2004).
There are several structural implications of right-handed hemisomes. The strong H3/H3 4-helix bundle at the dyad axis and the weak H4/H2B 4-helix bundles linking the central tetramer to flanking dimers precludes formation of H3/H4/H2B/H2A hemisomes, and indeed no stable H3 hemisomes have been observed. Therefore, the existence of CenH3 hemisomes suggests that CenH3 induces structural alterations that stabilize the tetrameric particle. The crossing of entry/exit DNA in the CenH3 hemisome may be an important feature, because it can potentially stabilize the hemisome. In contrast, the entry/exit DNA of H3 octameric nucleosomes does not cross most of the time, but rather is occupied by a linker histone (Bancaud et al., 2006; Prunell, 1998). Consistent with this difference, the H1 linker histone is depleted from centromeric chromatin (Maresca et al., 2005), and the H5 linker histone is incapable of associating with human CENP-A nucleosomes in vitro (Conde e Silva et al., 2007). In addition, surfaces involved in contacts within left-handed octameric nucleosomes will be exposed in right-handed hemisomes, such as the C-terminal docking domain of H2A and the N-terminal helix of H3 (Figure 5A–B). A right-handed configuration also changes the relative position of these domains (Figure 5C–D). The combination of additional exposed surfaces and altered presentation of the same surfaces might provide essential interaction domains for kinetochore proteins to assemble functional centromeres.
Our finding that CenH3 nucleosomes are right-handed also might help explain why key residues involved in H3/H3 4-helix bundle formation are invariant in CenH3s, despite considerable divergence elsewhere in the core. This observation suggests that the CenH3 dimerization interface is occupied under at least some circumstances. We suggest that this interface has been retained to permit CenH3/H3 hybrid formation (Foltz et al., 2006), which would result in left/right core particles that should be unable to stably wrap DNA. Misincorporation of CenH3 outside of centromeres occurs under many circumstances (Blower and Karpen, 2001; Henikoff et al., 2000; Tomonaga et al., 2003; Van Hooser et al., 2001), yet is potentially catastrophic, causing dicentric formation, chromosome loss and dominant lethality (Heun et al., 2006; Tomonaga et al., 2003). By retaining the ability to dimerize with H3, misincorporated CenH3s would predominantly form structurally defective nucleosomes, thus helping to maintain the extraordinary fidelity of centromere maintenance.
At the boundary between CenH3 and H3 nucleosomal arrays, the change in the direction of DNA around histones from left-handed to right-handed might also have profound implications for maintaining functional centromeres. The uniform packaging of H3 nucleosomes in pericentric heterochromatin, induced in part by the uniform size of centromeric satellite repeats, is expected to be disturbed by the sudden change in the direction of DNA wrapping around CenH3. This would result in a higher-order structural transition from near-crystalline rigid heterochromatin to less densely packaged centromeric chromatin as implied by the unusually long linker DNA found in Drosophila centromeric chromatin (Dalal et al., 2007b). The octameric form of canonical H3 nucleosomes is believed to represent a critical evolutionary leap in being able to more densely package the genome, whereas tetrameric archael nucleosomes fail to condense into a comparable higher order packaging (Pereira et al., 1997; Sandman et al., 1998). Therefore, the presence of a CenH3 hemisome array that packages DNA in a right-handed orientation and resists octameric packaging would provide a singular location that remains decondensed during mitosis and accessible to binding by kinetochore proteins. The mutual incompatibility of nucleosome cores that wrap DNA in opposite directions suggests a novel mechanism for perpetual maintenance of the centromere within a chromosomal landscape that is dominated by conventional chromatin.
Experimental Procedures
In vitro chromatin assembly
In vitro chromatin assembly reactions were performed as previously described (Furuyama et al., 2006). Some reactions were done using plasmids containing randomly cloned ~3kb Drosophila fragments (Supplemental Data). Topoisomers were resolved in agarose gels with or without 1µg/ml chloroquine.
Yeast genetic analysis
All yeast strains (Table S1) were cultured in a standard yeast synthetic media containing TRP dropout supplement (Sigma) and 2% glucose. Doubling times were determined as follows. Three independent transformants were selected at random and cultured to late log overnight. Cultures were diluted to OD600 = 0.2 and OD600 measurements were taken every hour. The data from early- to mid-log phase were fitted to an exponential curve, then the doubling time was calculated from the exponent of the best fit curve using Microsoft Excel. Copy numbers were detemined by Southern blot analysis, as described in the legend to Figure S6. Mitotic stability measurements were done essentially as previously described (Koshland et al., 1987) from three independent transformants.
Topological assays of minichromosomes
All strains were grown at 25°C to mid-log phase, then α-factor was added to 1µg/ml for strains in the wildtype SBY3 background and 10µg/ml for the others, depending on their genotypes at the bar1 locus. Cultures were incubated for 1.5–2 hr to arrest at the G1/S phase boundary, spun and cell pellets washed to remove α-factor, then resuspended in fresh media. Half of each culture was incubated at 25°C while the other half was incubated at 37°C for 2 hr. Total DNA was purified by standard methods, then electrophoresed on 1.5% Tris-Borate-EDTA gels containing 0.3µg/ml chloroquine. The value of 0.3 µg/ml was empirically determined for this minichromosome to obtain maximal resolution of all topoisomers. Two dimensional gel electrophoresis was also performed; however, we found that one dimensional gel was sufficient due to the relatively small number of topoisomers to be resolved (Figure S4). Approximately 5–10 fold less DNA was used for strains carrying the CEN3mut-CEN6mut construct because of its high copy number. Southern blotting was performed at 57°C using a radiolabeled oligonucleotide (ACTAGCAATTGTGAGCGGATAACAATT, which is present in multiple tandem copies on the minichromosomes). Blots were autoradiographed using X-ray film or scanned using a Typhoon PhosphorImager, then analyzed using ImageJ software to obtain densitometric traces of each lane and areas under each peak. The uppermost band in each lane, which includes topoisomers with zero net writhe in the chloroquine gel and nicked circles, was omitted from calculation because of varying amounts of nicked species in different DNA preparations; therefore, a small overestimation of mean negative supercoiling might have resulted, especially for samples with less negative mean supercoiling values.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kerry Bloom for describing previous work that encouraged our yeast minichromosome study, Sue Biggins for helping us with the yeast experiments, Yamini Dalal for discussions regarding positive supercoiling, and Sue Biggins, Bungo Akiyoshi and Toshi Tsukiyama for yeast plasmids and strains. We thank Sue Biggins, Yamini Dalal, Roger Deal, Paul Talbert, Toshi Tsukiyama and Danielle Vermaak for critical readings of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alilat M, Sivolob A, Revet B, Prunell A. Nucleosome dynamics. Protein and DNA contributions in the chiral transition of the tetrasome, the histone (H3-H4)2 tetramer-DNA particle. J Mol Biol. 1999;291:815–841. doi: 10.1006/jmbi.1999.2988. [DOI] [PubMed] [Google Scholar]
- Bancaud A, Conde e Silva N, Barbi M, Wagner G, Allemand JF, Mozziconacci J, Lavelle C, Croquette v, Victor JM, Prunell A, Viovy JL. Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat Struct Mol Biol. 2006;13:444–450. doi: 10.1038/nsmb1087. [DOI] [PubMed] [Google Scholar]
- Bates AD, Maxwell A. DNA Topology. Oxford: Oxford University Press; 2005. [Google Scholar]
- Bloom K, Amaya E, Yeh E. Molecular Biology of the cytoskeleton. Cold Spring Harbor: Cold Spring Harbor Laboratories; 1984. Centromeric DNA structure in yeast chromatin; pp. 175–184. [Google Scholar]
- Bloom KS, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Conde e Silva N, Black BE, Sivolob A, Filipski J, Cleveland DW, Prunell A. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- Dalal Y, Furuyama T, Vermaak D, Henikoff S. Structure, dynamics, and evolution of centromeric nucleosomes. Proc Natl Acad Sci U S A. 2007a;104:15974–15981. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007b;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci U S A. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Dalal Y, Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc Natl Acad Sci U S A. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore J, Bryant Z, Nollmann M, Le MU, Cozzarelli NR, Bustamante C. DNA overwinds when stretched. Nature. 2006;442:836–839. doi: 10.1038/nature04974. [DOI] [PubMed] [Google Scholar]
- Hamiche A, Carot v, Alilat M, De Lucia F, O'Donohue MF, Revet B, Prunell A. Interaction of the histone (H3–H4)2 tetramer of the nucleosome with positively supercoiled DNA minicircles: Potential flipping of the protein from a left- to a right-handed superhelical form. Proc Natl Acad Sci U S A. 1996;93:7588–7593. doi: 10.1073/pnas.93.15.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Platero JS, van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci U S A. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A, Bloom K. Genetic manipulation of centromere function. Mol Cell Biol. 1987;7:2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehn B, Niedenthal R, Hegemann JH. In vivo analysis of the Saccharomyces cerevisiae centromere CDEIII sequence: requirements for mitotic chromosome segregation. Mol Cell Biol. 1991;11:5212–5221. doi: 10.1128/mcb.11.10.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- Koshland D, Rutledge L, Fitzgerald-Hayes M, Hartwell LH. A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell. 1987;48:801–812. doi: 10.1016/0092-8674(87)90077-8. [DOI] [PubMed] [Google Scholar]
- Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci U S A. 2006;103:4186–4191. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Malcolm AD, Snounou G. Netropsin increases the linking number of DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):323–326. doi: 10.1101/sqb.1983.047.01.037. [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nat Struct Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- Marc F, Sandman K, Lurz R, Reeve JN. Archaeal histone tetramerization determines DNA affinity and the direction of DNA supercoiling. J Biol Chem. 2002;277:30879–30886. doi: 10.1074/jbc.M203674200. [DOI] [PubMed] [Google Scholar]
- Maresca TJ, Freedman BS, Heald R. Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J Cell Biol. 2005;169:859–869. doi: 10.1083/jcb.200503031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh AD, Tytell JD, Sorger PK. Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Musgrave D, Forterre P, Slesarev A. Negative constrained DNA supercoiling in archaeal nucleosomes. Mol Microbiol. 2000;35:341–349. doi: 10.1046/j.1365-2958.2000.01689.x. [DOI] [PubMed] [Google Scholar]
- Musgrave DR, Sandman KM, Reeve JN. DNA binding by the archaeal histone HMf results in positive supercoiling. Proc Natl Acad Sci U S A. 1991;88:10397–10401. doi: 10.1073/pnas.88.23.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RG, Fangman WL. Nucleosome organization of the yeast 2-micrometer DNA plasmid: a eukaryotic minichromosome. Proc Natl Acad Sci U S A. 1979;76:6515–6519. doi: 10.1073/pnas.76.12.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz J, Stemmann O, Rank S, Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O'Day K, Trong HL, Charbonneau H, Margolis RL. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci U S A. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Zarzar TR, Salmon ED, Bloom K. Yeast kinetochores do not stabilize Stu2p-dependent spindle microtubule dynamics. Mol Biol Cell. 2003;14:4181–4195. doi: 10.1091/mbc.E03-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira SL, Grayling RA, Lurz R, Reeve JN. Archaeal nucleosomes. Proc Natl Acad Sci U S A. 1997;94:12633–12637. doi: 10.1073/pnas.94.23.12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizzi C, Clarke L. The chromatin structure of centromeres from fission yeast: differentiation of the central core that correlates with function. J Cell Biol. 1991;112:191–201. doi: 10.1083/jcb.112.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A. A topological approach to nucleosome structure and dynamics: the linking number paradox and other issues. Biophys J. 1998;74:2531–2544. doi: 10.1016/S0006-3495(98)77961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick MA, Westmoreland J, Bloom K. Heterogeneity and maintenance of centromere plasmid copy number in Saccharomyces cerevisiae. Chromosoma. 1990;99:281–288. doi: 10.1007/BF01731704. [DOI] [PubMed] [Google Scholar]
- Saavedra RA, Huberman JA. Both DNA topoisomerases I and II relax 2 micron plasmid DNA in living yeast cells. Cell. 1986;45:65–70. doi: 10.1016/0092-8674(86)90538-6. [DOI] [PubMed] [Google Scholar]
- Sandman K, Pereira SL, Reeve JN. Diversity of prokaryotic chromosomal proteins and the origin of the nucleosome. Cell Mol Life Sci. 1998;54:1350–1364. doi: 10.1007/s000180050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci U S A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stray JE, Crisona NJ, Belotserkovskii BP, Lindsley JE, Cozzarelli NR. The Saccharomyces cerevisiae Smc2/4 condensin compacts DNA into (+) chiral structures without net supercoiling. J Biol Chem. 2005;280:34723–34734. doi: 10.1074/jbc.M506589200. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F, Bergman LW, Simpson RT. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol. 1984;177:715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–3516. [PubMed] [Google Scholar]
- van Holde KE, Lohr DE, Robert C. What happens to nucleosomes during transcription? J Biol Chem. 1992;267:2837–2840. [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- Welburn JP, Cheeseman IM. Toward a molecular structure of the eukaryotic kinetochore. Dev Cell. 2008;15:645–655. doi: 10.1016/j.devcel.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P. Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:6620–6630. doi: 10.1128/MCB.24.15.6620-6630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kilmartin JV. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol. 2001;152:349–360. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.