Abstract
Several studies suggest that highly skewed X chromosome inactivation (HSXI) is associated with recurrent spontaneous abortion. We hypothesized that this association reflects an increased rate of trisomic conceptions due to anomalies on the X chromosome that lead both to HSXI and to a diminished oocyte pool. We compared the distribution of X chromosome inactivation (XCI) skewing percentages (range: 50%–100%) among women with spontaneous abortions in four karyotype groups—trisomy (n = 154), chromosomally normal male (n = 43), chromosomally normal female (n = 38), nontrisomic chromosomally abnormal (n = 61)—to the distribution for age-matched controls with chromosomally normal births (n = 388). In secondary analyses, we subdivided the nontrisomic chromosomally abnormal group, divided trisomies by chromosome, and classified women by reproductive history. Our data support neither an association of HSXI with all trisomies nor an association of HSXI with chromosomally normal male spontaneous abortions. We also find no association between HSXI and recurrent abortion (n = 45).
Introduction
X chromosome inactivation (XCI), the process by which one of the two X chromosomes of a female embryo undergoes transcriptional silencing, occurs early in embryogenesis. This process occurs in all cells except the female germ cell; both X chromosomes are active in the oocyte. The inactive X chromosome undergoes histone deacetylation and demethylation, followed by DNA methylation, which is inherited by all descendant cells. In theory, 50% of cells will contain an active X chromosome of maternal origin, 50% one of paternal origin. However, because the number of progenitor cells for hematopoietic stem cells is small (estimates are 8–16),1–4 highly skewed XCI (HSXI) (usually defined as 90% or higher) may occur by chance. The probability (based on a simple binomial model) that a woman will, by chance alone, demonstrate HSXI in a blood sample falls between 0.01% and 2.1%, depending on the number of progenitor cells.
Rarely, familial skewing involves mutations in XIST or other loci that affect initial XCI.5 A second source of HSXI is initial inactivation that occurs in an unusually small pool of cells. This phenomenon is thought to explain the association between monozygotic twinning and manifesting carriers of X-linked recessive mutations,6,7 as well as the high rate of HSXI in fetuses with confined placental mosaicism and trisomy rescue.8
Most often, HSXI is thought to result from selection in a population of cells in which inactivation was initially random;9 the degree of selection can vary among tissue types.10–13 Several lines of evidence support this hypothesis: (1) the proportion of cells with HSXI is greater in women of reproductive age than in newborns1,10 and, among adults, greater after age 60 than during the reproductive years;13,14 (2) heterozygous carriers of X-linked diseases sometimes exhibit preferential inactivation of the X chromosome with the disease allele (e.g., Wengler et al.,15 Devriendt et al.16); (3) HSXI is increased in women with structurally abnormal X chromosomes (deletions or translocations) in a manner that preserves the normal X chromosome (or autosomal) dosage.17,18 Pegoraro et al.19 described a striking example: a family with many apparently healthy women with 100% skewing, ascertained through the presence of manifesting carriers of an X-linked condition. The authors identified an Xq microdeletion and inversion in all family members with skewing.20 The only phenotypic consequence identified was increased risk of miscarriage.
This family stimulated the hypothesis that HSXI might be increased among women with recurrent spontaneous abortion (RSA). Several subsequent reports confirmed the prediction. Thus, early reports from Pittsburgh,21–23 British Columbia,24,25 and Japan26 show odds ratios ranging from 1.83 to infinity26 (our computations) for HSXI (defined as ≥ 90%) in relation to RSA. Although the highest estimates23,26 probably reflect confounding by maternal age, most other estimates do not. All reports compare cases with recurrent loss, variously defined, to controls; most control groups comprise women with at least one birth and no pregnancy loss. Inferences are limited, however, by methodological concerns, such as inclusion of previously described cases in sequential reports and selection of controls from a population different than the population in which cases were identified.22–25,27 In series in which controls derive from different sources and time periods than cases, technical variables in the assay can contribute to case-control differences. Six later reports,27–32 including one from the Pittsburgh team,27 which includes cases described in earlier reports, provide inconsistent results, with odds ratios ranging from 0.230 to 13.2.28 The literature as a whole is difficult to interpret, given the probable publication bias for positive associations.
Most previous studies lack information on the karyotype of the abortus. Women with recurrent abortions have an increased risk of repeat chromosomally normal losses. This observation, however, is most apparent in women younger than 35 years. Among older women, because of the strong association of age with trisomy, chromosomal anomalies occur in at least 50% of losses, even among women with two or more previous losses.33,34 Given that maternal age at conception has been increasing over time in the United States,35 we expect that, in many settings, trisomies occur in a high proportion of losses to recurrent aborters. For example, in our sample, the index loss is trisomic in 44% of the 50 women with recurrent losses (mean age 35.6 years). Thus, a phenomenon due to an increased rate of trisomic conceptions could masquerade as an effect seen in recurrent aborters.
Lanasa et al.23 argued that HSXI is likely to be a marker of an X chromosome microdeletion or mutation that leads to loss of male conceptions carrying the mutant X chromosome. However, their study had no information on the karyotype of the losses. In current populations, karyotypically normal male conceptions constitute only 15%–20% of spontaneous abortions.36,37 It is therefore unlikely that losses of male conceptions could be responsible for the strong associations with skewing reported in early studies. For example, in order to double the rate of HSXI among all spontaneous abortions, the frequency of HSXI would need to be six-fold higher among normal male losses than among all other pregnancies. This calculation makes the assumptions that (1) HSXI occurs in 5% of women, (2) 15% of clinically recognized pregnancies abort spontaneously, (3) 20% of pregnancy losses have a normal male karyotype, and (4) all chromosomally normal male losses are due to an X chromosome abnormality (unlikely, but the assumption most favorable to the hypothesis).
On the other hand, trisomies account for one-third to one-half of all pregnancy losses. If a calculation similar to the one above is used, the frequency of HSXI would need to be 3.5-fold higher among women with trisomic losses than among all other pregnancies in order to double the rate of HSXI among spontaneous abortions. Our study has the power to distinguish these effect sizes.
Oocyte maintenance in the ovary depends upon the presence of two normal X chromosomes. In female germ cells, both X chromosomes remain transcriptionally active. The absence of all or part of an X chromosome is associated with primary amenorrhea or premature ovarian failure. Associations may reflect either fewer primordial follicles or accelerated atresia. A likely explanation is that at least several discrete genes must be expressed from both X chromosomes to ensure normal germ cell and/or follicle survival (reviewed by Simpson and Rajkovic38 and by Laml et al.39). HSXI may thus be an indicator of X chromosome mutations or chromosome abnormalities that could lead to a decreased oocyte pool. Two studies40,41 support this idea, one study41 showing an association between HSXI and premature ovarian failure.
We hypothesized that the maternal age association with trisomy reflects an association with the size of the oocyte pool, leading to increased trisomy risk among women who have diminished pools for their chronologic age.42,43 If HSXI is associated with a smaller oocyte pool (see above), the association of HSXI with repeat spontaneous abortions could be due to an increase in the frequency of trisomic conceptions. Preliminary data from British Columbia24,44 reported such an association. We tested the hypothesis in a case-control study by examining whether HSXI is associated with trisomic spontaneous abortion. Secondarily, we examined associations between HSXI and both chromosomally normal male spontaneous abortion and recurrent pregnancy loss.
Subjects and Methods
Details of selection of cases and controls, demographics, and statistical methods are given in the Appendix. Fieldwork took place from February 2003 to January 2007. The institutional review boards at our university and the study hospital approved the study. All subjects gave informed consent.
Selection of Cases and Controls
Case subjects were women aged 18 or older with singleton spontaneous abortions of developmental age < 18 wks, who gave permission for karyotyping of their products of conception. The specimens were collected from the pathology laboratory of a large suburban hospital in New Jersey, USA. Karyotype results were obtained from 498/517 (96%) specimens in which culture was attempted, by either chromosome analysis or FISH (see Appendix). Of the 498 women, 354 (71%) completed the protocol, which included two telephone interviews concerning demographics, obstetric history, medical history, and common exposures and a visit to the hospital for a blood draw and an updated interview. Final analyses exclude 30 women (see Appendix). The analytical sample consists of 169 women with aneuploid losses (referred to as “trisomic” but also including four hypertriploids, one hypertetraploid, and six autosomal monosomies), 46 women with normal male losses, 43 women with normal female losses, and 66 women with nontrisomic chromosomally abnormal losses. For convenience, throughout the rest of the paper we refer to women on the basis of the karyotype of their abortus (“trisomy cases” or “trisomies” rather than “women with trisomic spontaneous abortions”).
For each case individual who completed the study, we selected an age-matched control with a recent normal live birth in the same hospital and with no known chromosomally abnormal previous pregnancy. Of 678 women selected as controls, 491 (72%) completed the protocol. To ensure comparability of measures for cases and controls, we excluded 64 controls whose XCI skewing percentage was measured in an external laboratory. The analytic sample thus includes 427 controls.
Follow-up of Women with XCI Skewing
We asked all women with XCI skewing percentage ≥ 85% and a subset of women with XCI 50% to <75%, matched for karyotype group and date of first blood draw, to provide a second blood sample for cytogenetic analysis and buccal swabs from both left and right cheeks for measurement of the XCI skewing percentage in this tissue. Of the 90 women who provided buccal swabs (45 with blood XCI ≥ 85%, 45 with blood XCI 50% to <75%), 86 had sufficient DNA to carry out the HUMARA assay. Three women were homozygous on buccal smear analysis, and three women were excluded because the karyotype of the abortus was uncertain (46,XX without confirmation from FISH). Thus, the buccal smear analyses comprise 80 women.
The XCI Assay
Skewing of XCI was measured with the HUMARA assay, which takes advantage of the differential methylation of a CpG site close to a highly polymorphic CAG repeat in the first exon of the X-linked androgen receptor (AR [MIM 3137000]) gene. Approximately 90% of women have distinguishable alleles. Sham digestion by RsaI allows sizing of the two alleles after PCR; digestion by RsaI plus HpaII allows distinction between the methylated and the nonmethylated allele. We used a slight modification of the method described in Hatakeyama et al.14 to assess the XCI skewing percentage at the AR locus.
Among 751 samples analyzed, 91% were judged heterozygous at the AR locus. Reliability of the assay (see Appendix) was measured in a variety of ways, including comparisons with an external laboratory and measurements over time within the study laboratory. We used the intraclass correlation coefficient (ICC) because it provides a more robust measure of reliability and agreement than the Pearson product-moment correlation (r). The ICCs ranged from a low of 0.80 (when we remade PCR products and used an external laboratory) to a high of 0.96 (when the same PCR products were run within 35 days in the study laboratory). Thus, our reliability on the same sample ranges from substantial to excellent.
These ICCs exclude samples judged to be homozygous by one or both laboratories. Among ten samples judged to be homozygous by either laboratory, there was agreement on seven. All three disagreements concerned questionable heterozygosity for alleles differing by only three base pairs. Because the XCI skewing percentage may be underestimated when alleles differ by only one repeat, we repeated all categorical statistical analyses, excluding the 93 (14%) samples in which allele sizes differed by only one repeat. Odds ratios were unchanged for the first three significant digits.
Statistical Analyses
We carried out two complementary analyses to estimate associations of the XCI skewing percentage with each karyotype group—trisomy, chromosomally male, chromosomally normal female, nontrisomic chromosomally normal—in comparison to those of controls. In the first, we analyzed the folded XCI skewing percentage as a categorical variable (50 to <60, 60 to <70, 70 to <80, 80 to <85, ≥ 85). We defined highly skewed HSXI as ≥ 85% (6.1% of 684 heterozygotes), because the small proportion (2.3%) of women with XCI ≥ 90% would have limited statistical power. We used conditional logistic regression45,46 to test the null hypothesis that at any maternal age there is no difference in the XCI skewing percentage between cases and controls. The analysis was adjusted by stratification for age at blood draw in single years. We also report the results of the primary analysis, using ≥ 90% as the upper boundary to facilitate comparison with other studies.
In the second analysis, we used a parametric model to estimate associations with the folded XCI skewing percentage as a continuous variable following a symmetric beta distribution. This analysis permitted us to test the null hypothesis that the mean of the folded XCI skewing percentage is the same for each case group and controls. We used maximum likelihood to estimate regression coefficients. To aid interpretation, we report age-adjusted mean folded XCI skewing percentages based on the parameters of the regression model. Figure 1 uses data from controls to illustrate that the folded beta distribution fits the observations well (coalescing the two highest categories, chi-square goodness of fit = 5.69, df = 7, p = 0.58).
Figure 1.
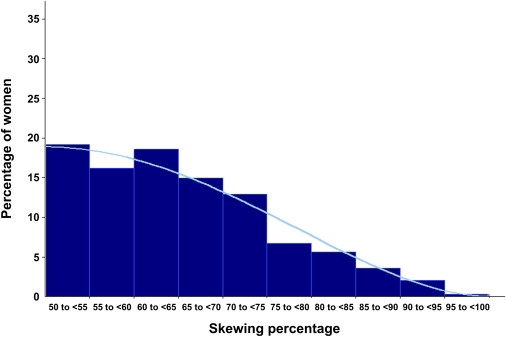
X Chromosome Inactivation Skewing Percentage among Controls with Age-Adjusted Fitted Beta Curve
We also carried out three secondary analyses: (1) dividing the nontrisomic chromosomally abnormal types into three groups: monosomy X, triploidy, other; (2) dividing trisomic groups into three subgroups: acrocentric, trisomy 16, other nonacrocentric; and (3) classifying women by reproductive history.
Finally, we present data bearing on the consistency of XCI measurements (1) over time for the 17 women who entered the study twice and (2) between blood and buccal mucosa for the 80 women with informative assays on both tissues.
Results
Primary Analysis: Four Karyotype Groups Versus Controls
The proportion of women with XCI ≥ 85% is 5.9% for controls and 5.8% for trisomies, the primary case group (Table 1). The odds of XCI ≥ 85% do not differ between any of the case groups and the controls (Table 2). Comparing trisomy cases and controls, the age-adjusted odds ratio for XCI ≥ 85% (versus 50% to <60%) is 1.2 (95% CI 0.5–2.8.). XCI ≥ 90% occurs in 2.0% of heterozygous trisomy cases and 2.3% of heterozygous controls; the age-adjusted odds ratio is 1.3 (95% CI 0.3–5.3). For chromosomally normal male cases, the age-adjusted odds ratio for XCI ≥ 85% is 0.3 (95% CI 0.04–2.5) (Table 2). There are no chromosomally normal male cases with XCI ≥ 90%. The folded beta regression analysis shows no statistically significant differences in the age-adjusted mean XCI skewing percentage between these three case groups and controls (Table 2).
Table 1.
Percentage of XCI Skewing among Live Birth Controls and among Spontaneous Abortion Cases Classified by Karyotype of the Abortus
| Pregnancy Type | Total Number | Number Heterozygous | XCI Skewing Percentage Distribution |
||||
|---|---|---|---|---|---|---|---|
| 50 to <60 | 60 to <70 | 70 to <80 | 80 to <85 | 85+ | |||
| Controls | |||||||
| Live births | 427 | 388 | 35.3 | 33.5 | 19.6 | 5.7 | 5.9 |
| Case groups for primary analysis | |||||||
| Trisomy | 169 | 154 | 32.5 | 27.3 | 24.0 | 10.4 | 5.8 |
| Chromosomally normal male | 46 | 43 | 41.9 | 20.9 | 23.3 | 11.6 | 2.3 |
| Chromosomally normal female | 43 | 38 | 50.0 | 23.7 | 15.8 | 7.9 | 2.6 |
| Nontrisomic chromosomal abnormality | 66 | 61 | 31.1 | 23.0 | 26.2 | 6.6 | 13.1 |
| Case groups for secondary analysis dividing nontrisomic chromosomal abnormalities | |||||||
| Monosomy X | 19 | 18 | 22.2 | 22.2 | 27.8 | 5.6 | 22.2 |
| Triploid | 30 | 28 | 35.7 | 21.4 | 28.6 | 3.6 | 10.7 |
| Other nontrisomic chromosomal abnormality | 17 | 15 | 33.3 | 26.7 | 20.0 | 13.3 | 6.7 |
| Case groups for secondary analysis dividing trisomies | |||||||
| Acrocentric trisomy | 78 | 70 | 30.0 | 32.9 | 24.3 | 10.0 | 2.9 |
| Trisomy 16 | 41 | 38 | 50.0 | 26.3 | 10.5 | 7.9 | 5.3 |
| Other nonacrocentric trisomy | 38 | 34 | 20.6 | 17.6 | 38.2 | 8.8 | 14.7 |
Table 2.
Percentage of XCI Skewing among Spontaneous Abortions, Classified by Karyotype, and Controls: Unadjusted and Age-Adjusted Odds Ratios for Folded XCI Skewing Percentages and Mean Folded XCI Skewing Percentages
| XCI Percentage | Unadjusted OR | Age-Adjusted ORa | 95% CI | Age-Adjusted Mean XCI Skewing Percentageb |
|---|---|---|---|---|
| Trisomy | ||||
| 50 to <60 | 1.0 | 1.0 | NA | |
| 60 to <70 | 0.9 | 0.9 | 0.6–1.5 | |
| 70 to <80 | 1.3 | 1.2 | 0.7–2.1 | |
| 80 to <85 | 2.0 | 1.7 | 0.8–3.6 | |
| 85+ | 1.1 | 1.2 | 0.5–2.8 | |
| Mean | 66.8 | |||
| Chromosomally normal male | ||||
| 50 to <60 | 1.0 | 1.0 | NA | |
| 60 to <70 | 0.5 | 0.5 | 0.2–1.1 | |
| 70 to <80 | 1.0 | 0.9 | 0.4–2.1 | |
| 80 to <85 | 1.7 | 1.5 | 0.5–4.8 | |
| 85+ | 0.3 | 0.3 | 0.04–2.5 | |
| Mean | 65.4 | |||
| Chromosomally normal female | ||||
| 50 to <60 | 1.0 | 1.0 | NA | |
| 60 to <70 | 0.5 | 0.5 | 0.2–1.1 | |
| 70 to <80 | 0.6 | 0.7 | 0.3–1.9 | |
| 80 to <85 | 1.0 | 1.2 | 0.3–4.4 | |
| 85+ | 0.3 | 0.3 | 0.03–2.0 | |
| Mean | 63.5 | |||
| Nontrisomic chromosomal abnormality | ||||
| 50 to <60 | 1.0 | 1.0 | NA | |
| 60 to <70 | 0.8 | 0.6 | 0.3–1.3 | |
| 70 to <80 | 1.5 | 1.5 | 0.7–3.1 | |
| 80 to <85 | 1.3 | 1.2 | 0.4–4.1 | |
| 85+ | 2.5 | 2.1 | 0.8–5.6 | |
| Mean | 68.6c | |||
| Control | ||||
| Live birth | reference | reference | NA | 65.5 |
NA denotes “not applicable.”
Adjusted for age at blood draw (in single years) by stratification. The conditional logistic regression analysis treats as uninformative one trisomic and two nontrisomic chromosomally abnormal spontaneous abortions (i.e., there are no controls or other spontaneous abortions in the age stratum).
Adjusted linearly for age at blood draw. Obtained from a beta regression model for the XCI proportion (XCI percentage/100). The expected value of a folded beta random variable is I0.5(θ,θ + 1), in which Ix(a,b) is the incomplete beta function from 0 to x and θ is the (estimated) age-adjusted shape parameter.
p = 0.02 for nontrisomic chromosomal abnormality compared with controls.
Among women with nontrisomic chromosomally abnormal spontaneous abortions, 13.1% showed XCI ≥ 85%. The age-adjusted odds ratio for XCI in this group is 2.1 (95% CI 0.8–5.6). Analysis of the XCI skewing percentage as a continuous variable indicates a statistically significant increase in the XCI skewing percentage in this case group as compared with controls (p = 0.02) (Table 2).
Secondary Analyses of Karyotype Groups
Dividing Nontrisomic Chromosomally Abnormal Cases
The odds ratios relating XCI ≥ 85% to monosomy X (n = 18) and triploid (n = 28) cases (each compared with controls) were 4.0 and 2.0, respectively, neither differing significantly from unity (Table 1). Analysis of the XCI skewing percentage as a continuous variable shows increased mean XCI skewing percentages for monosomy X, triploid, and other chromosomally abnormal cases, although only the increase with monosomy X was significant (p = 0.03) (Table 3). The means of the three karyotype groups included among other chromosomal abnormalities are not significantly different from each other (p = 0.51).
Table 3.
Percentage of XCI Skewing among Spontaneous Abortions, Dividing Nontrisomic Chromosomal Abnormality Cases, Trisomy Cases, and Controls: Age-Adjusted Odds Ratio for Folded XCI Skewing Percentage ≥ 85 and Mean XCI Skewing Percentage
| Comparison | Number | Age-Adjusted OR XCI 85%+ versus XCI 50% to <60%a | 95% CI | Age-Adjusted Mean XCI Skewing Percentageb |
|---|---|---|---|---|
| Nontrisomic chromosomal abnormalityc | ||||
| Controls | 388 | reference | NA | 65.5 |
| Monosomy X | 18 | 4.0 | 0.9–18.4 | 70.7d |
| Triploidy | 28 | 2.0 | 0.5–8.2 | 67.8 |
| Other nontrisomic abnormality | 15 | 0.8 | 0.1–7.5 | 66.9 |
| Trisomye | ||||
| Controls | 388 | reference | NA | 65.9 |
| Acrocentric trisomy | 70 | 0.6 | 0.1–2.7 | 65.9 |
| Trisomy 16 | 38 | 0.6 | 0.1–3.0 | 65.0 |
| Other nonacrocentric trisomy | 34 | 4.1 | 1.1–15.2 | 69.6f |
NA denotes “not applicable.”
Adjusted for age at blood draw (in single years) by stratification. The age-adjusted odds ratios are obtained from a conditional logistic regression analysis that includes all categories of the XCI skewing percentage.
Adjusted linearly for age at blood draw (see footnote to Table 4).
The analysis includes trisomy, chromosomally normal male, and chromosomally normal female spontaneous abortions (not shown). The conditional logistic regression analysis treats as uninformative the following number of spontaneous abortions: one trisomy, one monosomy X, and one other nontrisomic chromosomal abnormality (i.e., there are no controls or other pregnancy losses in the age stratum).
p = 0.03 for monosomy X in comparison with controls. Mean XCI skewing percentages do not differ among monosomy X, triploidy, and other nontrisomic chromosomal abnormality (2 df, p = 0.51).
The analysis included the three trisomy types and controls. The conditional logistic regression analysis treats as uninformative one acrocentric trisomy loss and 14 controls (i.e., there are no controls or other pregnancy losses in the age stratum).
p = 0.04 for other nonacrocentric trisomy in comparison with controls. Mean XCI skewing percentages do not differ among the trisomy types (2 df, p = 0.12).
Trisomy Type
We repeated the analyses, classifying trisomy cases by chromosome (acrocentric, trisomy 16, other nonacrocentric). The XCI skewing percentage, whether defined categorically or continuously, was unrelated to acrocentric trisomy and trisomy 16 cases. The odds ratio relating XCI ≥ 85% to other nonacrocentric trisomy cases (versus controls) was 4.1 (95% CI 1.1–15.2) (Table 1). The age-adjusted mean XCI skewing percentage was higher for this case group than for controls (p = 0.04) (Table 3). The three trisomy groups did not differ from each other (p = 0.12).
Secondary Analyses Related to Reproductive History
Division of the sample on the basis of the outcome of the index pregnancy and reproductive history shows no association between XCI skewing percentage and recurrent abortion (Table 4). The odds ratio for XCI ≥ 85% in recurrent aborters (defined as cases with 2+ spontaneous abortions < 20 wks and at least two more losses than live births; n = 45) versus multiparae (defined as controls with no spontaneous abortion < 20 wks and 2+ live births; n = 169) was 0.8 (95% CI 0.2–4.0). The age-adjusted mean XCI skewing percentage did not differ between women with RSAs and multiparae. Twenty-six of the 45 recurrent aborters had 3+ losses (including the index spontaneous abortion); none had XCI ≥ 85%.
Table 4.
Percentage of XCI Skewing among Groups Defined by Outcome of the Index Pregnancy: and Reproductive History
| Group | Total Number | Number Heterozygous | XCI Skewing Percentage Distribution |
||||
|---|---|---|---|---|---|---|---|
| 50 to <60 | 60 to <70 | 70 to <80 | 80 to <85 | 85+ | |||
| Index LB: multipara, reference groupa | 188 | 169 | 36.7 | 33.1 | 17.8 | 5.9 | 6.5 |
| Index SA: recurrent aborterb | 50 | 45 | 33.3 | 22.2 | 33.3 | 6.7 | 4.4 |
| Index SA: sporadic aborter, 2+ lossesc | 60 | 56 | 41.1 | 19.6 | 21.4 | 12.5 | 5.4 |
| Index SA: sporadic aborter, 1 lossd | 132 | 121 | 39.7 | 23.1 | 22.3 | 10.7 | 4.1 |
| Index SA: nullipara, 1 losse | 82 | 74 | 27.0 | 33.8 | 20.3 | 6.8 | 12.2 |
| Index LB: sporadic aborter, 1 lossf | 78 | 70 | 34.3 | 34.3 | 21.4 | 2.9 | 7.1 |
| Index LB: primipara, 0 lossesg | 141 | 129 | 32.6 | 34.1 | 21.7 | 7.0 | 4.7 |
| Index LB: otherh | 20 | 20 | 45.0 | 30.0 | 15.0 | 5.0 | 5.0 |
“Loss” indicates spontaneous abortion before 20 wks gestational age, SA indicates spontaneous abortion before 18 wks developmental age, and LB denotes “live birth.”
Loss = 0, live birth > 1.
Loss ≥ 2, loss − live birth ≥ 2.
Loss ≥ 2, loss − live birth < 2.
Loss = 1, live birth ≥ 1.
Loss = 1, live birth = 0.
Loss = 1, live birth ≥ 1.
Loss = 0, live birth = 1.
Loss ≥ 2 and loss − live birth ≥ 2 (n = 3) or loss ≥ 2 and loss − live birth < 2 (n = 17).
Consistency over Time and between Tissues
For 17 women who entered the study for two pregnancies and had blood drawn on both occasions, on average 286 days apart (range 147–836 days, median 198 days), the ICC for the XCI skewing percentage was 0.69; there was no significant difference between mean XCI skewing percentage for first and second study entrances. Skewing was in favor of the same allele in 82% of repeat samples (Table 5).
Table 5.
Agreement of XCI Skewing Percentages and Direction of Skewing among Samples and Tissues
| Comparison | Number | ICCa | Skewing in Favor of Same Allele |
|
|---|---|---|---|---|
| All Samples | Samples with HSXIb | |||
| Buccal tissue versus blood | ||||
| Left buccal smear versus blood | 80 | 0.41 | 56/80 (70%) | 35/40 (88%) |
| Right buccal smear versus blood | 80 | 0.39 | 59/80 (74%) | 33/39 (85%) |
| Both left and right versus blood | 80 | NA | 48/80 (60%) | 31/39 (79%) c |
| Buccal smear | ||||
| Left buccal smear versus right buccal smear | 80 | 0.48 | 61/80 (76%) | 8/9 (89%) |
| Blood samples | ||||
| Same laboratory, same PCR products, run 2 to 35 days apart | 70 | 0.96 | 69/70 (99%) | 13/13 (100%) |
| Same laboratory, same PCR products, run 4 to 9 months apart | 30 | 0.87 | 29/30 (97%) | 10/10 (100%) |
| Different laboratories, different PCR products | 19 | 0.80 | 19/19 (100%) | 8/8 (100%) |
| Repeat blood samples | 17 | 0.69 | 14/17 (82%) | 1/1 (100%) |
ICC denotes “intraclass correlation coefficient.”
Skewing ≥ 85% in at least one sample.
Among 39 women with skewing ≥ 85% in blood.
For 80 women for whom we were able to measure the XCI skewing percentage in both blood and buccal mucosa (Figure 2), the ICC between the XCI skewing percentage in blood and in buccal mucosa of the left cheek was 0.41, between blood and buccal mucosa of the right cheek was 0.39, and between buccal mucosa of the left and the right cheek was 0.48. Of the 39 women with blood XCI ≥ 85%, XCI ≥ 85% was detected in both buccal samples for three women and in one buccal sample for five. Of the 41 women with blood XCI 50% to <75%, one showed XCI ≥ 85% in one buccal sample. The mean folded XCI skewing percentage was lower in both left and right buccal mucosa than in blood (p < 0.0001).
Figure 2.
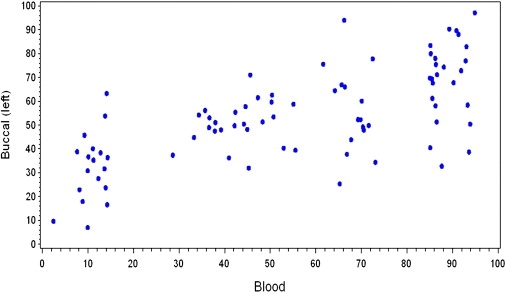
Unfolded XCI Skewing Percentage in Blood and Buccal Mucosa
Karyotypes of Women with a Second Blood Draw
Of the 90 women who had a second blood draw, 89 had chromosomally normal G-banded karyotypes at the 550 band level, and one had a balanced translocation; in the latter case, the abortus had an unbalanced translocation. Seven women showed > 5% X aneuploidy (four with 20 cells counted, three with 50 cells counted). X aneuploidy was unrelated to HSXI: it occurred in two of the 45 women with XCI ≥ 85% and five of the 45 women with XCI 50% to <75%.
Discussion
Association between HSXI and Trisomic Spontaneous Abortion
We hypothesized that an association between HSXI and RSA, if real, reflects an association with trisomic conception. Our proposed underlying mechanism was that abnormalities of the X chromosome are associated both with an increased rate of oocyte atresia and with skewed XCI in somatic cells as a result of selection. Several lines of evidence support the idea that the increasing rate of trisomy with increasing maternal age is related to the decreased size of the oocyte pool,43 although tests of the hypothesis yield conflicting results.37,47–49
Our data do not support our hypothesis, whether we define HSXI as ≥ 85% or ≥ 90% or analyze the XCI skewing percentage as a continuous variable. For XCI ≥ 85% in relation to trisomy, the age-adjusted odds ratio is 1.2 (95% CI 0.5–2.8), indicating that with 95% confidence we can rule out associations in excess of 2.8.
Our results contrast with two reports from British Columbia. The first report24 shows a 2.8 (95% CI 1.3–6.0, our computation) increase in the odds of XCI ≥ 90% among women with trisomic pregnancies compared with controls. The case group comprised women with RSAs, at least one of which was trisomic (n = 39), women with a trisomic spontaneous abortion but no RSA (n = 11), and women with a prenatal diagnosis of a mosaic trisomy of maternal origin (n = 53); the two control groups derive from several sources. The second report44 adds 16 trisomic cases, diagnosed prenatally or among losses of recurrent aborters, and mothers of children with maternal uniparental disomy for chromosome 15 (n = 21). Results for trisomy (but not uniparental disomy) are consistent with the first report. Combining all trisomy cases, the authors concluded that there is no shift in the distribution of XCI skewing percentage and that associations are primarily due to an excess of XCI ≥ 95%.
Our primary observation—that XCI skewing percentage is unrelated to trisomic pregnancy—is consistent with another study in which we were able to test another part of the hypothesis; namely, that HSXI is related to accelerated aging of the ovary. We analyzed XCI in a sample of women with recent pregnancies for whom we had measured levels of follicle-stimulating hormone and inhibin B, as well as the number of antral follicles, in one menstrual cycle. We found no association between HSXI and any of these measures of biologic aging of the ovary.50
Association between HSXI and Karyotypically Normal Male Spontaneous Abortion
Our data also show no hint of an association of XCI ≥ 85% with karyotypically normal male spontaneous abortion (age-adjusted OR 0.3; 95% CI 0.04–2.5); with 95% confidence, we can rule out associations in excess of 2.5. An association of abnormalities of the X chromosome with both XCI skewing percentage and chromosomal lethality may account for some cases of repeated male losses among women with recurrent abortions, but it is unlikely that such losses are common enough to produce detectable associations between HSXI and recurrent abortion.
Association between HSXI and Trisomy Divided by Chromosome Group
In the secondary analyses, we examined the relation of HSXI to predefined (e.g., Kline et al.37) classes of trisomy (Table 1 and Table 3). There is no association of XCI ≥ 85% with acrocentric trisomy or trisomy 16, although there is a significant association with nonacrocentric trisomies other than 16 (Table 3). Because these analyses were exploratory, the association with nonacrocentric trisomies, which is difficult to explain biologically, may be an artifact of multiple tests. However, the British Columbia series44 showed no association of HSXI with trisomies 13–15 or trisomy 16, but a positive association with other nonacrocentric trisomies and trisomies 21 and 22. Trisomies of different chromosomes vary in the patterns of increase with maternal age and the frequency or location of recombination sites,51 such that differences among chromosomal groups cannot be dismissed as implausible. Nonetheless, in light of the exploratory nature of both sets of analyses, observations related to specific classes of trisomy require confirmation.
Association between HSXI and Nontrisomic Chromosomally Abnormal Spontaneous Abortion
In the primary analysis, we detected an association of XCI skewing percentage, defined continuously, with nontrisomic chromosomally abnormal loss (p = 0.02). When we divided this karyotype group, the mean XCI skewing percentage was increased for monosomy X, triploid, and other chromosomally abnormal cases, although only the association with monosomy X was significant (p = 0.03). However, in light of the small number in each case group, the post hoc nature of this test, the multiple statistical tests performed, and biologic implausibility (each karyotype group has a different mode of origin), we think it likely that these observations represent a chance finding. We report them to alert others who may have relevant data with which to repeat the analysis.
Association between HSXI and Recurrent Spontaneous Abortion
Our study was not designed to test the association of HSXI with recurrent spontaneous abortion, but secondary analyses show no association. XCI ≥ 85% occurred in 4.4% of 45 recurrent aborters and 6.5% of 169 multiparae with live births only; age-adjusted mean XCI skewing percentages did not differ between the two groups. Recent studies in this area have been interpreted to cast doubt on a connection between HSXI and recurrent loss, mainly because differences between cases and controls are not statistically significant. Four studies27,29,31,32 show nonsignificant odds ratios of 2–3 relating HSXI (defined as ≥ 90%) to recurrent abortion, one28 shows a statistically significant 13-fold odds ratio, and one30 shows a nonsignificant inverse association. In summary, observations on a possible association of HSXI with recurrent abortion are inconclusive.
Strengths and Limitations
Our study has several strengths. First, we selected controls from the same population as cases over the same time period. Second, we measured the XCI skewing percentage blind to case-control and karyotype status. These design features eliminate the potential for bias in XCI measures due to technical or temporal factors. Moreover, selective participation is impossible given that XCI status was unknown to the women. Third, our study is the first to obtain karyotypes from a consecutive series of spontaneous abortions, providing greater generalizability than studies that draw on occasional karyotypes of losses to recurrent aborters. Fourth, the reliability of our assay ranged from substantial (ICC = 0.80), when we remade the PCR products, to excellent (ICCs 0.87–0.96), when we reused the same PCR products.
One potential limitation of our study is that for the categorical analysis we defined HSXI as ≥ 85% rather than as ≥ 90%, which is the more common cutpoint. We chose 85% because the proportion (2.3%) of women with XCI ≥ 90% limited statistical power. If the higher cutpoint identifies a group with a high proportion of X chromosome abnormalities, our ≥ 85% cutpoint might dilute an association between skewed XCI and trisomy. On the other hand, if X chromosome abnormalities shift the distribution of XCI skewing percentages toward higher ratios, the continuous analysis is most appropriate to our research question. Although the proportion with XCI ≥ 90% is lower in our control sample than in many early studies, it agrees with observations from later studies (e.g., Amos-Landgraf et al.,1 Hogge et al.,27 Bolduc et al.10). The wide variation in the proportion with XCI ≥ 90% reported among controls in the literature could easily be due to sampling variation in the studies with 100 women or fewer or to technical issues related to the HUMARA assay.1,13
Biologic Significance of HSXI in Blood
A difficulty with all studies of the XCI skewing percentage is how to interpret its biologic significance. An underlying assumption of our hypothesis is that HSXI is a good indicator of the presence of X chromosome abnormalities. None of the women in our sample with HSXI had abnormalities visible with standard G-banded karyotypes. It is not known, however, how often HSXI is an indicator of gene mutations or copy number changes below the level detectable with banded karyotypes. We are currently using high-resolution microarray analysis to examine whether or not HSXI is associated with copy number variation on the X chromosome.
A second difficulty relates to whether a single measure of the XCI skewing percentage (usually from a blood sample) is a valid measure of the true skewing percentage in a woman or, at a minimum, correctly identifies women with HSXI. Apart from the reproducibility of the HUMARA assay itself, which is good to excellent in our study, three other observations bear on this issue: (1) The ICC for measures of the XCI skewing percentage in blood samples taken from the same women at different times is lower than the ICC for repeat measurements on the same blood sample (Table 5). (2) The ICC for measures in buccal tissue versus blood is lower than the ICC for repeat measures in blood samples taken at different times. (3) The ICC for measures in left and right buccal smears is lower than the ICC for repeat measures in blood samples taken at different times.
Our data indicate that primary HSXI is rare: of 684 women with informative XCI skewing percentages, only the three (0.4%) who showed HSXI (≥85%) in all three samples are candidates. Even in the absence of HSXI in all tissues, however, samples taken from different tissues or at different times show a tendency for skewing toward the same allele, especially when HSXI is present in at least one sample (Table 5). This observation is compatible with the idea that HSXI is an indicator of genetic differences between the two X chromosomes that influence selection toward inactivation of the same chromosome—an inference consistent with the observation from studies of twin pairs,52–54 which also imply a large genetic component.
Our observations suggest that it is naive to think that the XCI skewing percentage in blood DNA measures a parameter that was set during fetal life and thus pertains to the entire individual. On the contrary, it appears that most women have differing degrees of XCI skewing in different tissues and even within the same tissue at different times. This observation suggests that when seeking to identify associations between XCI skewing (usually HSXI) and specific phenotypes, it is important to consider the possibility that an XCI skewing measure in blood may not be a reliable predictor of skewing in the tissues of interest.
Summary
We hypothesized that an association of HSXI with recurrent loss, if real, reflects an association with trisomic conception. Hence, we tested associations with trisomic losses irrespective of reproductive history. Our study does not support this hypothesis, nor does it support the hypothesis that the XCI skewing percentage is related to chromosomally normal male loss or a history of recurrent abortion.
Appendix: Detailed Subjects and Methods
Protocol
From February 25, 2003, to November 18, 2005, we identified women age 18 or older with singleton spontaneous abortions (developmental age less than 18 wks) whose products of conception were submitted to the Pathology Department of a hospital in NJ, USA. We asked permission to karyotype the abortus. If a woman's abortus was successfully karyotyped, we asked her to (1) complete a short telephone interview so that we could determine whether or not she was eligible for hormone measures (e.g., not taking a hormonal contraceptive) in addition to measures of XCI; (2) complete a more extensive telephone interview regarding demographic characteristics, obstetric and medical histories, and common exposures; and (3) make one visit to the hospital for a blood draw and a brief update interview about recent exposures. Women who were eligible for hormone measures provided samples timed to their menstrual periods. The original study goals did not include hormone measures; the samples have not been analyzed and are not relevant to this paper.
For each case individual who completed the study, we selected an age-matched control individual with a recent chromosomally normal live birth ≥ 1800 g, without a major anatomic malformation. Eligible controls had no pregnancy loss after the index pregnancy and no known prior chromosomally abnormal pregnancy. Candidates were women who delivered at the study hospital 6–12 months before the date of selection. They were selected from a roster of women who delivered between April 1, 2003, and May 31, 2006, and consented to be contacted about the research. Hospital staff asked 6505 women for permission to list them as candidates, and 5346 (82%) agreed.
Controls were matched to cases for projected age (±6 months) at the blood draw. Our computerized selection procedure was designed to alternate between selecting a control who was younger and a control who was older than the case. If a control did not complete the study, we replaced her, in order to obtain a comparison group as similar as possible in age to cases who completed the study. Because we view all controls of the same age as interchangeable, we used this procedure to ensure comparability in the age distributions, but we did not maintain the matches in the analysis. Rather, we controlled for age linearly or by stratification. If a case was eligible for hormone measures but her control was not, we used the same procedure to select and enroll a second control who was eligible for hormone measures.
Control recruitment lagged behind case recruitment because (1) we required that a case complete the protocol before we selected her control (so that we could match for age at blood draw) and (2) if a case was eligible for hormone measures, we required that the first selected control complete her intake interview so that we could determine whether or not a second control was needed. Control recruitment began on November 10, 2003. The protocol for controls was identical to the protocol for cases.
The interviewer knew the outcome of the index pregnancy, but she did not know the karyotype of the spontaneous abortion (except in the < 4% of instances when a participant revealed it). XCI was measured without knowledge of any participant characteristics, including the outcome of the index pregnancy.
We asked all women with an XCI skewing percentage ≥ 85 (hereafter, XCI ≥ 85%), as well as a subset of women with XCI 50% to <75%, matched for karyotype group and date of first blood draw, to provide a second blood sample for cytogenetic studies and buccal swabs for measurement of the XCI skewing percentage.
The study was approved by the institutional review boards at our university and the study hospital. All participants gave informed consent. Fieldwork ended in January 2007.
Women with Spontaneous Abortions
We identified 855 women with spontaneous abortions: 729 women were offered karyotype studies, and of those, 695 accepted the offer. The hospital pathology laboratory selected fetal material, almost always chorionic villi, obtained during suction curettage. Five hundred seventeen had sufficient fetal tissue to set up in culture after dissection and enzymatic digestion and/or to save a portion of the suspension for multiplex fluorescent in situ hybridization (FISH). We attempted FISH with probes for chromosomes 13, 15, 16, 18, 21, 22, X, and Y for specimens in which the cultured sample yielded a chromosomally normal female karyotype (because of the possibility of maternal cell growth) or did not yield a karyotype because the culture did not grow or was contaminated. Because the FISH probes that we used identify about 85% of abnormalities in spontaneous abortions,36 we classified cases with normal XX or XY complements by FISH only as normal female or male. FISH on uncultured material was informative for 91 of 110 specimens karyotyped as 46, XX: normal female in 59, normal male in 7, and abnormal in 25. FISH was informative for 49 of 68 specimens in which the culture failed: normal female in 15, normal male in 9, and abnormal in 25. We karyotyped 498 (96%) specimens by either chromosome analysis or FISH (Table 6).
Table 6.
Number of Women Identified or Selected Who Declined the Study, Were Ineligible, or Completed the Protocol
| Protocol Status | Spontaneous Abortion | Control |
|---|---|---|
| Identified or Selected | ||
| Total | 855 | 678a |
| Specimen received at research laboratory | 684 | NA |
| Tissue culture set up and/or analysis by FISHb | 517 | NA |
| Karyotyped | 498 | NA |
| Moved, not located, did not speak English | 11 | 47 |
| Declined the study or withdrew | 133 | 140 |
| Completed the protocol | ||
| Total | 354 | 491 |
| Trisomyc | 170 | NA |
| Chromosomally normal maled | 49 | NA |
| Chromosomally normal femalee | 48 | NA |
| Nontrisomic chromosomal abnormality | 74 | NA |
| Unknown karyotypef | 13 | NA |
| Analytic exclusions | ||
| Total | 30 | 64 |
| Nontrisomic loss with prior trisomy | 1 | NA |
| Repeat entrance | 17 | NA |
| Unknown karyotypef | 12 | NA |
| XCI percentage not assessedg | 2 | |
| XCI percentage measured by an external laboratoryh | 0 | 62 |
| Analytic sample | ||
| Total | 324 | 427i |
| Trisomy | 169 | NA |
| Chromosomally normal male | 46 | NA |
| Chromosomally normal female | 43 | NA |
| Nontrisomic chromosomal abnormality | 66 | NA |
NA denotes “not applicable.”
Excludes 214 women selected as second controls who were ineligible for the hormone component.
FISH denotes “fluorescent in situ hybridization.”
Includes single, double, and triple autosomal trisomies, autosomal monosomies, hypertriploids, and hypertetraploids.
Includes nine abortus specimens for which FISH analysis with probes for chromosomes 13, 15, 16, 18, 21, 22, X, and Y indicated a chromosomally normal male, and the karyotype from culture was not obtained.
Includes abortus specimens for which FISH analysis with probes for chromosomes 13, 15, 16, 18, 21, 22, X, and Y indicated a chromosomally normal female, and the karyotype from culture was either chromosomally normal female (n = 37) or not obtained (n = 11).
We classified the abortus karyotype as unknown if the karyotype from culture was 46,XX but the sample was not analyzed by FISH with probes for chromosomes 13, 15, 16, 18, 21, 22, X, and Y.
XCI denotes “X chromosome inactivation.”
For blood samples drawn on or after June 15, 2006, the XCI percentage was provided by an external laboratory.
Includes 334 first controls and 93 second controls.
Among the 498 women with karyotyped losses, 354 (71%) completed the protocol. The principal reasons for not completing the protocol were refusal (60%) and withdrawal (33%). Mean maternal age is similar for nonparticipants and participants. For the 70 nonparticipants who completed the first interview, ethnicity and mean number of prior pregnancies ending in live birth, spontaneous abortion, and prior induced abortion are similar to those of the 354 participants. Educational levels are significantly higher among participants; the association persisted when we adjusted for age, ethnicity, and obstetric history. Analyses exclude 30 women (detailed in Table 6). The analytic sample thus includes 324 women—169 with autosomal aneuploid losses (referred to as “trisomic,” including four hypertriploids, one hypertetraploid, and six autosomal monosomies), 46 with chromosomally normal male losses, 43 with chromosomally normal female losses, and 66 with nontrisomic chromosomally abnormal losses.
Live Birth Controls
Controls were sampled with replacement. In total, we selected 892 controls, including 214 women who were selected as second controls but were not invited to participate because they were ineligible for hormone measures. Of the remaining 678 women, 491 (72%) completed the protocol. The principal reasons for not completing the protocol were refusal (54%), withdrawal (21%), or our inability to locate the individuals (13%) (Table 6).
Mean maternal age is similar for the 491 participants and the 140 women who declined to participate or withdrew. The 47 other nonparticipants, principally women who no longer resided at the address provided on the consent form, were younger. Sixty-eight nonparticipants completed the first interview. Ethnicity and mean number of prior pregnancies ending in live birth, spontaneous abortion, or induced abortion are similar for these 68 nonparticipants and the 491 participants. Educational levels are significantly higher among participants; the association persisted when we adjusted for age, ethnicity, and obstetric history.
The analysis excludes data from 64 women, all controls, whose XCI skewing percentage was measured in an external laboratory (n = 62) or not measured (n = 2). The analytic sample thus includes 427 controls. Among women whose XCI assay was informative, the XCI skewing percentage did not differ between first (n = 304) and second (n = 84) controls (p = 0.41, Wilcoxon rank-sum test).
Comparison of Cases and Controls
As expected, trisomy case individuals are older than the other case individuals and older than control individuals (each of whom was matched to a case individual who completed the protocol) (Table 7). Adjusting for age, the mean number of pregnancies ending in live birth is higher and the mean number of spontaneous abortions is lower among controls than among cases. In addition, the four case groups and controls differ in the mean number of induced abortions, although no two-group comparison was significant at α = 0.05. The four case groups and controls did not differ in education or ethnicity. The proportion with informative XCI assays did not vary among the pregnancy-outcome groups.
Table 7.
Selected Characteristics of Women Who Completed the Protocol, Classified by the Outcome of the Index Pregnancy
| Selected Characteristics | Control | Spontaneous Abortion |
|||
|---|---|---|---|---|---|
| Trisomy | Chr. Normal Male | Chr. Normal Female | Nontrisomic Chr. Abnormality | ||
| Number of women | 427 | 169 | 46 | 43 | 66 |
| Age at blood drawa | 35.5 (4.6) | 37.1 (4.6) | 33.4 (5.5) | 33.1 (4.2) | 33.6 (4.2) |
| Gestation (days)b | NA | 64.8 (12.6) | 68.9 (23.4) | 64.7 (16.2) | 67.1 (14.9) |
| Live birthsc | 1.9 (0.9) | 1.1 (1.2) | 0.8 (1.2) | 0.8 (0.8) | 1.0 (0.8) |
| Spontaneous abortionsd | 0.3 (0.6) | 1.5 (0.9) | 1.6 (1.3) | 1.5 (1.0) | 1.5 (0.9) |
| Induced abortionse | 0.2 (0.7) | 0.3 (0.7) | 0.4 (0.6) | 0.1 (0.4) | 0.1 (0.3) |
| No college degreef | 25.1 | 27.2 | 37.8 | 20.9 | 22.7 |
| College degreeg | 51.3 | 45.6 | 37.8 | 51.2 | 48.5 |
| Postgraduate degreeh | 23.6 | 27.2 | 24.4 | 27.9 | 28.8 |
| White, non-Hispanici | 87.4 | 85.2 | 82.6 | 79.1 | 84.8 |
| Informative XCI percentagej | 90.9 | 91.1 | 93.5 | 88.4 | 92.4 |
NA denotes “not applicable.”
Data are given as mean (SD). Age varies significantly with the outcome of the index pregnancy (p < 0.0001). Because controls were matched to each woman with a spontaneous abortion, they are significantly older than women with nontrisomic losses and significantly younger than women with trisomic losses. As expected, women with trisomic losses are significantly older than women with nontrisomic losses.
Data are given as mean (SD). Mean gestation does not differ among the four spontaneous abortion groups (p = 0.36).
Pregnancies at blood draw; data are given as mean (SD). Adjusted for age, the mean number of live births differs among the five groups (p < 0.0001).
Pregnancies at blood draw; data are given as mean (SD). Adjusted for age, the mean number of spontaneous abortions (< 20 wks gestation) differs among the five groups (p < 0.0001).
Pregnancies at blood draw; data are given as mean (sd). Adjusted for age, the mean number of induced abortions differs among the five groups (p = 0.003).
Excludes one woman with a chromosomally normal male loss and an unknown education level. Adjusted for age, education does not differ among the five groups (p = 0.79).
Excludes one woman with a chromosomally normal male loss and an unknown education level. Adjusted for age, education does not differ among the five groups (p = 0.79).
Excludes one woman with a chromosomally normal male loss and an unknown education level. Adjusted for age, education does not differ among the five groups (p = 0.79).
Adjusted for age, ethnicity does not differ among the five groups (p = 0.58).
Adjusted for age, the proportion informative for the XCI percentage (i.e., heterozygous at the AR locus) does not differ among the five groups (p = 0.96).
The XCI Assay
We used a Flexigene kit (QIAGEN) to prepare DNA from peripheral blood samples that had been stored at −20°C. We used the HUMARA assay, which takes advantage of the differential methylation of a CpG site close to a highly polymorphic (approximately 90% of women have distinguishable alleles) CAG repeat in the first exon of the X-linked androgen receptor (AR) gene. We digested 1 μg of DNA with 5 U of RsaI alone and another 1 μg with 5 U of RsaI and 10 U of HpaII. After digestion, the DNA was desalted by passing it through a Performa DTR gel filtration cartridge (Edge Biosystems). PCR was performed on 10 ng of both digested samples with the use of primers amplifying the CAG repeat on the AR gene. The methylated CpG site of the inactive allele is not digested by HpaII, leaving it available for amplification by a primer flanking the repeat, whereas the CpG site of the active allele is digested and does not amplify. The forward primer was labeled with FAM for the single digest and with HEX for the double digest. After PCR, a portion of the sample was mixed with a labeled size marker and run on an ABI310 automated genetic sequencer (GeneScan Analysis software, version 3.12) for detection of fluorescence. From each batch of 10 to 12 samples, we tested two samples for the completeness of the HpaII digestion by PCR amplification of the 5′ region of the MIC2 gene, which is unmethylated on both X chromosomes and completely digested by HpaII.
We used a slight modification of the method described in Hatakeyama et al.14 to assess the XCI skewing percentage at the AR locus. The XCI skewing ratio was determined by comparing the ratio of allele peak heights in the HpaII digested sample (d1 and d2, for smaller and larger PCR product sizes, respectively) with the ratio in the sample digested by RsaI alone (u1 and u2). The method corrects for differences in amplification efficiency of the two alleles. The XCI skewing ratio equals (d1/u1)/(d2/u2). We converted the ratio to an XCI skewing percentage by computing proportion (P) as P = [(d1/u1)/{(d1/u1) + (d2/u2)}] × 100, which ranges from 0% to 100%. We refer to P as the unfolded XCI skewing percentage. In most analyses we analyze the folded XCI skewing percentage, 50 + |P − 50|, which ranges from 50% to 100%. Among the 751 samples analyzed, we determined the XCI skewing ratio in the 684 (91.1%) that were heterozygous at the AR locus.
Reliability of the XCI Assay
We carried out several reliability studies. We used the intraclass correlation coefficient (ICC) because it provides a more robust measure of reliability and agreement than does the Pearson product-moment correlation coefficient (r).
The first study comprised samples collected early in the study. We reassayed the PCR products from a stratified random sample of 30 samples (ten with XCI ≥ 85%, ten with XCI 75% to <85%, ten with XCI 50% to <75%), four times, twice in our laboratory, three months apart, and twice in an external laboratory, two months apart. The ICC among all five assays was 0.89; within our laboratory it was 0.87. Mean XCI skewing percentages differed significantly between assays. Toward the end of the study, we carried out three additional reliability studies: (1) We conducted a reliability study to determine whether or not to draw on assay results from an external laboratory for samples (all from controls) collected late in the study. Comparison of assay results with the use of the same PCR products of 24 randomly selected controls with XCI skewing percentages measured in both laboratories revealed that the mean XCI skewing percentage was significantly higher in our laboratory than in the external laboratory, despite excellent agreement (ICC = 0.93) between laboratories. Hence, analyses exclude samples assayed at the external laboratory. (2) The most stringent reliability study drew on a stratified random sample for which we remade PCR preparations to compare our assays with assays from the external laboratory. For 19 of 24 samples judged heterozygous by both laboratories, the ICC was 0.80; mean XCI skewing percentages did not differ between laboratories. (3) Finally, during the latter half of the study, we reassayed the PCR products of 70 samples in consecutive assays. The sample overrepresented specimens with high XCI skewing percentages (17% with XCI ≥ 85%). The ICC was 0.96; XCI skewing percentages did not differ between the first and second assays. In sum, reliability ranged from a low of 0.80 (when we remade PCR products) to a high of 0.96 (when the same PCR products were run within 35 days of each other in the same laboratory).
These ICCs exclude samples judged homozygous by one or both laboratories. Among ten samples judged homozygous by either laboratory, there was agreement on seven. Disagreements concerned questionable heterozygosity for alleles differing by three base pairs.
Buccal Swabs
Of the 90 women who provided buccal swabs (45 with blood XCI ≥ 85%, 45 with blood XCI 50% to <75%), 86 provided sufficient DNA from both cheeks, allowing us to obtain the XCI skewing percentage. Of these, the buccal mucosa of 83 women were heterozygous at the AR locus. Three of the 83 women were excluded from analysis because the karyotype of the abortus was uncertain (i.e., 46,XX without confirming FISH).
Statistical Analysis
We carried out two complementary analyses to estimate associations of the XCI skewing percentage with each case group—trisomy, chromosomally normal male, chromosomally normal female, nontrisomic chromosomally abnormal—each compared with controls.
The first analysis used conditional logistic regression45,46 to test the null hypothesis that, at any maternal age, there is no difference in the XCI skewing percentage between cases and controls. The analysis adjusted by stratification for age at blood draw in single years. We analyzed the folded XCI skewing percentage categorically (50 to <60, 60 to <70, 70 to <80, 80 to <85, ≥ 85), excluding homozygotes from the analysis. We defined highly skewed XCI as ≥ 85% (6.1% of 684 heterozygotes) because the small proportion (2.3%) of women with XCI ≥ 90% limited statistical power. We also report the results of the primary analysis using ≥ 90% as the upper cutpoint to facilitate comparison with other studies. We used maximum likelihood estimates of the odds ratios and 95% confidence intervals to estimate associations between karyotype group and the XCI skewing percentage, with XCI 50% to <60% as the reference group. This analysis has the advantage that adjusting for age by stratification requires no assumptions about the shape of the association between age and the XCI skewing percentage. Because the XCI skewing percentage may be underestimated when alleles differ by only one CAG repeat, we repeated the analysis, excluding the 93 (13.6%) samples in which allele sizes differed by only one repeat. Results were unchanged (data not shown).
The second analysis used a parametric model to estimate associations with the XCI proportion, P/100, which was assumed to follow a symmetric beta distribution. The beta distribution, Beta(a,b), with parameters a and b both greater than zero, provides a flexible family of density curves for proportions varying between 0 and 1. The mean of the Beta(a,b) distribution is μ = a / (a + b), and the variance is μ (1-μ) / (a + b + 1). Because, in theory, the mean of the unfolded XCI skewing percentage is 50, we specialize the model by letting a = b, denoting the common value by θ. The quantity a + b = 2θ is called the “shape parameter”: when θ is large, the density curve is highly peaked around the mean μ = 0.5; when θ = 1, the density curve is flat, corresponding to a uniform distribution; when θ < 1, the density curve is U-shaped. For convenience, hereafter, we refer to θ itself as the shape parameter, rather than 2θ.
When the XCI proportion is modeled as a Beta(θ,θ) distribution, the likelihood function is exactly the same whether one uses the observed folded or unfolded XCI proportions. The analytic model specifies that the logarithm of θ is a linear function of the explanatory factors (e.g., age, karyotype group). This analysis permitted us to test the null hypothesis that the mean of the folded XCI skewing percentage is the same for each case group and controls. In these analyses, we adjusted for maternal age linearly. We used maximum likelihood to estimate the regression coefficients.
We also carried out three secondary analyses: (1) dividing the nontrisomic chromosomally abnormal cases into three groups: monosomy X, triploidy, other; (2) dividing trisomic cases into three groups: acrocentric, trisomy 16, other nonacrocentric; and (3) classifying women by reproductive history.
Finally, we present data bearing on the consistency of measures of XCI over time for women who entered the study twice (n = 17) and between blood and buccal mucosa for the women (n = 80) with informative assays on both tissues.
Web Resources
The URL for data presented herein is as follows:
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim/
Acknowledgments
This work was supported by a grant from the National Institutes on Child Health and Development (R01 HD 42725). We thank Martin Hochberg and his colleagues at The Valley Hospital in Ridgewood, NJ, USA, for providing access to their patients. We especially thank Arthur Christiano in Pathology, who facilitated our work and advised on diagnostic issues. We thank Larry Bologna, Denise Campbell, Gina Chavez, Lois Deyo, Cheryl Dulaff, Diane Gerardi, Nancy Librera, Deborah Manente, Mary Reiner, Louis Rizzo, Donna Rochette, and Marriett Trentacoste, who facilitated our work at the study hospital. We thank Richard Buchsbaum, whose programming and data management expertise facilitated both the day-to-day fieldwork and the statistical analysis. We gratefully acknowledge Project Director L. Perry Brothers and Fieldworkers Melissa Bieliecki, Kathleen Carstens, Beth Fishner, and Renee Davenport, who assisted in tasks too countless to list. Most of all, we thank the women who participated, without whom this research would not have been possible.
References
- 1.Amos-Landgraf J.M., Cottle A., Plenge R.M., Friez M., Schwartz C.E., Longshore J., Willard H.F. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am. J. Hum. Genet. 2006;79:493–499. doi: 10.1086/507565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fialkow P.J. Primordial cell pool size and lineage relationships of five human cell types. Ann. Hum. Genet. 1973;37:39–48. doi: 10.1111/j.1469-1809.1973.tb01813.x. [DOI] [PubMed] [Google Scholar]
- 3.Puck J.M., Stewart C.C., Nussbaum R.L. Maximum-likelihood analysis of human T-cell X chromosome inactivation patterns: Normal women versus carriers of X-linked severe combined immunodeficiency. Am. J. Hum. Genet. 1992;50:742–748. [PMC free article] [PubMed] [Google Scholar]
- 4.Tonon L., Bergamaschi G., Dellavecchia C., Rosti V., Lucotti C., Malabarba L., Novella A., Vercesi E., Frassoni F., Cazzola M. Unbalanced X-chromosome inactivation in haemopoietic cells from normal women. Br. J. Haematol. 1998;102:996–1003. doi: 10.1046/j.1365-2141.1998.00867.x. [DOI] [PubMed] [Google Scholar]
- 5.Plenge R.M., Hendrich B.D., Schwartz C., Arena J.F., Naumova A., Sapienza C., Winter R.M., Willard H.F. A promoter mutation in the XIST gene in two unrelated families with skewed X-chromosome inactivation. Nat. Genet. 1997;17:353–356. doi: 10.1038/ng1197-353. [DOI] [PubMed] [Google Scholar]
- 6.Redonnet-Vernhet I., Ploos van Amstel J.K., Jansen R.P., Wevers R.A., Salvayre R., Levade T. Uneven X inactivation in a female monozygotic twin pair with Fabry disease and discordant expression of a novel mutation in the alpha-galactosidase A gene. J. Med. Genet. 1996;33:682–688. doi: 10.1136/jmg.33.8.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schroder W., Wulff K., Wollina K., Herrmann F.H. Haemophilia B in female twins caused by a point mutation in one factor IX gene and nonrandom inactivation patterns of the X-chromosomes. Thromb. Haemost. 1997;78:1347–1351. [PubMed] [Google Scholar]
- 8.Lau A.W., Brown C.J., Penaherrera M., Langlois S., Kalousek D.K., Robinson W.P. Skewed X-chromosome inactivation is common in fetuses or newborns associated with confined placental mosaicism. Am. J. Hum. Genet. 1997;61:1353–1361. doi: 10.1086/301651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Migeon B.R. Non-random X chromosome inactivation in mammalian cells. Cytogenet. Cell Genet. 1998;80:142–148. doi: 10.1159/000014971. [DOI] [PubMed] [Google Scholar]
- 10.Bolduc V., Chagnon P., Provost S., Dube M.P., Belisle C., Gingras M., Mollica L., Busque L. No evidence that skewing of X chromosome inactivation patterns is transmitted to offspring in humans. J. Clin. Invest. 2008;118:333–341. doi: 10.1172/JCI33166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gale R.E., Wheadon H., Boulos P., Linch D.C. Tissue specificity of X-chromosome inactivation patterns. Blood. 1994;83:2899–2905. [PubMed] [Google Scholar]
- 12.Knudsen G.P., Pedersen J., Klingenberg O., Lygren I., Orstavik K.H. Increased skewing of X chromosome inactivation with age in both blood and buccal cells. Cytogenet. Genome Res. 2007;116:24–28. doi: 10.1159/000097414. [DOI] [PubMed] [Google Scholar]
- 13.Sharp A., Robinson D., Jacobs P. Age- and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum. Genet. 2000;107:343–349. doi: 10.1007/s004390000382. [DOI] [PubMed] [Google Scholar]
- 14.Hatakeyama C., Anderson C.L., Beever C.L., Penaherrera M.S., Brown C.J., Robinson W.P. The dynamics of X-inactivation skewing as women age. Clin. Genet. 2004;66:327–332. doi: 10.1111/j.1399-0004.2004.00310.x. [DOI] [PubMed] [Google Scholar]
- 15.Wengler G.S., Parolini O., Fiorini M., Mella P., Smith H., Ugazio A.G., Notarangelo L.D. A PCR-based non-radioactive X-chromosome inactivation assay for genetic counseling in X-linked primary immunodeficiencies. Life Sci. 1997;61:1405–1411. doi: 10.1016/s0024-3205(97)00686-3. [DOI] [PubMed] [Google Scholar]
- 16.Devriendt K., Matthijs G., Legius E., Schollen E., Blockmans D., van Geet C., Degreef H., Cassiman J.J., Fryns J.P. Skewed X-chromosome inactivation in female carriers of dyskeratosis congenita. Am. J. Hum. Genet. 1997;60:581–587. [PMC free article] [PubMed] [Google Scholar]
- 17.Brown L.Y., Alonso M.L., Yu J., Warburton D., Brown S. Prenatal diagnosis of a familial Xq deletion in a female fetus: A case report. Prenat. Diagn. 2001;21:27–30. doi: 10.1002/1097-0223(200101)21:1<27::aid-pd971>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt M., Du Sart D., Kalitsis P., Fraser N., Leversha M., Voullaire L., Foster D., Davies J., Hills L., Petrovic V. X chromosome inactivation in fibroblasts of mentally retarded female carriers of the fragile site Xq27.3: Application of the probe M27 beta to evaluate X inactivation status. Am. J. Med. Genet. 1991;38:411–415. doi: 10.1002/ajmg.1320380252. [DOI] [PubMed] [Google Scholar]
- 19.Pegoraro E., Whitaker J., Mowery-Rushton P., Surti U., Lanasa M., Hoffman E.P. Familial skewed X inactivation: A molecular trait associated with high spontaneous-abortion rate maps to Xq28. Am. J. Hum. Genet. 1997;61:160–170. doi: 10.1086/513901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanasa M.C., Hogge W.A. X chromosome defects as an etiology of recurrent spontaneous abortion. Semin. Reprod. Med. 2000;18:97–103. doi: 10.1055/s-2000-13480. [DOI] [PubMed] [Google Scholar]
- 21.Lanasa M.C., Hogge W.A., Hoffman E. The X chromosome and recurrent spontaneous abortion: The significance of transmanifesting carriers. Am. J. Hum. Genet. 1999;64:934–938. doi: 10.1086/302352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanasa M.C., Hogge W.A., Kubik C., Blancato J., Hoffman E.P. Highly skewed X-chromosome inactivation is associated with idiopathic recurrent spontaneous abortion. Am. J. Hum. Genet. 1999;65:252–254. doi: 10.1086/302441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanasa M.C., Hogge W.A., Kubik C.J., Ness R.B., Harger J., Nagel T., Prosen T., Markovic N., Hoffman E.P. A novel X chromosome-linked genetic cause of recurrent spontaneous abortion. Am. J. Obstet. Gynecol. 2001;185:563–568. doi: 10.1067/mob.2001.117670. [DOI] [PubMed] [Google Scholar]
- 24.Beever C.L., Stephenson M.D., Penaherrera M.S., Jiang R.H., Kalousek D.K., Hayden M., Field L., Brown C.J., Robinson W.P. Skewed X-chromosome inactivation is associated with trisomy in women ascertained on the basis of recurrent spontaneous abortion or chromosomally abnormal pregnancies. Am. J. Hum. Genet. 2003;72:399–407. doi: 10.1086/346119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sangha K.K., Stephenson M.D., Brown C.J., Robinson W.P. Extremely skewed X-chromosome inactivation is increased in women with recurrent spontaneous abortion. Am. J. Hum. Genet. 1999;65:913–917. doi: 10.1086/302552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uehara S., Hashiyada M., Sato K., Sato Y., Fujimori K., Okamura K. Preferential X-chromosome inactivation in women with idiopathic recurrent pregnancy loss. Fertil. Steril. 2001;76:908–914. doi: 10.1016/s0015-0282(01)02845-x. [DOI] [PubMed] [Google Scholar]
- 27.Hogge W.A., Prosen T.L., Lanasa M.C., Huber H.A., Reeves M.F. Recurrent spontaneous abortion and skewed X-inactivation: Is there an association? Am J Obstet Gynecol. 2007;196:e381–e386. doi: 10.1016/j.ajog.2006.12.012. discussion e386–e388. [DOI] [PubMed] [Google Scholar]
- 28.Bagislar S., Ustuner I., Cengiz B., Soylemez F., Akyerli C.B., Ceylaner S., Ceylaner G., Acar A., Ozcelik T. Extremely skewed X-chromosome inactivation patterns in women with recurrent spontaneous abortion. Aust. N. Z. J. Obstet. Gynaecol. 2006;46:384–387. doi: 10.1111/j.1479-828X.2006.00622.x. [DOI] [PubMed] [Google Scholar]
- 29.Dasoula A., Kalantaridou S., Sotiriadis A., Pavlou M., Georgiou I., Paraskevaidis E., Makrigiannakis A., Syrrou M. Skewed X-chromosome inactivation in Greek women with idiopathic recurrent miscarriage. Fetal Diagn. Ther. 2008;23:198–203. doi: 10.1159/000116741. [DOI] [PubMed] [Google Scholar]
- 30.Kim J.W., Park S.Y., Kim Y.M., Kim J.M., Han J.Y., Ryu H.M. X-chromosome inactivation patterns in Korean women with idiopathic recurrent spontaneous abortion. J. Korean Med. Sci. 2004;19:258–262. doi: 10.3346/jkms.2004.19.2.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo P.L., Huang S.C., Chang L.W., Lin C.H., Tsai W.H., Teng Y.N. Association of extremely skewed X-chromosome inactivation with Taiwanese women presenting with recurrent pregnancy loss. J. Formos. Med. Assoc. 2008;107:340–343. doi: 10.1016/S0929-6646(08)60096-0. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan A.E., Lewis T., Stephenson M., Odem R., Schreiber J., Ober C., Branch D.W. Pregnancy outcome in recurrent miscarriage patients with skewed X chromosome inactivation. Obstet. Gynecol. 2003;101:1236–1242. doi: 10.1016/s0029-7844(03)00345-4. [DOI] [PubMed] [Google Scholar]
- 33.Stephenson M.D., Awartani K.A., Robinson W.P. Cytogenetic analysis of miscarriages from couples with recurrent miscarriage: A case-control study. Hum. Reprod. 2002;17:446–451. doi: 10.1093/humrep/17.2.446. [DOI] [PubMed] [Google Scholar]
- 34.Warburton D. Chromosomal causes of fetal death. Clin. Obstet. Gynecol. 1987;30:268–277. doi: 10.1097/00003081-198706000-00006. [DOI] [PubMed] [Google Scholar]
- 35.Matthews M.S., Hamilton B.E. Mean age of mother, 1970–2000. Natl. Vital Stat. Rep. 2002;51:1–13. [PubMed] [Google Scholar]
- 36.Jobanputra V., Sobrino A., Kinney A., Kline J., Warburton D. Multiplex interphase FISH as a screen for common aneuploidies in spontaneous abortions. Hum. Reprod. 2002;17:1166–1170. doi: 10.1093/humrep/17.5.1166. [DOI] [PubMed] [Google Scholar]
- 37.Kline J., Kinney A., Reuss M.L., Kelly A., Levin B., Ferin M., Warburton D. Trisomic pregnancy and the oocyte pool. Hum. Reprod. 2004;19:1633–1643. doi: 10.1093/humrep/deh310. [DOI] [PubMed] [Google Scholar]
- 38.Simpson J.L., Rajkovic A. Ovarian differentiation and gonadal failure. Am. J. Med. Genet. 1999;89:186–200. doi: 10.1002/(sici)1096-8628(19991229)89:4<186::aid-ajmg3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 39.Laml T., Preyer O., Umek W., Hengstschlager M., Hanzal H. Genetic disorders in premature ovarian failure. Hum. Reprod. Update. 2002;8:483–491. doi: 10.1093/humupd/8.5.483. [DOI] [PubMed] [Google Scholar]
- 40.Bretherick K.L., Metzger D.L., Chanoine J.P., Panagiotopoulos C., Watson S.K., Lam W.L., Fluker M.R., Brown C.J., Robinson W.P. Skewed X-chromosome inactivation is associated with primary but not secondary ovarian failure. Am. J. Med. Genet. A. 2007;143A:945–951. doi: 10.1002/ajmg.a.31679. [DOI] [PubMed] [Google Scholar]
- 41.Sato K., Uehara S., Hashiyada M., Nabeshima H., Sugawara J., Terada Y., Yaegashi N., Okamura K. Genetic significance of skewed X-chromosome inactivation in premature ovarian failure. Am. J. Med. Genet. A. 2004;130:240–244. doi: 10.1002/ajmg.a.30256. [DOI] [PubMed] [Google Scholar]
- 42.Kline J., Levin B. Trisomy and age at menopause: Predicted associations given a link with rate of oocyte atresia. Paediatr. Perinat. Epidemiol. 1992;6:225–239. doi: 10.1111/j.1365-3016.1992.tb00763.x. [DOI] [PubMed] [Google Scholar]
- 43.Warburton D. Biological aging and the etiology of aneuploidy. Cytogenet. Genome Res. 2005;111:266–272. doi: 10.1159/000086899. [DOI] [PubMed] [Google Scholar]
- 44.Bretherick K., Gair J., Robinson W.P. The association of skewed X chromosome inactivation with aneuploidy in humans. Cytogenet. Genome Res. 2005;111:260–265. doi: 10.1159/000086898. [DOI] [PubMed] [Google Scholar]
- 45.Breslow N.E., Day N.E. The Analysis of Case-Control Studies. IARC Scientific Publications; Lyon: 1980. Statistical Methods in Cancer Research, Volume I. [PubMed] [Google Scholar]
- 46.Fleiss J., Levin B., Paik M. Wiley; New York: 2003. Statistical Methods for Rates and Proportions. [Google Scholar]
- 47.Freeman S.B., Yang Q., Allran K., Taft L.F., Sherman S.L. Women with a reduced ovarian complement may have an increased risk for a child with Down syndrome. Am. J. Hum. Genet. 2000;66:1680–1683. doi: 10.1086/302907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kline J., Kinney A., Levin B., Warburton D. Trisomic pregnancy and earlier age at menopause. Am. J. Hum. Genet. 2000;67:395–404. doi: 10.1086/303009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Montfrans J.M., Dorland M., Oosterhuis G.J., van Vugt J.M., Rekers-Mombarg L.T., Lambalk C.B. Increased concentrations of follicle-stimulating hormone in mothers of children with Down's syndrome. Lancet. 1999;353:1853–1854. doi: 10.1016/s0140-6736(99)00936-8. [DOI] [PubMed] [Google Scholar]
- 50.Kline J., Kinney A., Levin B., Kelly A., Chih-Yu J., Brown S., Warburton D. X-chromosome inactivation and ovarian age during the reproductive years. Fertil. Steril. 2006;85:1488–1495. doi: 10.1016/j.fertnstert.2005.10.059. [DOI] [PubMed] [Google Scholar]
- 51.Hassold T., Hall H., Hunt P. The origin of human aneuploidy: Where we have been, where we are going. Hum Mol Genet. 2007;16:R203–R208. doi: 10.1093/hmg/ddm243. Spec No. 2. [DOI] [PubMed] [Google Scholar]
- 52.Christensen K., Kristiansen M., Hagen-Larsen H., Skytthe A., Bathum L., Jeune B., Andersen-Ranberg K., Vaupel J.W., Orstavik K.H. X-linked genetic factors regulate hematopoietic stem-cell kinetics in females. Blood. 2000;95:2449–2451. [PubMed] [Google Scholar]
- 53.Kristiansen M., Knudsen G.P., Bathum L., Naumova A.K., Sorensen T.I., Brix T.H., Svendsen A.J., Christensen K., Kyvik K.O., Orstavik K.H. Twin study of genetic and aging effects on X chromosome inactivation. Eur. J. Hum. Genet. 2005;13:599–606. doi: 10.1038/sj.ejhg.5201398. [DOI] [PubMed] [Google Scholar]
- 54.Vickers M.A., McLeod E., Spector T.D., Wilson I.J. Assessment of mechanism of acquired skewed X inactivation by analysis of twins. Blood. 2001;97:1274–1281. doi: 10.1182/blood.v97.5.1274. [DOI] [PubMed] [Google Scholar]


