Abstract
The tumor suppressor p53 has evolved a MDM2-dependent feedback loop that promotes p53 protein degradation through the ubiquitin–proteasome system. MDM2 is an E3-RING containing ubiquitin ligase that catalyzes p53 ubiquitination by a dual-site mechanism requiring ligand occupation of its N-terminal hydrophobic pocket, which then stabilizes MDM2 binding to the ubiquitination signal in the DNA-binding domain of p53. A unique pseudo-substrate motif or “lid” in MDM2 is adjacent to its N-terminal hydrophobic pocket, and we have evaluated the effects of the flexible lid on the dual-site ubiquitination reaction mechanism catalyzed by MDM2. Deletion of this pseudo-substrate motif promotes MDM2 protein thermoinstability, indicating that the site can function as a positive regulatory element. Phospho-mimetic mutation in the pseudo-substrate motif at codon 17 (MDM2S17D) stabilizes the binding of MDM2 towards two distinct peptide docking sites within the p53 tetramer and enhances p53 ubiquitination. Molecular modeling orientates the phospho-mimetic pseudo-substrate motif in equilibrium over a charged surface patch on the MDM2 at Arg97/Lys98, and mutation of these residues to the MDM4 equivalent reverses the activating effect of the phospho-mimetic mutation on MDM2 function. These data highlight the ability of the pseudo-substrate motif to regulate the allosteric interaction between the N-terminal hydrophobic pocket of MDM2 and its central acidic domain, which stimulates the E3 ubiquitin ligase function of MDM2. This model of MDM2 regulation implicates an as yet undefined lid-kinase as a component of pro-oncogenic pathways that stimulate the E3 ubiquitin ligase function of MDM2 in cells.
Keywords: MDM2, p53, Allostery, Kinase, Ubiquitination
Introduction
Reconstitution of a multicomponent ubiquitin–enzyme reaction mechanism, including the role of E1, E2, and E3 subcomponents as well as the substrate, is a fundamental goal in understanding the molecular events that regulate the ubiquitin–proteasome pathway [32]. The proteins within the E3 class of ubiquitination catalysts include among others the HECT domain and the RING domains. Unlike the HECT domain containing ubiquitin ligases, the E3-RING domain containing proteins do not form a covalent bond with ubiquitin, but catalyze the transfer of ubiquitin from the activated E2 to the substrate by as yet undefined mechanisms. How E3-RING domain containing ubiquitin ligases alter substrate conformation and drive E2 recognition of substrate are critical mechanistic goals. The pro-oncogenic protein MDM2 is such a RING domain containing E3 ubiquitin ligase that can promote the ubiquitination of the p53 tumor suppressor protein [44, 49], and MDM2 represents a model E3 ubiquitin ligase to evaluate the dynamics of a multiprotein ubiquitination system.
The E3-RING domain containing ubiquitin ligase MDM2 itself has been dissected into multiple domains with specific biochemical functions. The C-terminal RING domain in MDM2 co-ordinates the activity of MDM2 in E2-mediated ubiquitin transfer [22] and contains a C-terminal peptide tail that maintains RING-domain conformation [36, 46]. The RING domain also promotes MDM2-dependent stimulation of p53 protein synthesis through interactions with p53 mRNA during translation [7], and RNA itself can stimulate MDM2:p53 interactions and substrate ubiquitination [5, 6, 33]. There also is an ATP-binding motif imbedded within the RING domain that regulates the molecular chaperone functions of MDM2 [41, 51]. Thus, the RING domain of MDM2 mediates biochemical functions, including E2 binding, ATP binding, and RNA binding.
In addition to the C-terminal RING domain, MDM2 also contains an N-terminal allosteric hydrophobic pocket, which interacts with a specific linear peptide motif in proteins such as p53 and interferon-responsive transcription factors [20, 33]. This hydrophobic pocket was the first region on MDM2 that was shown to bind directly to p53 [8]. Adjacent to this N-terminal domain is an “acidic” domain that interacts at a second binding site within a flexible motif in the DNA binding domain of p53 [38]. The N-terminal hydrophobic binding pocket has been targeted by small molecules like Nutlins that can interact with MDM2 and activate p53 function by releasing p53 from MDM2-mediated transrepression [47]. However, such ligands do not block p53 ubiquitination [50] as ligand occupation of this N-terminal hydrophobic pocket forms a positive role in MDM2 function as an E3 ubiquitin ligase. MDM2 hydrophobic pocket occupation promotes a more stable interaction between the internal acidic domain of MDM2 and a ubiquitination signal in the DNA-binding domain of p53 [50].
These data have formed a “dual-site” model of MDM2-mediated ubiquitination of p53. Accordingly, small peptides derived from the BOX-I domain of p53 (containing the primary MDM2 binding site) or small molecules like Nutlin do not block p53 ubiquitination. By contrast, peptides derived from the BOX-V domain of p53 (containing the ubiquitination signal in the DNA-binding domain of p53) do inhibit MDM2 ubiquitination of p53 [50]. Allosteric interactions within full-length MDM2 protein has been recently examined through structure-function studies demonstrating that mutation of selected amino acids in the RING domain promotes a conformation change both in the N-terminal domain of MDM2 as defined by proteolytic cleavage susceptibility and in the acidic domain as defined by elevated intrinsic tryptophan fluorescence [52]. These latter data provided the first biophysical evidence for allosteric interactions among the various sub-domains in full-length MDM2 protein.
This dual-site model for MDM2-mediated ubiquitination of p53 presumably operates due to the striking flexibility of the N-terminal p53-binding domain in the presence of distinct peptide ligands [37, 40, 45]. A predominant feature of this ubiquitination reaction mechanism is that there is an induced stabilization of the acidic domain of MDM2 to the conformationally flexible region in the DNA-binding domain of p53 [38, 55] when the MDM2 hydrophobic pocket is occupied by substrate [50]. This linear peptide motif in the DNA-binding domain of p53 (named BOX-V motif) is normally cryptic in the folded p53 tetramer, but it is exposed on mutant p53 in human cancers as defined using a monoclonal antibody that binds to this peptide-epitope on denatured p53 protein [13, 48].
This flexible BOX-V motif in the DNA-binding domain of p53 is also a multiprotein docking site; many protein kinases, including CHK1/2, DAPK, and CK1, interact with this region on p53 to catalyze phosphorylation of the transactivation domain of p53 at Ser20 [9, 23]. Destabilizing mutations in p53 that “unfold” p53 protein at the conformationally flexible region of p53 in the BOX-V domain enhance mutant p53 protein ubiquitination in cell systems [39] and can induce mutant p53 degradation via MDM2 function in murine transgenes [42]. The enhanced degradation of mutant p53 protein by an MDM2-dependent pathway has important implications for regulating the pro-oncogenic functions of mutant p53 in human cancers; indeed, most human cancers appear to have a form of mutant p53 protein with enhanced steady-state levels [30] and therefore has presumably evaded the normal mutant protein degradation machinery.
In order to define further the nature of the allosteric regulation of MDM2, we have examined the function of a flexible and unstructured N-terminal peptide motif adjacent to the hydrophobic pocket of MDM2. Such small flexible peptide motifs have been postulated to be important in a wide range of signaling proteins [27–29]. This flexible motif in MDM2 has been called a pseudo-substrate motif or lid. Pseudo-substrate motifs operate in a range of allosterically regulated enzymes, including protein kinases and metabolic enzymes, and have been termed intrasteric regulatory motifs [1, 16, 17]. The pseudo-substrate motif of MDM2 harbors a phosphorylation site whose phospho-mimetic mutation can alter the conformation of the N-terminal domain of MDM2 protein as defined by NMR studies [25, 40]. This phospho-mimetic mutation was proposed to stabilize the pseudo-substrate motif over the hydrophobic pocket and occlude p53 binding. However, this has not been tested experimentally. As ligands that fill the hydrophobic pocket like Nutlin can prime MDM2 and stimulate the MDM2 E3 ubiquitin ligase function toward p53 [50], the role of the unphosphorylated or phosphorylated lid is not necessarily evident—would it function as a positive or negative cofactor in regulating the stability of the MDM2:p53 complex, and therefore, would it ultimately stimulate or attenuate p53 protein ubiquitination?
In this report, we show that the flexible lid is a positive regulatory motif that maintains the thermostability of MDM2 and that a phospho-mimetic mutation in the MDM2 lid stabilizes the MDM2:p53 complex. We propose a cis-acting intrasteric mechanism to explain how phospho-mimetic mutation of the MDM2 lid might open the hydrophobic pocket. This results in enhanced binding of MDM2 to the first site on p53, which in turn enhances the allosteric interaction between the acidic domain of MDM2 and the ubiquitination signal in the DNA-binding domain of p53. These data also highlight the growing realization that flexible and unstructured linear peptide motifs play critical roles in controlling protein function in signal transduction pathways [29] and provide a biochemical foundation to study how changes in the rates of MDM2 phosphorylation at this flexible motif regulates the E3 ubiquitin ligase function of MDM2.
Results
A phospho-mimetic mutation in the pseudo-substrate motif stimulates the E3 ubiquitin ligase function of MDM2
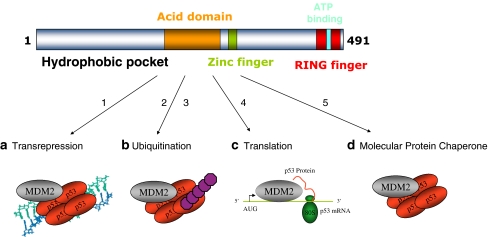
MDM2 is composed of distinct ligand binding domains that regulate its multiple functions (Fig. 1). The integration of our current study on the MDM2 pseudo-substrate motif to the known biochemical functions of MDM2 is also summarized in Fig. 1. The MDM2 oncoprotein is known to have diverse molecular functions in (1) transcriptional repression of p53, (2) ubiquitination and degradation of p53, (3) synthesis of p53 protein, and (4) ATP-dependent molecular chaperone functions on p53 protein (Fig. 1). It is not known if these four functions are integrated, function in parallel, or are coordinately switched on or off. However, human and mouse genetic studies have established a specific role for the RING domain and hydrophobic pocket in the control of MDM2 function as a transrepressor and as an E3 ubiquitin ligase [2, 44].
Fig. 1.
Four distinct modes of MDM2 function. The top panel highlights the domain structure of MDM2, including the hydrophobic pocket in the N-terminus [20], the acidic central domain [55], a zinc-binding motif [56], the RING domain [18], and ATP-binding site imbedded within the RING domain [35]. MDM2 was identified as a protein that regulates the p53 tumor suppressor via (1) inhibition of p53-dependent transcription [26, 43]. Germline mutations in the mdm2 promoter that result in MDM2 over-production result in enhanced MDM2 protein in chromatin fractions, which mediates suppression of p53 function as a transcription factor [2, 52]. The second function identified for MDM2 involved its ability to (2, 3) catalyze p53 degradation through a ubiquitin-dependent pathway [19] [15]. This ubiquitination function of MDM2 also involves ubiquitination and degradation of ribosomal proteins that can normally function to stimulate p53 protein synthesis [31]. According to the dual-site mechanism of MDM2-mediated ubiquitination of p53 [50], two structural changes can enhance p53 ubiquitination by MDM2: one structural change involves (2) destabilizing missense mutations in p53 that unfolds the p53 tetramer and that can expose the ubiquitination signal in the DNA-binding domain [38], thus enhancing p53 ubiquitination in cells [39]; the second structural change involves (3) the lid of MDM2 whose phospho-mimetic mutation increases MDM2-mediated ubiquitination of p53 tetramers by stabilizing the MDM2:p53 complex (this study). Small molecules that bind to the N-terminal hydrophobic pocket of MDM2 (Nutlins) can activate p53 [47] presumably by inhibiting the chromatin-bound pool of MDM2 since Nutlins cannot inhibit MDM2 function as an E3 ubiquitin ligase [50]. Rather, to date, peptide mimetics that bind to the MDM2 acidic domain provide a lead for inhibiting MDM2 function as a E3 ubiquitin ligase [50]. Additional research has highlighted novel molecular functions of MDM2. A cellular function was assigned to the RNA-binding activity of MDM2 [12, 21, 24], which involves (4) an interaction of MDM2 with p53 mRNA at ribosomes to stimulate p53 protein synthesis [7]. A function was also assigned to the ATP-binding motif of MDM2 [35], which involves (5) the ability of MDM2 to exhibit the property of a molecular chaperone by catalyzing the ATP-dependent folding of the p53 tetramer to enhance p53 function as a transcription factor [41, 51]
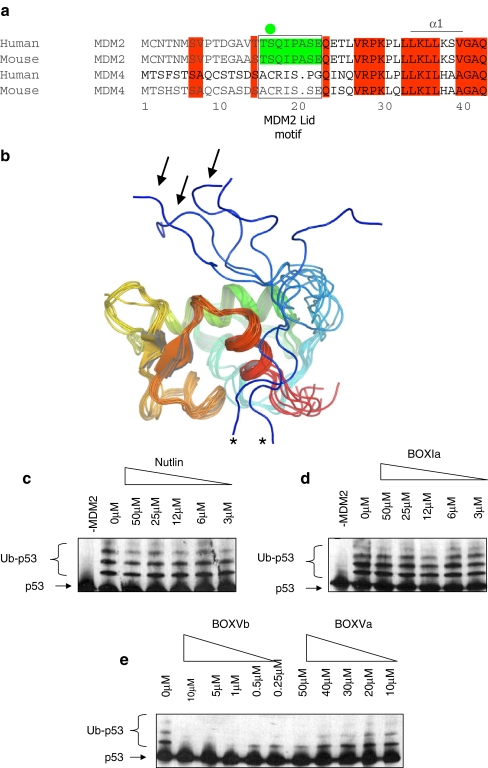
In order to dissect further the dual-site model of MDM2-mediated ubiquitination of p53, we focused our analysis on the effects of the pseudo-substrate motif (Fig. 2a) on the in vitro E3 ubiquitin ligase function of MDM2. An NMR study of the unliganded apo-form of the N-terminal domain of MDM2 revealed the pseudo-substrate motif to be largely unstructured and in a dynamic equilibrium (Fig. 2b). Further, ligands that occupy the hydrophobic pocket can alter the conformation of MDM2 [25, 37, 40, 45], highlighting the conformational flexibility of MDM2. However, ligands such as Nultin or a p53 peptide (BOX-I domain peptide) do not inhibit MDM2-mediated ubiquitination (Fig. 2c and d), while peptides that bind to the acidic domain of MDM2 (BOX-V domain homology peptides) can inhibit MDM2 function (Fig. 2e; as in [33, 50, 52]). As the conformational flexibility of this N-terminal pseudo-substrate motif of MDM2 could have significant regulatory effects on MDM2 conformation, this motif also provides a novel tool with which to evaluate how perturbation of the hydrophobic pocket of MDM2 might regulate allosterically the E3 ubiquitin ligase function of MDM2.
Fig. 2.
The N-terminal pseudo-substrate motif of MDM2. a Conservation of amino acids 1–44, including the pseudo-substrate motif (aka, the lid) within MDM2 homologues and absence of the pseudo-substrate motif in the orthologue MDM4. The pseudo-substrate motif is surrounded by a box, and the phosphorylation site at Ser17 is highlighted with a green circle. b The NMR structure of the non-liganded N-terminal p53 binding domain of MDM2 (PDB ID 1Z1M, residues 10–109; apo-MDM2 [45]) reveals a large degree of conformational heterogeneity in the N-terminal 20 residues (colored from blue (N-terminal) to red (C-terminal)). Pseudo-substrate motif conformations acting like a lid to cover the MDM2 hydrophobic pocket are highlighted with arrows, and conformations with the pseudo-substrate motif interacting with the MDM2 surface are highlighted with asterisks. c–e The effects of small ligands on MDM2 function as an E3 ubiquitin ligase. A ubiquitination assay was assembled with purified E1, E2, MDM2, and p53 proteins without or with increasing concentrations of c Nutlin, dBOX-I peptide derived from the N-terminus of p53, or eBOX-V homology peptides derived from Rb (BOX-Vb) and p53 DNA binding domain (BOX-Va). Ubiquitination was assayed as indicated in the “Materials and methods”. The position of the un-ubiquitinated p53 substrate is highlighted with arrows and p53-ubiquitin adducts with brackets
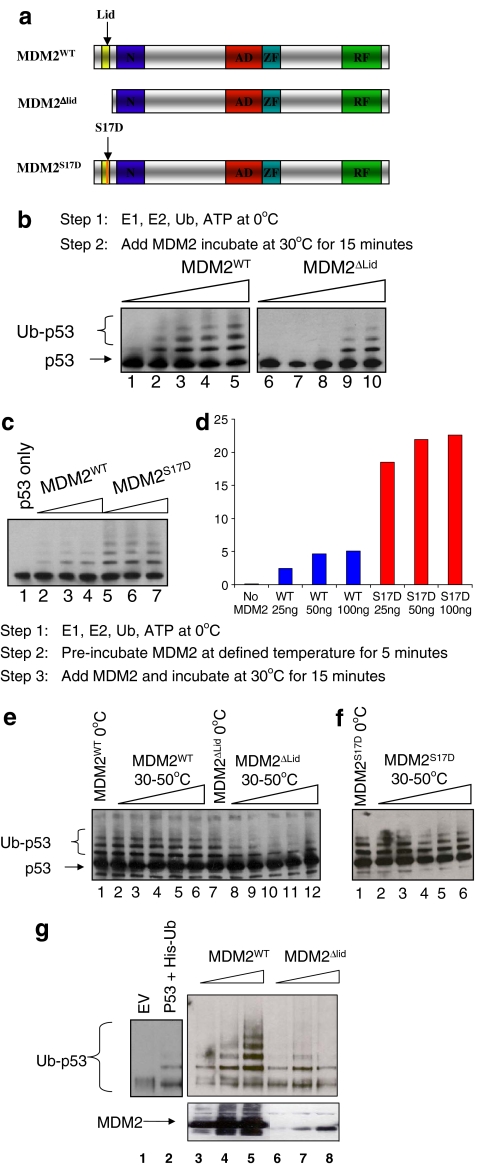
To define a role for this flexible pseudo-substrate motif or lid, purified untagged wild-type MDM2 and MDM2 mutants were generated, including one with an N-terminal flexible pseudo-substrate motif deletion (MDM2ΔLID; Fig. 3a). A titration of MDM2 (Fig. 3b, lanes 1–5) or MDM2ΔLID (Fig. 3b, lanes 6–10) in ubiquitination reactions demonstrated that MDM2ΔLID has a lower specific activity, although it can still catalyze ubiquitination. The lowered specific activity of MDM2ΔLID suggests the pseudo-substrate motif normally has an intrinsic positive regulatory effect on MDM2 function.
Fig. 3.
A phospho-mimetic substitution in the MDM2 lid stimulates the E3 ubiquitin ligase function of MDM2. a Diagram of the functional domains of MDM2, including (1) wt-MDM2; (2) MDM2ΔLID, which has a deletion of the flexible motif; and (3) MDM2S17D, which has a phospho-mimetic aspartate mutation at Ser17 (S17D). b MDM2-pseudo-substrate motif deletion reduces the specific activity of MDM2. Purified wt-MDM2 or MDM2ΔLID was titrated directly into ubiquitination reactions using p53 as a substrate. After 10 min, the reactions were stopped and processed for immunoblotting to measure extents of p53 ubiquitination. c The S17D mutation in the MDM2 pseudo-substrate motif stimulates its E3 ubiquitin ligase function. Purified wt-MDM2 (lanes 2–4) or MDM2S17D (lanes 5–7) were titrated directly into ubiquitination reactions using p53 as a substrate. After 10 min, the reactions were stopped and processed for immunoblotting to measure extents of p53 ubiquitination, which is quantified in d. e MDM2-pseudo-substrate motif deletion promotes thermoinstability in MDM2. Purified wt-MDM2 or MDM2ΔLID was added directly to ubiquitination reactions (lanes 1 and 7) or was preincubated at increasing temperatures (0–50 °C) in ubiquitination buffer (lanes 2–6 and 8–12) followed by titration in ubiquitination reactions using p53 as a substrate. After 10 min, the reactions were stopped and processed for immunoblotting to measure extents of p53 ubiquitination. f S17D mutation in the MDM2 pseudo-substrate motif does not destabilize MDM2. Purified MDM2S17D was preincubated at increasing temperatures (0–50 °C) in ubiquitination buffer followed by titration in ubiquitination reactions using p53 as a substrate. After 10 min, the reactions were stopped and processed for immunoblotting to measure extents of p53 ubiquitination. g MDM2-pseudo-substrate motif deletion promotes MDM2 instability in cells. P53 was co-transfected into cells with his-tagged ubiquitin and vector control (lane 2) or increasing amounts of MDM2 (lanes 3–5) or MDM2ΔLID (lanes 6–8). The reactions were processed to measure p53 ubiquitination as indicated [39]
The pseudo-substrate motif also has a SQ phospho-acceptor site whose phospho-mimetic mutation (S to D) induces a conformational change in the N-terminal domain of MDM2 as defined using NMR [25] (Fig. 2a and b). This mutation was predicted to stabilize the lid equilibrium into a position that partially occludes ligands like p53 and that this mutation might therefore block the MDM2:p53 complex. However, the biochemical effects of this mutation on MDM2 function in vitro and in vivo have not actually been defined experimentally. As such, we also evaluated the effect of the phospho-mimetic pseudo-substrate motif mutant (MDM2S17D; Fig. 3a) on MDM2:p53 interactions. Three possibilities include (1) that the phospho-mimetic motif could act like a lid and partially cover the hydrophobic pocket, thus destabilizing the MDM2–p53 interaction [25]; (2) the phospho-mimetic motif could act like a lid and partially cover the hydrophobic pocket, thus stabilizing MDM2 acid domain interactions with p53 via the dual site allosteric model [50]; and (3) the phospho-mimetic mutation might stabilize the lid in equilibrium in a distinct conformation and “open” the hydrophobic pocket (Fig. 2b). The phospho-mimetic mutation in MDM2 in fact did not inhibit the ubiquitination function of MDM2; rather, the Asp17 mutation increased the specific activity of MDM2 as an E3 ubiquitin ligase (Fig. 3c, lanes 5–7 vs 2–4 and d).
These data are consistent with the lid functioning as a positive rather than as a negative regulatory motif. Regulatory motifs often alter the thermostability of an enzyme or protein [11], and we evaluated whether the pseudo-substrate motif deletion alters thermostability of MDM2. The preincubation of MDM2ΔLID at the indicated temperatures completely inactivated the E3 ubiquitin ligase function of the protein (Fig. 3e, lanes 8–12 vs 7). MDM2S17D was not thermosensitive when preincubated at elevated temperatures compared to MDM2ΔLID (Fig. 3f). These data suggest that the pseudo-substrate motif contributes positively to maintaining the thermostability of MDM2 protein. Consistent with this, transfection of the plasmid encoding MDM2ΔLID into cells destabilizes the protein and accordingly reduces its specific activity as an E3 ubiquitin ligase toward p53 (Fig. 3g, lanes 6–8 vs 3–5).
A phospho-mimetic mutation in the pseudo-substrate motif stabilizes MDM2:p53 interactions
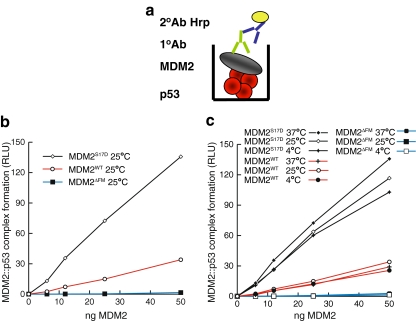
We next analyzed whether the elevated specific activity of MDM2S17D as a E3 ubiquitin ligase could be attributed to enhanced binding of MDM2 to p53 protein resulting in a more stable MDM2:p53 protein complex. This enhanced stability of the MDM2:p53 complex would in turn translate to enhanced p53 ubiquitination. A titration of MDM2S17D, wt-MDM2, or MDM2ΔLID provided a correlation between specific activity of an E3 ubiquitin ligase and enhanced stability of the MDM2:p53 protein complex (Fig. 4b). Although MDM2ΔLID was essentially unable to form a stable contact with the p53 tetramer presumably as a result of pseudo-substrate motif deletion, MDM2S17D exhibited a striking increase in its binding for p53 protein as defined by the stability of the MDM2:p53 complex (Fig. 4b). The preincubation of MDM2S17D, wt-MDM2, or MDM2ΔLID at distinct temperatures had no effect in altering wt-MDM2 protein or MDM2S17D interactions with p53 (Fig. 4c). This elevated stability of the MDM2:p53 complex as a result of the Asp17 mutation in MDM2 is consistent with the enhanced E3 ubiquitin ligase activity of MDM2S17D (Fig. 3). However, these data are not compatible with the hypothesis that this phospho-mimetic substitution would destabilize the MDM2:p53 interactions [25].
Fig. 4.
A phospho-mimetic substitution in the MDM2 lid stabilizes MDM2:p53 tetramer complex formation. a Strategy for measuring the stability of the MDM2:p53 complex. Tetrameric forms of p53 protein were adsorbed onto the solid phase (in red) followed by MDM2 titration (gray). The stability of MDM2 bound to p53 tetramers was measured using a monoclonal antibody specific for MDM2 by chemiluminescence. b The S17D mutation in the MDM2 pseudo-substrate motif stabilizes the MDM2:p53 tetrameric complex. Increasing amounts of the indicated MDM2 protein (wt-MDM2, MDM2ΔLID, or MDM2S17D) were titrated into reactions where fixed amounts of tetrameric p53 were on solid phase as described previously [38]. The extent of MDM2 binding was quantified using an anti-MDM2 monoclonal antibody and binding stability depicted using enhanced chemiluminescence in relative light units (RLU). c Elevated temperature does not alter the activating effect of S17D mutation on MDM2:p53 complex stability. MDM2 was preincubated at distinct temperatures, as indicated, and binding activity to the p53 tetramer was measured as described above in RLU
The data presented above showing that the phospho-mimetic S17D substitution of MDM2 stabilizes the MDM2:p53 interaction could be explained by two models consistent with the dual site docking model of MDM2 function [50]; (1) the phospho-mimicking pseudo-substrate motif is indeed partially occluding the hydrophobic binding pocket [25], but is acting in a positive auto-allosteric manner (i.e., like Nutlin [50]), or (2) the S17D mutation orientates the pseudo-substrate motif in a position that stabilizes the N-terminal domain of MDM2 in an open conformation with the pseudo-substrate motif in equilibrium at a position outwith the hydrophobic pocket (Fig. 2b, asterisks). The striking conformational flexibility of MDM2 when peptide ligands of differing length occupy the pocket [37] or when the lid is in distinct conformations [40] is consistent with either of these two models. We carried out further biochemical characterization of MDM2S17D in order to distinguish between these two models.
One method to examine whether the Asp17 mutation stabilizes the pseudo-substrate motif over the hydrophobic pocket of MDM2 or whether the mutation opens the hydrophobic pocket by shifting lid equilibrium would be to determine whether MDM2S17D is sensitive or resistant to Nutlin. It could be predicted that MDM2S17D cannot interact with Nutlin (or the BOX-I domain of p53) if the Asp17 modified pseudo-substrate motif is stabilized over the hydrophobic pocket. However, Nutlin destabilizes the MDM2S17D:p53 complex as well as the wt-MDM2:p53 complex (Fig. 5b and c). Although these data might not be compatible with the model that the Asp17 substituted pseudo-substrate motif is partially stabilized over the hydrophobic pocket of MDM2, modeling data have shown that if the phospho-mimetic lid does occlude the hydrophobic pocket, it does so only in the entry region of the N-terminal p53 peptide [40]. Thus, we evaluated whether the MDM2S17D has a higher or lower binding activity for the p53 peptide itself derived from the BOX-I domain.
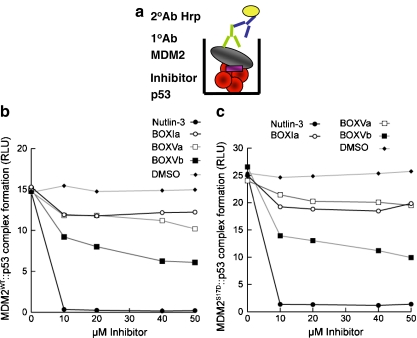
Fig. 5.
Nutlin destabilizes the MDM2S17D:p53 tetramer complex. a Strategy for measuring the stability of the MDM2:p53 complex in the presence of small ligands. Tetrameric forms of p53 protein were adsorbed onto the solid phase (in red) followed by MDM2 titration (gray) in the absence or presence of the indicated inhibitors, including (1) Nutlin-3, (2) BOX-Ia peptides, (3) BOX-Va or BOX-Vb peptides, and (4) DMSO carrier. The stability of MDM2 bound to p53 tetramers was measured using a monoclonal antibody specific for MDM2 by chemiluminescence. b and c MDM2S17D binding to p53 is not inhibited by Nutlin. MDM2 (b) or MDM2S17D (c) was assembled into reactions containing the indicated ligand: BOX-I peptide, BOX-V peptides, or Nutlin. The mixture was added to tetrameric p53 protein on the solid phase, and the amount of MDM2 bound stably to p53 was quantified using an MDM2 monoclonal antibody. The stability of the MDM2:p53 complex is depicted in relative light units
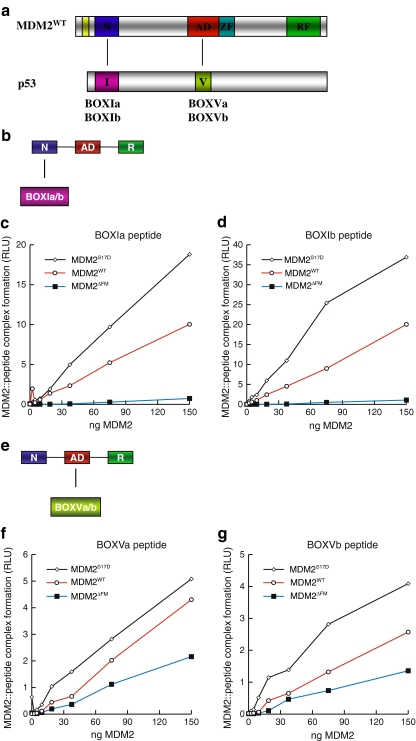
As seen with the full-length p53 tetramers, MDM2S17D also binds more stably to the BOX-I peptides, relative to wt-MDM2 (Fig. 6c and d). The BOX-I peptide “a” is the naturally occurring peptide from p53, while BOX-I peptide “b” is the optimized higher affinity peptide named 12.1 identified using combinatorial peptide libraries [4]. This enhanced binding of MDM2S17D to the BOX-I peptide from p53 again is not compatible with the model that the Asp17 pseudo-substrate motif is stabilized over the hydrophobic pocket, and if this were the case, then MDM2S17D should have a lower binding activity for the p53 BOX-I peptide. As a control, MDM2ΔLID cannot form a stable complex with the BOX-I peptide (Fig. 6c and d), although MDM2ΔLID can form a stable complex with the ubiquitination signal within the BOX-V peptide from p53 (Fig. 6f and g). These latter data indicate that the integrity of the acidic domain of MDM2 is maintained to a significant degree in MDM2ΔLID. Furthermore, the MDM2S17D is also more active in binding to the BOX-V peptide that wt-MDM2 (Fig. 6f and g), which together explains in part why MDM2S17D binds better to the p53 tetramer (Fig. 4b and c). These data also suggest that conformational changes in the N-terminal hydrophobic pocket of MDM2 as a result of the phospho-mimetic lid mutation are transferred to the acidic domain of MDM2, which is consistent with the dual-site model of MDM2-mediated ubiquitination of p53.
Fig. 6.
Phospho-mimetic substitution in the MDM2 lid motif enhances MDM2 binding activity for BOX-I and BOX-V peptide ligands. a The domain structure of MDM2 minidomains, relative to p53 minidomains containing the BOX-I and BOX-V docking regions is highlighted. The panel highlights (a) an interaction between the N-terminal MDM2 domain (N) and the p53 BOX-I motif (I) and (b) an interaction between the acid domain of MDM2 (AD) and the BOX-V motif of p53 (V). b The assay was designed to measure the interaction between the N-terminal domain of full-length MDM2 and the BOX-I-derived p53 peptide. c and d MDM2S17D has an enhanced binding activity for the BOX-I domain of p53. wt-MDM2, MDM2ΔLID, or MDM2S17D proteins were titrated into reaction buffer and incubated onto the solid phase containing BOX-I a peptide (left panel) and BOX-I b peptide (right panel). The amount of MDM2 bound stably to the p53 peptides was quantified using an MDM2 monoclonal antibody. The stability of the MDM2:p53 complex is depicted in relative light units. e The assay was designed to measure the interaction between the central acidic domain of full-length MDM2 and the BOX-V-derived peptides. f and g MDM2S17D has an enhanced binding activity for the BOX-V domain of p53. wt-MDM2, MDM2ΔLID, or MDM2S17D proteins were titrated into reaction buffer and incubated onto the solid phase containing BOX-V a peptide (left panel) and BOX-V b peptide (right panel). The amount of MDM2 bound stably to the p53 peptides was quantified using an MDM2 monoclonal antibody. The stability of the MDM2:p53 complex is depicted in relative light units
A single missense mutation of the RING domain of MDM2 changes the conformation of both the hydrophobic pocket and the acidic domain [52]. We evaluated further this intra/interdomain allosteric interaction in MDM2S17D protein by examining its binding activity for BOX-I- and BOX-V-derived p53 peptides in the absence and presence of Nutlin. A Nutlin titration into reactions containing wt-MDM2 or MDM2S17D indicated that Nutlin is able to compete with MDM2 binding to the BOX-I-derived peptide (Fig. 7b and c), as expected. However, quantitative differences are observed in this reaction. MDM2S17D protein is more sensitive in its binding to the BOX-I peptide in the presence of Nutlin relative to wt-MDM2, consistent with the model that the phospho-mimetic mutation opens the hydrophobic pocket. Another prediction of this allosteric model would be that Nutlin would alter the binding activity of MDM2S17D to the BOX-V-domain-derived peptides. This is observed experimentally; although the BOX-I peptide can partially destabilize MDM2S17D:BOX-V peptide complexes (Fig. 7e), Nutlin completely destabilizes this interaction (Fig. 7f). The acute sensitivity of the MDM2:p53 tetrameric complex to Nutlin (Fig. 5) can therefore be explained because not only does Nutlin destabilize the MDM2:BOX-I peptide complex by direct competition but it can also destabilize the MDM2:BOX-V peptide complex by allosteric effects as suggested previously [52].
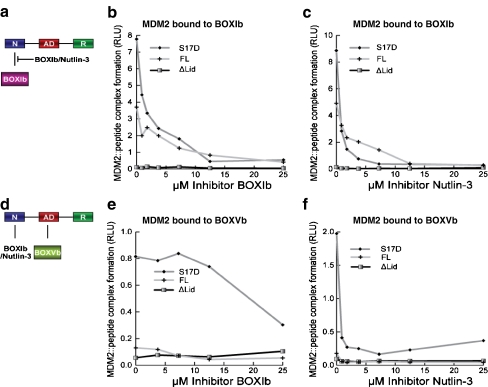
Fig. 7.
Nutlin destabilizes the MDM2:p53 BOX-I and BOX-V peptide complex. a Strategy for measuring the stability of the MDM2:p53 BOX-I peptide complex in the presence of small ligands. The biotinylated BOX-Ib peptide was adsorbed onto the streptavidin-coated solid phase followed by wt-MDM2, MDM2ΔLID, or MDM2S17D proteins titration in the absence or presence of the indicated inhibitors, including b the BOX-Ib peptide and c Nutlin-3. The stability of MDM2 bound to the BOX-I peptide was measured using a monoclonal antibody specific for MDM2 by chemiluminescence and is depicted in relative light units. d Strategy for measuring the stability of the MDM2:p53 BOX-V peptide complex in the presence of small ligands. The biotinylated BOX-Vb peptide was adsorbed onto the streptavidin-coated solid phase followed by wt-MDM2, MDM2ΔLID, or MDM2S17D proteins titration in the absence or presence of the indicated inhibitors, including e the BOX-Ib peptide and f Nutlin-3. The stability of MDM2 bound to the BOX-V peptide was measured using a monoclonal antibody specific for MDM2 by chemiluminescence and is depicted in relative light units
Mutation of basic residues in the hydrophobic domain is dominant over Asp17 and inactivates MDM2S17D
Together, the data allow the formation of a model, whereby the Asp17 substitution does change the conformation of MDM2 [25], but it does so by “opening” the hydrophobic pocket. One model to explain why the phospho-mimetic mutation in the MDM2 lid stabilizes MDM2:p53 interactions is that the phospho-amino acid is stabilized by interactions with a second binding site on MDM2 (as in Fig. 2b, asterisks). NMR analysis shows that, in comparison to the peptide bound form, the N-terminal domain of apo-MDM2 is more unstructured and flexible, especially the regions surrounding the peptide binding cleft [45]. The unliganded structure shows a narrow, shallow binding groove as a result of the closer association of the two sub-domains. As suggested previously [25], some conformers show MDM2 residues 18–24 can have helical character and partially occlude the shallow end of the p53 binding cleft (Fig. 8b). The remainder of the binding pocket, however, is not occupied by the remainder of the pseudo-substrate motif [25], and further, conformers show this region unwound and more displaced from the binding pocket (Fig. 2b, asterisks). This conformational heterogeneity of the pseudo-substrate motif eludes to a dynamic equilibrium between more structured conformations occupying part of the binding groove and less ordered states removed from binding pocket. The mutual occupancy of the p53 peptide and the pseudo-substrate domain is excluded (Fig. 8b), and p53 would preferentially bind to the N-terminal domain of MDM2 when the D17-pseudo-substrate domain was displaced from the binding site (Fig. 8c and d).
Fig. 8.
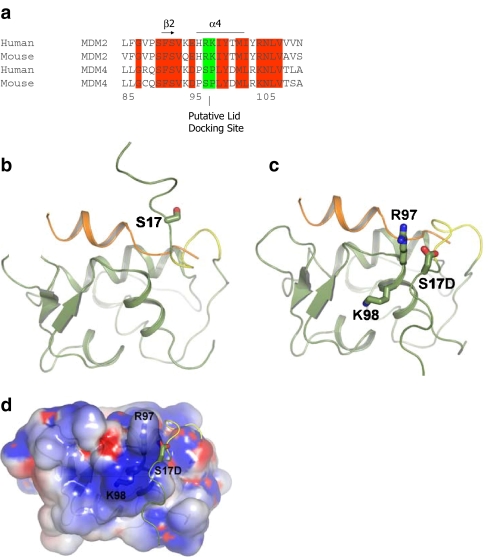
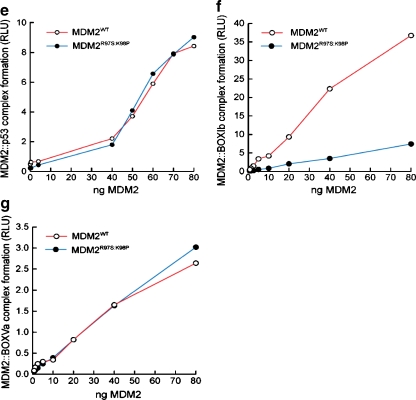
A model of lid function through stabilization of the phospho-mimetic motif on the surface of MDM2. a Multiple sequence alignment of MDM2 and MDM4 highlighting the evolutionary divergence between MDM2 and MDM4 at the potential pseudo-substrate motif basic docking site for acidic residues (i.e., Asp, phosphate) at amino acid residues 97–98. b The C-terminal region of the pseudo-substrate domain (residues 18–24; colored yellow) can have helical character and occupy the shallow end of the hydrophobic binding pocket as reported previously [25]. The p53 peptide from PDB structure 1YCR [20] is shown for comparison (colored orange). The N-terminal p53 peptide sequence and the lid cannot occupy the hydrophobic pocket simultaneously. c The pseudo-substrate motif can also exist displaced from the binding pocket with Ser17 (S17D mutation shown for illustration) in proximity to a basic region (see d) at the N-terminal of helix α2′ composed of Arg97/Lys98. Based on this, it is postulated that the S17D (mimicking phosphorylation of Ser17) mutation could stabilize the N-terminal domain of MDM2 in a conformation primed for p53 binding by forming electrostatic interactions with residues Arg97/Lys98. e–g Effects of Arg97/Lys98 mutation on wild-type MDM2 activity. e MDM2 codon 97–98 residue mutation to the MDM4 equivalent does not destabilize the MDM2:p53 tetramer complex. Increasing amounts of the indicated MDM2 protein (wt-MDM2 or MDM2R97S:K98P) were titrated into reactions where fixed amounts of tetrameric p53 were on solid phase as described previously above. The extent of MDM2 binding was quantified using an anti-MDM2 monoclonal antibody and binding stability depicted using enhanced chemiluminescence in relative light units (RLU). f The effects of MDM2 codon 97–98 residue mutation to the MDM4 equivalent on MDM2:BOX-I peptide complex stability. Increasing amounts of MDM2 protein (wt-MDM2 or the double mutant MDM2R97S:K98P) were titrated into reactions with fixed amounts of the BOX-I peptide on solid phase, as described previously above. The extent of MDM2 binding was quantified using an anti-MDM2 monoclonal antibody and binding stability depicted using enhanced chemiluminescence in relative light units (RLU). g The effects of MDM2 codon 97–98 residue mutation to the MDM4 equivalent on MDM2:BOX-V peptide complex stability. Increasing amounts of MDM2 protein (wt-MDM2 or the double mutant MDM2R97S:K98P) were titrated into reactions with fixed amounts of the BOX-V peptide on solid phase, as described previously above. The extent of MDM2 binding was quantified using an anti-MDM2 monoclonal antibody and binding stability depicted using enhanced chemiluminescence in relative light units (RLU)
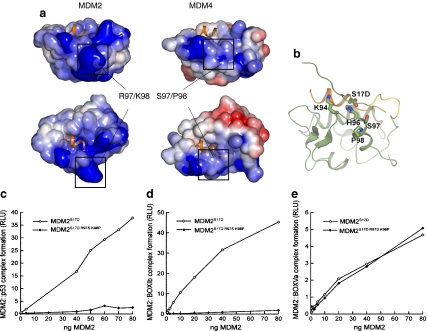
The increased ligand binding activity of MDM2S17D could thus be explained by the mutation causing a shift in equilibrium toward a more open conformation primed for peptide binding. In fact, the phospho-pseudo-substrate motif peptide of MDM2S17D has a higher binding activity for MDM2 compared to wild-type peptide [25], and the phospho-mimetic mutation may preferentially bind to a cationic region on the MDM2 surface. Inspection of the MDM2 apo-structure NMR ensemble reveals that the conformationally heterogeneous pseudo-substrate motif can be in proximity to a positively charged region at the N-terminal part of helix α2′ composed of His96, Arg97, and Lys98, which could complex acidic residues (Fig. 8c and d). Binding of Asp17 to this positively charged region may help stabilize the pseudo-substrate motif away from the binding groove and shift the equilibrium toward a conformation favorable for peptide binding by MDM2.
In regard to this equilibrium model, it is interesting to note that the His96, Arg97, and Lys98 equivalent residues of MDM4 are altered, thus removing a potentially cationic interface (Fig. 9a). This might give clues to residues that have co-evolved with the MDM2 pseudo-substrate motif, which is also absent in MDM4. To disrupt this positively charged patch (Fig. 9a), we created an MDM2 mutant which contains the Arg97Lys98 mutated to the MDM4 equivalent of Ser97Pro98 (Fig. 8a). MDM4 has been shown to have a conserved structure with MDM2 and bind p53 peptides in the same manner and with comparable affinities [34]. A titration of the wt-MDM2 and MDM2R97S:K98P demonstrated that the basic substitutions maintain an equivalent ability of MDM2 to bind to tetrameric p53 (Fig. 8e), reduced interactions with the p53 BOX-I peptide (Fig. 8f), and maintain equivalent binding activity of MDM2 for the p53 BOX-V peptide (Fig. 8g). These data indicate that the Ser97Pro98 substitutions do not disrupt fundamentally the core folding of the hydrophobic domain, not the allosteric interactions with the acidic domain of MDM2.
Fig. 9.
Inactivation of MDM2S17D by mutation of surface basic residues of MDM2. a Electrostatic potential mapped onto the solvent accessible surface. Positive- and negative-charged regions colored blue and red, respectively. Alignment figure generated with ESPript [14]. Structural figures generated with PyMol (www.pymol.org). The electrostatic calculations were performed with APBS [3] and highlighted is (left) wild-type MDM2 and (right) MDM2 with R97/K98 residues substituted with the S97/P98 MDM4 residues. b When the basic patch on MDM2 is mutated to Ser97/Pro98, the phospho-mimetic pseudo-substrate motif may bind other basic regions of the MDM2 surface such as Lys94/His96, which line the hydrophobic binding pocket and explain why the MDM2 triple mutant (MDM2S17D:R97S:K98P) binds p53 with a substantially lower activity than does the MDM2 double mutant (MDM2R97S:K98P; see c–e). c MDM2 codon 97–98 residue mutation to the MDM4 equivalent inactivates MDM2S17D as a p53 binding protein. Increasing amounts of the indicated MDM2 protein (MDM2S17D or the triple mutant MDM2S17D:R97S:K98P) were titrated into reactions where fixed amounts of tetrameric p53 were on solid phase as described previously above. The extent of MDM2 binding was quantified using an anti-MDM2 monoclonal antibody and binding stability depicted using enhanced chemiluminescence in relative light units (RLU). d The effects of MDM2 codon 97–98 residue mutation to the MDM4 equivalent on MDM2S17D:BOX-I peptide complex stability. Increasing amounts of MDM2 protein (MDM2S17D or the triple mutant MDM2S17D:R97S:K98P) were titrated into reactions with fixed amounts of the BOX-I peptide on solid phase, as described previously above. The extent of MDM2 binding was quantified using an anti-MDM2 monoclonal antibody and binding stability depicted using enhanced chemiluminescence in relative light units (RLU). e The effects of MDM2 codon 97–98 residue mutation to the MDM4 equivalent on MDM2S17D:BOX-V peptide complex stability. Increasing amounts of MDM2 protein (MDM2S17D or the triple mutant MDM2S17D:R97S:K98P) were titrated into reactions with fixed amounts of the BOX-V peptide on solid phase, as described previously above. The extent of MDM2 binding was quantified using an anti-MDM2 monoclonal antibody and binding stability depicted using enhanced chemiluminescence in relative light units (RLU)
By contrast, the enhanced binding of MDM2S17D to the p53 tetramer is completely eliminated in the MDM2S17D:R97S:K98P triple mutant (Fig. 9c). Further, the MDM2S17D:R97S:K98P triple mutant is unable to form a stable complex with the p53 BOX-I peptide (Fig. 9d), but can still bind to the BOX-V peptide (Fig. 9e). Thus, the R97S:K98P double mutations can convert MDM2S17D protein from an activated to an inhibited form, under conditions where the MDM2 R97S:K98P double mutant remains active. This inactivated triple mutant is consistent with the model that mutation of the basic patch stabilizes the phospho-mimetic lid over the hydrophobic pocket, which is not compatible with p53 binding (Fig. 9b).
Discussion
MDM2 is a multidomain E3 RING-finger ubiquitin ligase and represents a model protein with which to define mechanisms of substrate ubiquitination. The recent dual-site model for MDM2-mediated ubiquitination of p53 suggested an allosteric component to the E3 ubiquitin ligase function, which invokes MDM2 docking to two distinct sites on p53: the BOX-I transactivation domain and a conformationally flexible motif in the BOX-V domain of p53. The allostery in the N-terminal domain that operates toward the acidic domain [50] can be propagated presumably via the striking conformational flexibility of the N-terminal domain of MDM2 as defined by NMR [25, 37, 45]. The unexpected feature of the dual site model was that the N-terminal hydrophobic pocket of MDM2 can act as an allosteric ligand binding site and that ligands like Nutlin can prime MDM2 and “activate” the E3 ubiquitin ligase function of MDM2 [50]. The allosteric interactions have been further linked to the RING domain, as certain mutation in the RING can induce conformational changes in both the hydrophobic pocket and the central acidic domain [52].
A novel model can be developed that incorporates the various biochemical and biophysical studies of the flexible N-terminal domain of MDM2 and which supports an equilibrium model for pseudo-substrate motif function. As originally suggested [25] and later confirmed in the NMR structure of apo-MDM2 [40, 45], the N-terminal segment of MDM2 can partially occlude the shallow end of the p53-binding cleft (Fig. 8b); however, this motif is not well structured, and the remainder of the binding pocket remains empty, at least in the non-phosphorylated state. In this conformation, Ile19 occupies much of the space taken up by Pro27 of the p53 peptide chain [20] and as such excludes the mutual occupancy of the pseudo-substrate motif and p53. Interestingly, Ile19 is the only residue in the N-terminal region that exhibits any significant interaction with the rest of apo-MDM2 [45], forming hydrophobic contacts with His96, Arg97, and Tyr100 in the N-terminal part of α2′. In some NMR conformers, Ile19 is displaced from the site occupied by Pro27, forming a more intimate association with the N-terminal region of helix α2′. Based on this, we have proposed that the pseudo-substrate motif exists in dynamic equilibrium between states that are incompatible with or compatible with p53 peptide binding; p53 can only bind when the pseudo-substrate motif has dissociated from the hydrophobic binding site. In favor of this equilibrium, both NMR studies indicated several residues within the pseudo-substrate domain and the region surrounding helix α2′ that behaved as though in slow conformational exchange [25, 45]. Furthermore, it is interesting to note that the shorter p53 peptide, lacking residues 27–29, which share an overlapping binding site with the pseudo-substrate motif, binds to MDM2 with a ten-fold higher affinity [37].
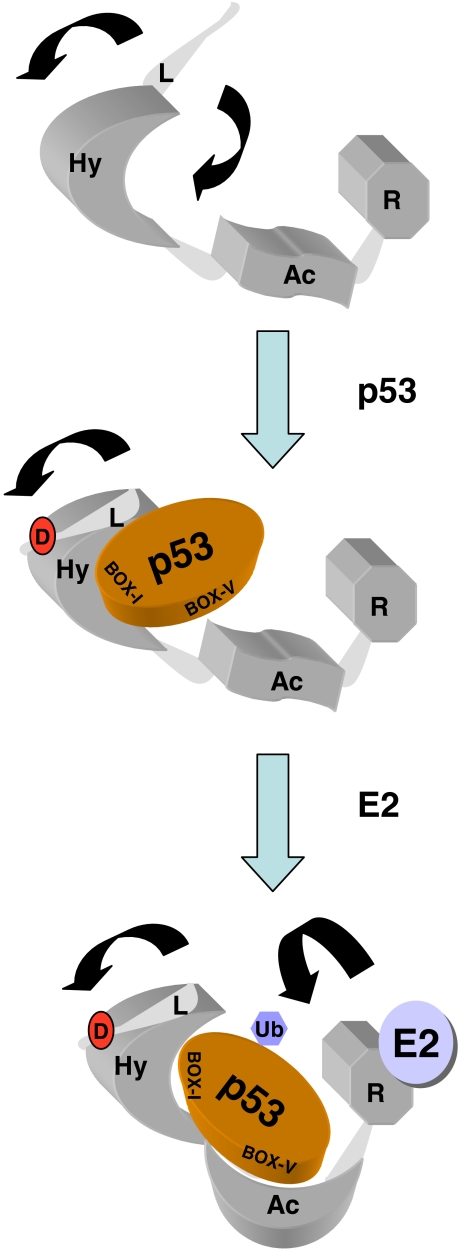
Thus, in order for p53 to bind to MDM2, the following events need to occur possibly in a concerted fashion; residues 19–25 forming the pseudo-substrate motif must dissociate from one end of the groove and be replaced by segments 27–29 of the incoming p53; the two MDM2 subdomains swing apart from one another by 3–4 Å; during the process of binding, strand β3′ is formed, completing the terminal (β1, β2, β3′) sheet that caps one end of the groove and helps to hold the two subdomains in their new more rigid conformation. However, deletion of the flexible lid destabilizes the N-terminal domain of MDM2 (this study), so this motif also has a positive role to play in the MDM2-p53 interaction. In addition to these ordered and sequential changes in MDM2-substrate binding, our current study suggests an activating model for the function of the flexible pseudo-substrate motif upon phosphorylation, which enhances displacement of the pseudo-substrate domain by altering its equilibrium, thus opening the groove to stabilize p53-peptide binding to MDM2 (Fig. 10). This conformational change upon hydrophobic pocket occupation by its ligand then appears to be propagated to the central acidic domain of MDM2 that results in enhanced MDM2 interactions with p53 (Fig. 10).
Fig. 10.
Effects of the MDM2 lid on allosteric regulation of its E3 ubiquitin ligase function. (a) MDM2 is composed of three key functional domains, including the hydrophobic pocket in the N-terminus (Hy), the acidic domain in the central portion of MDM2 (Ac), and the C-terminal RING domain (R). The N-terminus also has a flexible and unstructured pseudo-substrate motif (L), which is in equilibrium over the hydrophobic pocket (like a lid) or outwith the pocket (arrows). (b) Upon generation of the phospho-mimetic mutation in the pseudo-substrate motif, the flexible lid chain is stabilized in an equilibrium outwith the hydrophobic pocket, allowing enhanced binding to p53 at the BOX-I motif. (c) A conformational change ensues, which stabilizes the interaction between the Ac domain of MDM2 and the BOX-V motif in the DNA-binding domain of p53. This allows the E2-mediated ubiquitin transfer to the p53 substrate. The mechanisms whereby the E3:E2 interaction drives p53 substrate recognition and ubiquitin transfer is not defined nor is it known whether interactions between MDM2 and p53 occur intra or intermolecularly within the p53 tetramer. The model does not incorporate the p53 tetrameric structure since it is not known if MDM2 docking to p53 tetramers is an intra- or intermolecular mechanism
MDM2 regulation in cis by a pseudo-substrate peptide motif highlights the growing realization that many signal transduction events are modulated by relatively small and unstructured polypeptide motifs [27–29]. Although there are many well-defined globular protein domains that form folded independent compact structures, these globular domains represent only a fraction of the cellular polypeptide sequence repertoire. The remaining peptide sequences are intrinsically disordered and comprise linear motifs with weaker binding kinetics. Thus, signal transduction among many components interacting via linear peptide motifs with weaker binding kinetics can provide specific and sensitive regulation of cellular signal transduction processes. The regulation of MDM2 by such a flexible motif highlights how these motifs can impact signal transduction. Further perturbations in these linear interaction motifs, for example by covalent modifications like phosphorylation, have the potential to drive signaling changes that mediate changes in the protein–protein interaction dynamics central to signal transduction. The ability of MDM2 to be modulated by a flexible peptide motif opens the door to identify the physiological signals that regulate this conformational switch in MDM2 as well as potentially novel pathways that respond to such an activated MDM2 conformation. For example, if the MDM2-lid kinase were to be identified, it is possible that this would promote p53 protein ubiquitination and degradation in cells, and this lid-kinase pathway would therefore function as a pro-oncogenic signal in cancers. By contrast, protein phosphatases that antagonize this kinase would be predicted to function as co-tumor suppressor of p53 function by attenuating MDM2-mediated ubiquitination of p53.
Conclusions: perspectives on potential drug leads in the MDM2 pathway based on protein–protein interactions in the ubiquitination system
The molecular and structural mechanisms of MDM3 E3-RING domain-E2-mediated ubiquitination of p53 are only beginning to be defined. Complex and dynamic protein–protein interactions can be anticipated a priori. These include firstly the allosteric intradomain interactions in MDM2 itself, for example, highlighted by the fact that the RING domain plays a role in regulating the conformation of the N-terminal hydrophobic domain and the acidic domain [52]. It is striking that specific mutations in the RING domain appear to open the conformation of the acidic domain of MDM2 based on elevated intrinsic tryptophan fluorescence. This suggests that ligands that bind to the RING domain (like RNA, ATP, and E2 ubiquitin–conjugating protein) would regulate the interconversion between open and closed conformations of MDM2 within the acidic domain. This might in turn modify the type of small molecules acquired in screens for ligands that bind to the acidic domain of MDM2 and inhibit p53 ubiquitination, i.e., reconstitution of the complete E2–E3–p53 tetramer ubiquitination reaction using highly purified proteins and with specific RING-domain co-factors will likely affect the dynamics of the acidic domain conformation. The ability of the phospho-mimetic MDM2 lid to stabilize the MDM2:BOX-V peptide interaction (Fig. 10) further puts focus on the acidic domain as a key site for developing small ligands that disrupt MDM2 ubiquitination of p53.
The second protein–protein interaction to consider is the undefined effect of E3-RING domain on the E2-mediated enzymatic transfer of ubiquitin to the p53 substrate. Small molecule first generation leads have already been identified that disrupt substrate ubiquitination possibly by effecting E2 activity as well as E3 functions [54]. We do not know how E2 protein conformation is altered by E3-RING domain binding and in turn how ligand binding at the RING domain of MDM2 would alter the catalytic function of the E2 proteins. The definition of E2–E3 interfaces using purified minidomains and then using the reconstituted ubiquitination system would likely shed light on novel allosteric stages in this multicomponent ubiquitination reaction. The third protein–protein dynamic is the fact that the p53 protein is tetrameric, and although it has at least two main binding sites for MDM2, it is not clear how quaternary structure of p53 contributes to the valency of the MDM2:p53 interaction and in turn what changes occur in p53 tetramers that allow E2-dependent ubiquitination transfer to p53.
Although the N-terminal hydrophobic pocket of MDM2 is a drug binding site, drug occupation at this site does not inhibit p53 ubiquitination due to the allosteric nature of the E3 ubiquitin ligase function of MDM2. However, the three scenarios reviewed above provide biochemical approaches with which to develop peptide-mimetic leads for disrupting protein–protein interactions in the p53 substrate–E3–E2 enzymatic ubiquitination system. Additionally, if phosphorylation in vivo of the MDM2 lid will be proven to stabilize the interaction between the acidic domain of MDM2 and the BOX-V peptide in the p53 DNA-binding domain, this would drive p53 protein ubiquitination and degradation in cells. The lid-kinase pathway itself would in turn provide a novel approach for developing drug leads that attenuate MDM2-mediated ubiquitination and degradation of p53 protein.
Materials and methods
Plasmids and site-directed mutagenesis
MDM2WT and MDM2ΔLid were cloned into the Invitrogen Gateway system entry vector pDONR221 (Invitrogen) using the following primers: full-length MDM2 Fwd 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCAT GTGCAATACCAACATGTCTGTACCTACT-3′ and Rev 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAGGGGAAATAAGTTAGCACAATCAT-3′: lid deletion MDM2 Fwd 5′- GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGACCCTGGTTAGACCAAAGCCATTGCTT-3′ and Rev—same as full length MDM2. This vector was used as a template to perform site-directed mutagenesis using the in vitro mutagenesis system QuikChange (Stratagene) as directed by the manufacturer. MDM2 serine 17 was mutated to an aspartic acid using the following primers (bases which introduce amino acid change are in bold): amino acid 17 S>D Fwd 5′-GGTGCTGTAACCACCGACCAGATTCCAGCTTCG-3′ and Rev 5′CGAAGCTGGAATCTGGTCGGTGGTTACAGCACC-3′. For native expression of protein in Escherichia coli, pDONR221-MDM2 vectors were recombined with the pDEST14 vector (Invitrogen) as recommended by the manufacturer. For expression in mammalian cells, the pDONR-MDM2 vectors were recombined with the pDEST3.2 vector (Invitrogen). pDONR221-MDM2 was used as a template to PCR clone MDM2ΔLid into pCDNA3.1 with EcoR1 and Xho1 restriction sites. EcoR1 Fwd primer 5′- GCCTCGAATTCATGACCCTGGTTAGACCAAAGCCATTGCTT-3′ and Xho1 Rev 5′- GCCTCGAGCTCCTAGGGGAAATAAGTTAGCACAATCAT-3′. pCDNA3.1 full length MDM2 was subjected to site-directed mutagenesis as described before to introduce mutation at serine 17 (S>D).
Protein expression and purification
pDEST14-MDM2 constructs were overexpressed in BL-21 arabinose-inducible E. coli for 3 h at room temperature. Cells were harvested by centrifugation at 6,000×g for 10 min and frozen in liquid nitrogen. Bacterial pellet was lysed in 10% sucrose, 50 mM Tris–HCl (pH 8), 150 mM NaCl, and 150 μg/ml lysozyme and left on ice for 45 min before sonication. After sonication, 2 mM Pefabloc, 5 mM DTT, and 1 mM benzamidine was added to lysate before centrifugation at 30,000×g for 20 min. Lysate was loaded onto a fast flow SP column (GE Healthcare) equilibrated with buffer A (25 mM Hepes, pH 7.5, 10% glycerol, 1 mM benzamidine, 5 mM DTT, 50 mM KCl, and 2 mM Pefabloc). Bound protein was eluted with increasing salt concentration using buffer B (same as buffer A but 1M KCl) and MDM2 function measured in E3 ubiquitin ligase assays.
MDM2-peptide and p53 protein binding activity assays
Recombinant human MDM2 protein, ubiquitination assays, and p53 protein binding assays were developed as described previously [38, 50]. For p53-peptide binding assays, the plate was adsorbed with streptavidin overnight and washed six times with PBS-T, and biotinylated peptides were added for 1 h followed by titrating increasing amounts of MDM2 (from 3 to 200 ng). Following further washes for six times with PBS-T, wells were incubated with secondary rabbit anti-mouse horseradish peroxidase antibodies followed by further washing and ECL. The results were quantified using Fluoroskan Ascent FL equipment (Labsystems) and analyzed with Ascent Software version 2.4.1 (Labsystems). Peptides BOXIa, BOXIb, BOXVa, and BOXVb were from Chiron Mimetopes and Nutlin3a from Alexis Biochemicals. Peptide sequences are as follows: BOXIa - Biotin-SGSGPPLSQETFSDLWKLLP; BOXIb-Biotin-SGSGMPRFMDYWEGLN; BOXVa-Biotin-SGSGRNSFEVRVCACPGRD; BOXVb-Biotin-SGSGDQIMMCSMYGICKVKNIDLK.
In vitro ubiquitination assay
Reactions contained 25 mM HEPES (pH 8.0), 10 mM MgCl2, 4 mM ATP, 0.5 mM DTT, 0.05% (v/v) Triton X-100, 0.25 mM benzamidine, 10 mM creatine phosphate, 3.5 units/ml creatine kinase, ubiquitin (2 μg), E1 (100 nM), E2 (1 μM), and p53 (0.5 μg). Reactions were assembled on ice by adding, last, purified MDM2, MDM2Δlid, or MDM2S17D at various concentrations (3–200 ng), followed by incubations for 15 min at 30 °C, and analyzed with 4–12% NuPAGE gels in a MOPS buffer system (Invitrogen) followed by immunoblot. For temperature gradient, MDM2 was subjected to heat treatment for 5 min prior to addition to ubiquitination reaction.
Cell culture and purification of his-ubiquitin conjugates
H1299 cells were grown at 37 °C in RPMI with 10% FBS and 5% CO2. p53−/−MDM2−/− mouse embryonic fibroblasts were grown in DMEM with 10% (v/v) FBS and 10% CO2. Transient transfections were carried out as described [10]. Cells were lysed in NP40 buffer (25 mM HEPES, pH 7.5, 0.1% NP40, 150 mM KCl, 5 mM DTT, and 50 mM NaF) and analyzed by 4–12% NuPAGE/immunoblot. His-ubiquitin conjugates were purified as previously described [53].
Acknowledgments
This work was supported by Programme Grants to KLB (C377/A6355) and TRH (C483/A6354) from the Cancer Research UK. EW is funded by a Cancer Research UK PhD studentship (C483/A5547).
References
- 1.Adams J, Chen ZP, Denderen BJ, Morton CJ, Parker MW, Witters LA, Stapleton D, Kemp BE. Protein Sci. 2004;13:155. doi: 10.1110/ps.03340004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arva NC, Gopen TR, Talbot KE, Campbell LE, Chicas A, White DE, Bond GL, Levine AJ, Bargonetti J. J Biol Chem. 2005;280:26776. doi: 10.1074/jbc.M505203200. [DOI] [PubMed] [Google Scholar]
- 3.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc Natl Acad Sci U S A. 2001;98:10037. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottger V, Bottger A, Howard SF, Picksley SM, Chene P, Garcia-Echeverria C, Hochkeppel HK, Lane DP. Oncogene. 1996;13:2141. [PubMed] [Google Scholar]
- 5.Burch LR, Midgley CA, Currie RA, Lane DP, Hupp TR. FEBS Lett. 2000;472:93. doi: 10.1016/S0014-5793(00)01427-7. [DOI] [PubMed] [Google Scholar]
- 6.Burch L, Shimizu H, Smith A, Patterson C, Hupp TR. J Mol Biol. 2004;337:129. doi: 10.1016/j.jmb.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 7.Candeias MM, Malbert-Colas L, Powell DJ, Daskalogianni C, Maslon MM, Naski N, Bourougaa K, Calvo F, Fahraeus R. Nat Cell Biol. 2008;10:1098. doi: 10.1038/ncb1770. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Marechal V, Levine AJ. Mol Cell Biol. 1993;13:4107. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig AL, Chrystal JA, Fraser JA, Sphyris N, Lin Y, Harrison BJ, Scott MT, Dornreiter I, Hupp TR. Mol Cell Biol. 2007;27:3542. doi: 10.1128/MCB.01595-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dornan D, Hupp TR. EMBO Rep. 2001;2:139. doi: 10.1093/embo-reports/kve025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyson HJ, Wright PE. Curr Opin Struct Biol. 2002;12:54. doi: 10.1016/S0959-440X(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 12.Elenbaas B, Dobbelstein M, Roth J, Shenk T, Levine AJ. Mol Med. 1996;2:439. [PMC free article] [PubMed] [Google Scholar]
- 13.Gannon JV, Greaves R, Iggo R, Lane DP. EMBO J. 1990;9:1595. doi: 10.1002/j.1460-2075.1990.tb08279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouet P, Courcelle E, Stuart DI, Metoz F. Bioinformatics. 1999;15:305. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 15.Haupt Y, Maya R, Kazaz A, Oren M. Nature. 1997;387:296. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 16.Kobe B, Kemp BE. Nature. 1999;402:373. doi: 10.1038/46478. [DOI] [PubMed] [Google Scholar]
- 17.Kobe B, Heierhorst J, Kemp BE. Adv Second Messenger Phosphoprotein Res. 1997;31:29. doi: 10.1016/s1040-7952(97)80006-7. [DOI] [PubMed] [Google Scholar]
- 18.Kostic M, Matt T, Martinez-Yamout MA, Dyson HJ, Wright PE. J Mol Biol. 2006;363:433. doi: 10.1016/j.jmb.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 19.Kubbutat MH, Jones SN, Vousden KH. Nature. 1997;387:299. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 20.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Science. 1996;274:948. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 21.Lai Z, Freedman DA, Levine AJ, McLendon GL. Biochemistry. 1998;37:17005. doi: 10.1021/bi980596r. [DOI] [PubMed] [Google Scholar]
- 22.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Cell Death Differ. 2008;15:841. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 23.Maclaine NJ, Oster B, Bundgaard B, Fraser JA, Buckner C, Lazo PA, Meek DW, Hollsberg P, Hupp TR. J Biol Chem. 2008;283:28563. doi: 10.1074/jbc.M804433200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marechal V, Elenbaas B, Piette J, Nicolas JC, Levine AJ. Mol Cell Biol. 1994;14:7414. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCoy MA, Gesell JJ, Senior MM, Wyss DF. Proc Natl Acad Sci U S A. 2003;100:1645. doi: 10.1073/pnas.0334477100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. Cell. 1992;69:1237. doi: 10.1016/0092-8674(92)90644-R. [DOI] [PubMed] [Google Scholar]
- 27.Neduva V, Russell RB. FEBS Lett. 2005;579:3342. doi: 10.1016/j.febslet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Neduva V, Russell RB. Nucleic Acids Res. 2006;34:W350. doi: 10.1093/nar/gkl159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neduva V, Russell RB. Curr Opin Biotechnol. 2006;17:465. doi: 10.1016/j.copbio.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Nenutil R, Smardova J, Pavlova S, Hanzelkova Z, Muller P, Fabian P, Hrstka R, Janotova P, Radina M, Lane D, Coates P, Vojtesek B. J Pathol. 2005;207:251. doi: 10.1002/path.1838. [DOI] [PubMed] [Google Scholar]
- 31.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mol Cell. 2008;32:180. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Passmore LA, Barford D. Biochem J. 2004;379:513–525. doi: 10.1042/BJ20040198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersson S, Kelleher M, Pion E, Wallace M, Ball KL. Role of MDM2 acid domain interactions in recognition and ubiquitination of the transcription factor IRF-2. Biochem J. 2009;418:575–585. doi: 10.1042/BJ20082087. [DOI] [PubMed] [Google Scholar]
- 34.Popowicz GM, Czarna A, Rothweiler U, Sszwagierczak A, Krajewski M, Weber L, Holak TA. Cell Cycle. 2007;6:2386. doi: 10.4161/cc.6.19.4740. [DOI] [PubMed] [Google Scholar]
- 35.Poyurovsky MV, Jacq X, Ma C, Karni-Schmidt O, Parker PJ, Chalfie M, Manley JL, Prives C. Mol Cell. 2003;12:875. doi: 10.1016/S1097-2765(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 36.Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, Prives C. EMBO J. 2007;26:90. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schon O, Friedler A, Freund S, Fersht AR. J Mol Biol. 2004;336:197. doi: 10.1016/j.jmb.2003.11.051. [DOI] [PubMed] [Google Scholar]
- 38.Shimizu H, Burch LR, Smith AJ, Dornan D, Wallace M, Ball KL, Hupp TR. J Biol Chem. 2002;277:28446. doi: 10.1074/jbc.M202296200. [DOI] [PubMed] [Google Scholar]
- 39.Shimizu H, Saliba D, Wallace M, Finlan L, Langridge-Smith PR, Hupp TR. Biochem J. 2006;397:355. doi: 10.1042/BJ20051521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Showalter SA, Bruschweiler-Li L, Johnson E, Zhang F, Bruschweiler R. J Am Chem Soc. 2008;130:6472. doi: 10.1021/ja800201j. [DOI] [PubMed] [Google Scholar]
- 41.Stevens C, Pettersson S, Wawrzynow B, Wallace M, Ball K, Zylicz A, Hupp TR. FEBS J. 2008;275:4875. doi: 10.1111/j.1742-4658.2008.06627.x. [DOI] [PubMed] [Google Scholar]
- 42.Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, Pelt CS, Lozano G. Genes Dev. 2008;22:1337–1344. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thut CJ, Goodrich JA, Tjian R. Genes Dev. 1997;11:1974. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toledo F, Wahl GM. MDM2 and MDM4: p53 regulators as targets in anticancer therapy. Int J Biochem Cell Biol. 2007;39:1476. doi: 10.1016/j.biocel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uhrinova S, Uhrin D, Powers H, Watt K, Zheleva D, Fischer P, McInnes C, Barlow PN. J Mol Biol. 2005;350:587. doi: 10.1016/j.jmb.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 46.Uldrijan S, Pannekoek WJ, Vousden KH. EMBO J. 2007;26:102. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. Science. 2004;303:844. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 48.Vojtesek B, Dolezalova H, Lauerova L, Svitakova M, Havlis P, Kovarik J, Midgley CA, Lane DP. Oncogene. 1995;10:389. [PubMed] [Google Scholar]
- 49.Vousden KH, Lane DP. Nat Rev Mol Cell Biol. 2007;8:275. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 50.Wallace M, Worrall E, Pettersson S, Hupp TR, Ball KL. Mol Cell. 2006;23:251. doi: 10.1016/j.molcel.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 51.Wawrzynow B, Zylicz A, Wallace M, Hupp T, Zylicz M. J Biol Chem. 2007;282:32603. doi: 10.1074/jbc.M702767200. [DOI] [PubMed] [Google Scholar]
- 52.Wawrzynow B, Pettersson S, Zylicz A, Bramham J, Worrall E, Hupp TR, Ball KL (2009) J Biol Chem 284:11517–11530 [DOI] [PMC free article] [PubMed]
- 53.Xirodimas D, Saville MK, Edling C, Lane DP, Lain S. Oncogene. 2001;20:4972. doi: 10.1038/sj.onc.1204656. [DOI] [PubMed] [Google Scholar]
- 54.Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV, Safiran YJ, Oberoi P, Kenten JH, Phillips AC, Weissman AM, Vousden KH. Cancer Cell. 2005;7:547. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 55.Yu GW, Rudiger S, Veprintsev D, Freund S, Fernandez-Fernandez MR, Fersht AR. Proc Natl Acad Sci U S A. 2006;103:1227. doi: 10.1073/pnas.0510343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu GW, Allen MD, Andreeva A, Fersht AR, Bycroft M. Protein Sci. 2006;15:384. doi: 10.1110/ps.051927306. [DOI] [PMC free article] [PubMed] [Google Scholar]