Abstract
The host innate immune response, including the production of type-I IFN, represents the primary line of defense against invading viral pathogens. Of the hundreds of IFN-stimulated genes (ISGs) discovered to date, ISG15 was one of the first identified and shown to encode a ubiquitin-like protein that functions, in part, as a modifier of protein function. Evidence implicating ISG15 as an innate immune protein with broad-spectrum antiviral activity continues to accumulate rapidly. This review will summarize recent findings on the innate antiviral activity of ISG15, with a focus on the interplay between ubiquitination and ISGylation pathways resulting in modulation of RNA virus assembly/budding. Indeed, ubiquitination is known to be proviral for some RNA viruses, whereas the parallel ISGylation pathway is known to be antiviral. A better understanding of the antiviral activities of ISG15 will enhance our fundamental knowledge of host innate responses to viral pathogens and may provide insight useful for the development of novel therapeutic approaches designed to enhance the immune response against such pathogens.
Key Words: Antiviral, Budding, Innate immunity, Interferon-stimulated gene 15, ISGylation, Ubiquitin, Ubiquitination
Introduction
The host innate immune response represents a critical, initial line of defense against invading pathogens, and the magnitude of this response can influence disease progression. The IFN system is a key component and complex regulator of innate immunity. Type I IFNs function primarily to promote an antiviral state in an infected host by inducing signaling through the IFN receptor to stimulate expression of a plethora of gene products that function in host defense. One of the early induced IFN-stimulated genes (ISGs), first identified more than 20 years ago, is a small ubiquitin-like protein termed ISG15 [1,2,3]. This review will summarize several recent reports that have contributed to our fundamental understanding of ISG15 antiviral activity, and which have provided novel insights into the mechanism(s) of the antiviral action exhibited by this broad-spectrum, IFN-induced, innate immune protein.
Ubiquitination versus ISGylation
Ubiquitin and ubiquitin-like proteins (e.g. ISG15 and SUMO) are small molecules expressed in eukaryotes that are conjugated to target proteins, often resulting in modulation of the target protein's stability/function/localization [4]. The mechanisms by which proteins are ubiquitinated and ISGylated have many common features, and these protein modifiers regulate a diverse array of biological processes. These protein modification pathways have been reviewed in detail elsewhere [5, 6] (fig. 1), and thus will only be summarized here. Both ubiquitination and ISGylation result in covalent modification of target proteins following a series of enzymatic reactions that occur in a stepwise fashion (fig. 1). The process of ubiquitination begins with protease cleavage of precursor ubiquitin molecules at their C-termini to reveal the di-glycine motif. In the presence of ATP, the Gly-Gly motif is then adenylated by the ubiquitin activating enzyme (E1). The ubiquitin moiety is then transferred to a cysteine residue on a ubiquitin conjugating enzyme (E2). Finally, the E2 enzyme in conjunction with a ubiquitin ligase (E3), which recognizes the specific target protein, transfers the ubiquitin moiety typically onto the ∊-NH2 group of a lysine residue of the substrate. In addition to lysine residues, other amino acids (e.g. serine/threonine) and the extreme N-termini of target proteins can potentially serve as target sites for ubiquitination and/or influence the ubiquitination process [7,8,9]. Proteins that are modified by poly-ubiquitination are often destined for degradation by the 26S proteasome. For example, this process is crucial in immune cells where protein ubiquitination regulates protein signaling pathways, innate and adaptive immune responses to pathogens, and expression of surface receptors [4, 5, 10, 11]. In contrast, mono-ubiquitination of proteins does not result in degradation, but rather can serve to modulate protein function, localization and/or sorting within the cell [4, 5, 11].
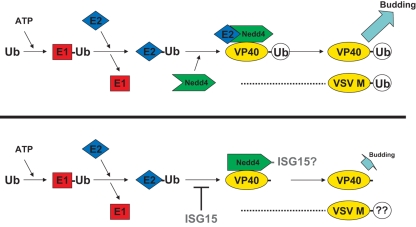
Fig. 1.
Schematic diagram of the enzymatic cascade leading to ubiquitination of Ebola VP40 by Nedd4 E3 ligase (top half), and impairment of this pathway by ISG15 at the stage of ubiqutin (Ub) transfer from E2 to Nedd4 (bottom half). E1 = Ub activating enzyme; E2 = Ub conjugating enzyme; E3 = Ub ligase (Nedd4). Efficient VP40-monoUb leads to optimal budding of VP40 VLPs (top), whereas an impairment of VP40-monoUb by expression of ISG15 leads to decreased budding of VP40 VLPs (bottom). Since VSV buds in a manner similar to that of Ebola, and since VSV M is also mono-ubiquitinated by host Nedd4, it will be of interest to determine whether ISG15 can inhibit budding of VSV by a similar mechanism.
Unlike ubiquitination, ISGylation is an IFN-stimulated and regulated process that appears to mimic mono-ubiquitination functions, such as modulation of enzymatic activity, rather than the poly-ubiquitination function and protein degradation [12,13,14,15,16]. Like ubiquitination, ISGylation involves a cascade of enzymatic reactions involving E1, E2, and E3 enzymes. Briefly, ISG15 is synthesized as precursor protein (165 aa) that is like ubiquitin, subsequently cleaved to reveal di-glycine residues at its C-terminus in the form of a LRLRGG motif. This GG motif is adenylated in the presence of ATP, and ISG15 is then transferred sequentially from E1, E2 and E3 enzymes to its final destination on a lysine residue within the target substrate [11]. Because of the many similarities of the ubiquitination and ISGylation pathways, it is not surprising that there is some degree of overlap and competition between these pathways [17,18,19,20,21]. While the E1 enzymes are specific for ISGylation and ubiquitination, some of the enzymes that function in the ubiquitination pathway (e.g. E2 enzyme UbcH8) have a similar function in the ISGylation pathway [19, 21]. One of the features that is common to both pathways is that both ubiquitination and ISGylation are reversible. De-ubiquitinating enzymes and de-ISGylation enzymes (e.g. UBP43) function to cleave ubiquitin and/or ISG15 moieties from conjugated proteins [22,23,24,25]. De-ubiquitination and de-ISGylation provide an additional level of regulation in these pathways to maintain homeostasis [25].
Ubiquitination and Virus Budding
Over the last decade, numerous reports have demonstrated that the host ubiquitination machinery is linked to the process of budding or egress for many RNA virus families [for review see [26]]. For example, ubiquitin was first implicated in regulating Gag-mediated budding of retroviruses, and subsequently linked to VP40-mediated budding of Ebola virus-like particles (VLPs) [27,28,29,30]. Several lines of evidence linked ubiquitin with virus egress, including: (1) the presence of free, excess ubiquitin within budding virions [31], (2) detection of mono-ubiquitinated forms of viral matrix proteins [30,32,33,34,35,36,37,38] and (3) disruption of virus budding in the presence of proteasome inhibitors that deplete free ubiquitin in the cell [27,28,29, 34, 35, 39, 40]. In addition, the PPxY-type L-domain conserved within the matrix proteins of several RNA viruses is known to mediate interactions with WW-domains present within HECT E3 ubiquitin ligases, such as Nedd4, to facilitate budding either by mono-ubiquitination of the viral matrix protein, or by ubiquitination and/or recruitment of other host proteins (e.g. endosomal sorting complex required for transport (ESCRT) pathway) involved in virus budding [30, 35, 36,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58]. In sum, ubiquitination has been linked or at least implicated to play a role in the budding process of retroviruses, rhabdoviruses, filoviruses, arenaviruses, paramyxoviruses and reoviruses [26]. Thus, small molecule inhibitors designed to block the ubiquitination process and/or viral L-domain function may have broad-spectrum antiviral activity.
ISG15 Has Broad-Spectrum Antiviral Activity
While ubiquitin appears to be a broad-spectrum enhancer of virus production, ISG15 on the other hand appears to be a broad-spectrum inhibitor of virus production. Antiviral activity associated with protein ISGylation in vitro and/or in vivo has been reported for both DNA and RNA viruses, including influenza A and B, Sindbis, hepatitis B, herpes simplex type-1, murine γ-herpesvirus, vesicular stomatitis virus, lymphocytic choriomeningitis virus, respiratory syncytial virus, HIV-1 and Ebola virus [59,60,61,62,63,64]. Recent studies have provided some new and important insights into the molecular mechanisms by which ISG15 functions as an innate antiviral protein with broad-spectrum activity in vitro and in vivo. Results from several of the most recent studies will be summarized below.
ISG15 and Sindbis Virus
First, an elegant paper by Lenschow et al. [65] was of significance as it helped to prove that protein ISGylation and ISG15 possessed broad-spectrum antiviral activity in an animal model of infection, and the findings underscored the importance of employing both in vitro and in vivo models for analyses of antiviral mechanisms. One of the major findings was that ISG15–/– mice were more susceptible to infection with influenza A/WSN/33, influenza B/Lee/40, Sindbis, herpes simplex type 1, and murine γ-herpesvirus [65]. Importantly, the ability of ISG15 to be conjugated to target proteins appeared to be necessary for the observed antiviral effect, since Sindbis virus infection could be recovered by overexpressing wild-type ISG15, but not by overexpressing a nonconjugating mutant of ISG15 [65]. Although ISG15 conjugation was important for the observed antiviral effect, the target substrate(s) for ISGylation was not identified. Since ISG15 is now known to have more that 150 targets [11], the antiviral effect may be due to ISGylation of either a viral protein, a host protein, or both. Knowing the identity of the target protein(s) will be instrumental in elucidating the mechanism of action of ISG15 and for the possible development of future antiviral strategies.
In a more recent report, the same group identified arginine-151 in mouse ISG15 as playing a key role in protein ISGylation by mediating an interaction with UbE1L [66]. Indeed, an R151A mutation attenuated the ability of ISG15 to inhibit replication of Sindbis virus in IFN-α/βR(–/–) mice, and mice lacking this the UbE1L enzyme were highly susceptible to Sindbis virus infection [66]. In sum, their results demonstrated that protein conjugation is key for the antiviral effects of ISG15 in this animal model of Sindbis virus infection [66].
ISG15 and HIV-1
IFN was shown to inhibit retroviral and lentiviral virion release [67, 68], and ISG15 was implicated in IFN-mediated inhibition of HIV-1 [69, 70]. As ubiquitination of Gag protein was shown to play a role in HIV-1 assembly and budding, it seemed logical to hypothesize that ISG15 could interfere with ubiquitination of Gag and consequently disrupt this late stage of virus replication. Indeed, recent findings indicated that this hypothesis was correct. Okumura et al. [71] were the first to clearly demonstrate that expression of ISG15 mimicked the antiviral properties of IFN and blocked egress of HIV-1 from infected cells. The authors found that expression of ISG15 appeared to block the ubiquitination process that normally enhances HIV-1 Gag-mediated release of infectious virus [71]. Interestingly, ISG15 did not affect synthesis of viral proteins in the infected cell and, unlike the findings with other viral systems [65, 72], conjugation of ISG15 to either viral (HIV-1 Gag), or host (tsg101) proteins involved in budding was not evident [71]. However, ISG15 expression did disrupt the interaction between host tsg101 and the p6 domain of HIV-1 Gag. This virus-host interaction has been shown previously to be crucial for efficient budding of HIV-1. To further prove that the inhibitory effect on budding was specific for ISG15, the authors demonstrated that ISG15-specific siRNAs reversed the IFN-mediated inhibitory effect on HIV-1 release [71]. While the precise mechanism by which ISG15 interferes with the budding process of HIV-1 remains to be defined, the authors postulated that the decrease in ubiquitination of Gag and tsg101 may modify the binding affinity of this interaction or destabilize this complex. In addition, the authors noted that since components of the ubiquitination pathway were known to be targets for ISGylation [20, 73, 74], ISG15 may target one or more of the E1, E2, or E3 enzymes involved in HIV-1 egress. In sum, these findings were of significance in that: (1) they revealed an IFN-induced innate immune response that could target a late stage of HIV-1 replication, and (2) they suggested that ISG15 may have broad-spectrum antiviral activity against other RNA viruses that utilize L-domain/ubiquitination pathways for efficient egress.
ISG15 and Ebola VLP Budding
Most recently, independent studies from two groups revealed that ISG15 could inhibit budding mediated by the VP40 matrix protein of Ebola virus, and the mechanism of inhibition involved disruption of Nedd4-mediated ubiquitination of Ebola VP40 [75, 76] (fig. 1). Like HIV-1, the host ubiquitination pathway and E3 ubiquitin ligase Nedd4 have been linked to budding of negative-sense RNA virus such as Ebola virus and vesicular stomatitis virus (VSV) [30, 35, 39, 53, 55, 56, 77]. In a prior publication, Malakhov et al. [23] identified Nedd4 as a binding partner for Ubp43, an ISG15-deconjugating enzyme. Thus, Malakhova and Zhang [76] sought to determine whether Nedd4 could be targeted by ISG15 and whether this potential interaction would negatively regulate Nedd4 ligase activity. Indeed, the authors found that expression of ISG15 and the ISGylation system led to an overall decrease in Nedd4-ligase activity and Nedd4-mediated ubiquitination [76]. They demonstrated further that free ISG15 was sufficient to impair Nedd4 ligase activity both in vitro and in vivo [76]. Importantly, they determined that the molecular mechanism of ISG15-mediated inhibition of Nedd4 function was due to an abrogation of ubiquitin transfer from the E2 enzyme to the active site of Nedd4 [76], thus preventing Nedd4-mediated ubiquitination of target proteins.
As further proof of this novel mechanism, Malakhova and Zhang [76] utilized the Ebola VP40 VLP budding assay as a model system to assess Nedd4 function in the absence or presence of ISG15 and the ISGylation system. Indeed, they found that efficient budding of Ebola VP40 VLPs, which is dependent in part on Nedd4 function, was impaired in the presence of ISG15. The decrease in VP40 VLP release correlated well with an observed decrease in the detection of ubiquitinated forms of VP40. Lastly, budding of Ebola VP40 VLPs was shown to be enhanced in ISG15–/– knockout cells compared to that from normal murine embryonic fibroblasts [76], suggesting that endogenous levels of ISG15 can limit budding in unmanipulated cells.
In a simultaneous and independent report, our studies reached many of the same conclusions described above using both similar and distinct experimental approaches [75]. Prior to this study, we were one of the first groups to suggest that the ubiquitination process and Nedd4 ligase may be important for budding of Ebola VP40 VLPs, and that the PPxY-type L-domain within the VP40 protein was necessary for mediating interactions with the various WW-domains present within Nedd4 [30, 56]. Our main objective in this area of investigation was to elucidate the mechanism of Ebola VP40 VLP budding and host interactions at the molecular level in order to develop novel strategies or antiviral therapies to inhibit the budding process. In agreement with results of Malakhova and Zhang [76], we found that expression of ISG15 in the absence or presence of the ISGylation system (UbE1L and UbcH8) was able to inhibit budding of Ebola VP40 VLPs [75]. We have demonstrated not only that ISG15 interacted with Nedd4 ligase to inhibit ubiquitination of VP40, but also that ISG15-mediated inhibition of VP40 budding was dependent on the presence of VP40 L-domains. Indeed, budding of an L-domain deletion mutant of VP40 known to be insensitive to Nedd4-mediated ubiquitination, was also insensitive to ISG15-mediated inhibition of budding [75]. Lastly, inhibition of Ebola VP40 VLP budding by ISG15 was evident only in the presence of wild-type Nedd4, and not in the presence of a dominant-negative, enzymatically inactive mutant of Nedd4 [75]. Together, these findings revealed a previously undescribed mechanism of antiviral activity for ISG15.
Interestingly, like Malakhova and Zhang [76], our group was able to demonstrate that expression of ISG15 also had an inhibitory effect on budding of VSV; a rhabdovirus that possesses a similar PPxY-type L-domain that interacts with Nedd4 ligase to promote efficient virion budding [75, 76]. Thus, ISG15-mediated inhibition of budding may not be limited to Ebola. We found that overexpression of ISG15 resulted in titers of wild-type VSV that were on average 10-fold lower than those measured in the absence of ISG15 [75]. Moreover, overexpression of ISG15 was found to have virtually no inhibitory effect on budding of VSV mutant PY>A4, a L-domain mutant of VSV that is insensitive to Nedd4-mediated ubiquitination [75]. Thus, viruses that utilize the host ubiquitination machinery for efficient egress may be susceptible to the inhibitory effect of ISG15.
ISG15 and Influenza Virus
As the above studies highlight, the antiviral activity of ISG15 against different viruses can be both dependent or independent of ISG15 conjugation. In a recent study by Lai et al. [78], the authors demonstrate that antiviral activity of ISG15 against influenza B virus requires conjugation of ISG15 to target proteins. Although these authors had shown previously that mice lacking ISG15 were highly susceptible to influenza B virus infection, it was unclear whether ISG15 conjugation to target proteins was important for this antiviral activity. Using mice lacking the E1 activating enzyme for ISG15 conjugation (UbE1L–/– mice), the authors observed a 2–3 log increase in titers of influenza B virus in the lungs of these mice at 3 and 6 days after infection compared to those observed in wild-type mice [78]. In addition, the kinetics of lethality and survival observed in the UbE1L–/– mice infected with influenza B virus were essentially identical to those observed in ISG15–/– mice [78]. The authors concluded that the antiviral activity of ISG15 against influenza B virus in this in vivo model of infection is dependent on ISG15 conjugation; however, the identity of proteins targeted for ISGylation remains to be determined.
Viruses Fight Back
Viruses have evolved many elegant strategies to counteract host immune responses, particularly IFN-related pathways. Interestingly, several reports suggest that some viruses possess the ability to specifically counteract the antiviral effects of ISG15 and/or ISGylation. For example, one of the earliest reports by Yuan and Krug [72], demonstrated that the NS1 protein of influenza B virus prevents the activation of ISG15 by the E1 enzyme UbE1L, thus preventing ISG15 conjugation to target proteins. More recently, a unique strategy to inhibit both ubiquitination and ISGylation pathways was identified in nairoviruses and arteriviruses [79]. These viruses were shown to encode ovarian tumor domain-containing proteases capable of hydrolyzing both ubiquitin and ISG15 from target proteins [79]. Thus, the nairo- and arteriviruses appear to have developed a strategy to antagonize the antiviral effects of ISG15 [79]. Finally, in a comprehensive report by Guerra et al. [80], the authors not only demonstrate that ISG15 possesses antiviral activity against vaccinia virus, but also show that viral E3 protein can bind to ISG15 and disrupt its antiviral activity. Thus, a vaccinia virus strain lacking E3 (VVDeltaE3L) was unable to replicate in ISG15+/+ cells, but was able to replicate in ISG15-deficient cells [80]. Moreover, infection of ISG15–/– mice with VVDeltaE3L resulted in significant disease and mortality, which was not observed in ISG15+/+ mice infected with VVDeltaE3L [80]. The ability of other viruses to specifically target and abrogate ISGylation pathways as a means of immune evasion is of interest and awaits further investigation.
Summary and Future Directions
The host innate immune response is a critical first line of defense against invading viral pathogens. A better understanding of the distinct innate immune responses and the interplay between the virus and host is important for our understanding of immunopathogenesis, vaccine design, and development of new therapeutic strategies. The ISGs represent a vast array of proteins with a complex series of functions and outcomes. However, rapid progress has been made into this dynamic area of innate immunity, and the reports summarized here highlight some recent and insightful findings on the antiviral activity of ISG15. Despite these advancements, many gaps in our knowledge of the molecular mechanisms of ISG15 antiviral activity remain to be addressed. Some areas for further investigation include: (1) identifying the viral and/or host target proteins for ISG15 in viral infections where ISG15 conjugation is required for its antiviral activity; (2) determining whether expression and/or conjugation of ISG15 impairs replication, budding, and pathogenesis of infectious Ebola virus in cell culture and in the ISG15 animal models of infection; (3) determining whether ISG15 inhibits assembly/budding of additional L-domain-containing RNA viruses (e.g. Marburg virus) that likely utilize ubiquitination pathways for efficient replication and spread; (4) development of novel strategies to modulate the ISGylation system in an effort to enhance the innate immune response and more effectively combat viral infections, and (5) investigating the functional overlap of cellular ubiquitination and ISGylation pathways, the potential regulatory cross talk between these pathways, and their role in immune responses to invading pathogens.
Acknowledgements
This work was supported in part by NIH grant AI-077014-02 to R.N.H and by AI-19737-23 to P.M.P. The authors thank Leslie King for critically reviewing the manuscript.
References
- 1.Korant BD, Blomstrom DC, Jonak GJ, Knight E., Jr Interferon-induced proteins. Purification and characterization of a 15,000-dalton protein from human and bovine cells induced by interferon. J Biol Chem. 1984;259:14835–14839. [PubMed] [Google Scholar]
- 2.Blomstrom DC, Fahey D, Kutny R, Korant BD, Knight E., Jr Molecular characterization of the interferon-induced 15-kDa protein: molecular cloning and nucleotide and amino acid sequence. J Biol Chem. 1986;261:8811–8816. [PubMed] [Google Scholar]
- 3.Haas AL, Ahrens P, Bright PM, Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J Biol Chem. 1987;262:11315–11323. [PubMed] [Google Scholar]
- 4.Herrmann J, Lerman LO, Lerman A. Ubiquitin and ubiquitin-like proteins in protein regulation. Circ Res. 2007;100:1276–1291. doi: 10.1161/01.RES.0000264500.11888.f0. [DOI] [PubMed] [Google Scholar]
- 5.Petroski MD. The ubiquitin system, disease, and drug discovery. BMC Biochem. 2008;9(suppl 1):S7. doi: 10.1186/1471-2091-9-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staub O. Ubiquitylation and isgylation: overlapping enzymatic cascades do the job. Sci STKE. 2004;2004:pe43. doi: 10.1126/stke.2452004pe43. [DOI] [PubMed] [Google Scholar]
- 7.Amir RE, Haecker H, Karin M, Ciechanover A. Mechanism of processing of the NF-kappa B2 p100 precursor: identification of the specific polyubiquitin chain-anchoring lysine residue and analysis of the role of NEDD8-modification on the SCF(beta-TrCP) ubiquitin ligase. Oncogene. 2004;23:2540–2547. doi: 10.1038/sj.onc.1207366. [DOI] [PubMed] [Google Scholar]
- 8.Dada LA, Welch LC, Zhou G, Ben-Saadon R, Ciechanover A, Sznajder JI. Phosphorylation and ubiquitination are necessary for Na,K-ATPase endocytosis during hypoxia. Cell Signal. 2007;19:1893–1898. doi: 10.1016/j.cellsig.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadeh R, Breitschopf K, Bercovich B, Zoabi M, Kravtsova-Ivantsiv Y, Kornitzer D, Schwartz A, Ciechanover A. The N-terminal domain of MyoD is necessary and sufficient for its nuclear localization-dependent degradation by the ubiquitin system. Proc Natl Acad Sci USA. 2008;105:15690–15695. doi: 10.1073/pnas.0808373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bibeau-Poirier A, Servant MJ. Roles of ubiquitination in pattern-recognition receptors and type I interferon receptor signaling. Cytokine. 2008;43:359–367. doi: 10.1016/j.cyto.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 11.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 12.Dao CT, Zhang DE. ISG15: a ubiquitin-like enigma. Front Biosci. 2005;10:2701–2722. doi: 10.2741/1730. [DOI] [PubMed] [Google Scholar]
- 13.Kim KI, Zhang DE. ISG15, not just another ubiquitin-like protein. Biochem Biophys Res Commun. 2003;307:431–434. doi: 10.1016/s0006-291x(03)01216-6. [DOI] [PubMed] [Google Scholar]
- 14.Ritchie KJ, Zhang DE. ISG15: the immunological kin of ubiquitin. Semin Cell Dev Biol. 2004;15:237–246. doi: 10.1016/j.semcdb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Martensen PM, Justesen J. Small ISGs coming forward. J Interferon Cytokine Res. 2004;24:1–19. doi: 10.1089/107999004772719864. [DOI] [PubMed] [Google Scholar]
- 16.Andersen JB, Hassel BA. The interferon regulated ubiquitin-like protein, ISG15, in tumorigenesis: friend or foe? Cytokine Growth Factor Rev. 2006;17:411–421. doi: 10.1016/j.cytogfr.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Zou W, Zhang DE. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J Biol Chem. 2006;281:3989–3994. doi: 10.1074/jbc.M510787200. [DOI] [PubMed] [Google Scholar]
- 18.Zou W, Papov V, Malakhova O, Kim KI, Dao C, Li J, Zhang DE. ISG15 modification of ubiquitin E2 Ubc13 disrupts its ability to form thioester bond with ubiquitin. Biochem Biophys Res Commun. 2005;336:61–68. doi: 10.1016/j.bbrc.2005.08.038. [DOI] [PubMed] [Google Scholar]
- 19.Zhao C, Beaudenon SL, Kelley ML, Waddell MB, Yuan W, Schulman BA, Huibregtse JM, Krug RM. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc Natl Acad Sci USA. 2004;101:7578–7582. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeuchi T, Iwahara S, Saeki Y, Sasajima H, Yokosawa H. Link between the ubiquitin conjugation system and the ISG15 conjugation system: ISG15 conjugation to the UbcH6 ubiquitin E2 enzyme. J Biochem. 2005;138:711–719. doi: 10.1093/jb/mvi172. [DOI] [PubMed] [Google Scholar]
- 21.Kim KI, Giannakopoulos NV, Virgin HW, Zhang DE. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol Cell Biol. 2004;24:9592–9600. doi: 10.1128/MCB.24.21.9592-9600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catic A, Fiebiger E, Korbel GA, Blom D, Galardy PJ, Ploegh HL. Screen for ISG15-crossreactive deubiquitinases. PLoS ONE. 2007;2:e679. doi: 10.1371/journal.pone.0000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277:9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 24.Nicholson B, Leach CA, Goldenberg SJ, Francis DM, Kodrasov MP, Tian X, Shanks J, Sterner DE, Bernal A, Mattern MR, Wilkinson KD, Butt TR. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 2008;17:1035–1043. doi: 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singhal S, Taylor MC, Baker RT. Deubiquitylating enzymes and disease. BMC Biochem. 2008;9(suppl 1):S3. doi: 10.1186/1471-2091-9-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strack B, Calistri A, Accola MA, Palu G, Gottlinger HG. A role for ubiquitin ligase recruitment in retrovirus release. Proc Natl Acad Sci USA. 2000;97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schubert U, Ott DE, Chertova EN, Welker R, Tessmer U, Princiotta MF, Bennink JR, Krausslich HG, Yewdell JW. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc Natl Acad Sci USA. 2000;97:13057–13062. doi: 10.1073/pnas.97.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patnaik A, Chau V, Wills JW. Ubiquitin is part of the retrovirus budding machinery. Proc Natl Acad Sci USA. 2000;97:13069–13074. doi: 10.1073/pnas.97.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc Natl Acad Sci USA. 2000;97:13871–13876. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Putterman D, Pepinsky RB, Vogt VM. Ubiquitin in avian leukosis virus particles. Virology. 1990;176:633–637. doi: 10.1016/0042-6822(90)90035-p. [DOI] [PubMed] [Google Scholar]
- 32.Heidecker G, Lloyd PA, Fox K, Nagashima K, Derse D. Late assembly motifs of human T-cell leukemia virus type 1 and their relative roles in particle release. J Virol. 2004;78:6636–6648. doi: 10.1128/JVI.78.12.6636-6648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott DE, Coren LV, Copeland TD, Kane BP, Johnson DG, Sowder RC, 2nd, Yoshinaka Y, Oroszlan S, Arthur LO, Henderson LE. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J Virol. 1998;72:2962–2968. doi: 10.1128/jvi.72.4.2962-2968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott DE, Coren LV, Sowder RC, 2nd, Adams J, Nagashima K, Schubert U. Equine infectious anemia virus and the ubiquitin-proteasome system. J Virol. 2002;76:3038–3044. doi: 10.1128/JVI.76.6.3038-3044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harty RN, Brown ME, McGettigan JP, Wang G, Jayakar HR, Huibregtse JM, Whitt MA, Schnell MJ. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J Virol. 2001;75:10623–10629. doi: 10.1128/JVI.75.22.10623-10629.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vana ML, Tang Y, Chen A, Medina G, Carter C, Leis J. Role of Nedd4 and ubiquitination of Rous sarcoma virus Gag in budding of virus-like particles from cells. J Virol. 2004;78:13943–13953. doi: 10.1128/JVI.78.24.13943-13953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jager S, Gottwein E, Krausslich HG. Ubiquitination of human immunodeficiency virus type 1 Gag is highly dependent on Gag membrane association. J Virol. 2007;81:9193–9201. doi: 10.1128/JVI.00044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gottwein E, Jager S, Habermann A, Krausslich HG. Cumulative mutations of ubiquitin acceptor sites in human immunodeficiency virus type 1 gag cause a late budding defect. J Virol. 2006;80:6267–6275. doi: 10.1128/JVI.02177-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor GM, Hanson PI, Kielian M. Ubiquitin depletion and dominant-negative VPS4 inhibit rhabdovirus budding without affecting alphavirus budding. J Virol. 2007;81:13631–13639. doi: 10.1128/JVI.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ott DE, Coren LV, Sowder RC, 2nd, Adams J, Schubert U. Retroviruses have differing requirements for proteasome function in the budding process. J Virol. 2003;77:3384–3393. doi: 10.1128/JVI.77.6.3384-3393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calistri A, Del Vecchio C, Salata C, Celestino M, Celegato M, Gottlinger H, Palu G, Parolin C. Role of the feline immunodeficiency virus L-domain in the presence or absence of Gag processing: involvement of ubiquitin and Nedd4-2s ligase in viral egress. J Cell Physiol. 2009;218:175–182. doi: 10.1002/jcp.21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medina G, Pincetic A, Ehrlich LS, Zhang Y, Tang Y, Leis J, Carter CA. Tsg101 can replace Nedd4 function in ASV Gag release but not membrane targeting. Virology. 2008;377:30–38. doi: 10.1016/j.virol.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Usami Y, Popov S, Popova E, Gottlinger HG. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J Virol. 2008;82:4898–4907. doi: 10.1128/JVI.02675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chung HY, Morita E, von Schwedler U, Muller B, Krausslich HG, Sundquist WI. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J Virol. 2008;82:4884–4897. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhadina M, McClure MO, Johnson MC, Bieniasz PD. Ubiquitin-dependent virus particle budding without viral protein ubiquitination. Proc Natl Acad Sci USA. 2007;104:20031–20036. doi: 10.1073/pnas.0708002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heidecker G, Lloyd PA, Soheilian F, Nagashima K, Derse D. The role of WWP1-Gag interaction and Gag ubiquitination in assembly and release of human T-cell leukemia virus type 1. J Virol. 2007;81:9769–9777. doi: 10.1128/JVI.00642-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Urata S, Noda T, Kawaoka Y, Yokosawa H, Yasuda J. Cellular factors required for Lassa virus budding. J Virol. 2006;80:4191–4195. doi: 10.1128/JVI.80.8.4191-4195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segura-Morales C, Pescia C, Chatellard-Causse C, Sadoul R, Bertrand E, Basyuk E. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J Biol Chem. 2005;280:27004–27012. doi: 10.1074/jbc.M413735200. [DOI] [PubMed] [Google Scholar]
- 49.Klinger PP, Schubert U. The ubiquitin-proteasome system in HIV replication: potential targets for antiretroviral therapy. Expert Rev Anti Infect Ther. 2005;3:61–79. doi: 10.1586/14787210.3.1.61. [DOI] [PubMed] [Google Scholar]
- 50.Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol. 2005;168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blot V, Perugi F, Gay B, Prevost MC, Briant L, Tangy F, Abriel H, Staub O, Dokhelar MC, Pique C. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J Cell Sci. 2004;117:2357–2367. doi: 10.1242/jcs.01095. [DOI] [PubMed] [Google Scholar]
- 52.Sakurai A, Yasuda J, Takano H, Tanaka Y, Hatakeyama M, Shida H. Regulation of human T-cell leukemia virus type 1 (HTLV-1) budding by ubiquitin ligase Nedd4. Microbes Infect. 2004;6:150–156. doi: 10.1016/j.micinf.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 53.Yasuda J, Nakao M, Kawaoka Y, Shida H. Nedd4 regulates egress of Ebola virus-like particles from host cells. J Virol. 2003;77:9987–9992. doi: 10.1128/JVI.77.18.9987-9992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gottwein E, Bodem J, Muller B, Schmechel A, Zentgraf H, Krausslich HG. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J Virol. 2003;77:9474–9485. doi: 10.1128/JVI.77.17.9474-9485.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Timmins J, Schoehn G, Ricard-Blum S, Scianimanico S, Vernet T, Ruigrok RW, Weissenhorn W. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J Mol Biol. 2003;326:493–502. doi: 10.1016/s0022-2836(02)01406-7. [DOI] [PubMed] [Google Scholar]
- 56.Licata JM, Simpson-Holley M, Wright NT, Han Z, Paragas J, Harty RN. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J Virol. 2003;77:1812–1819. doi: 10.1128/JVI.77.3.1812-1819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yasuda J, Hunter E, Nakao M, Shida H. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 2002;3:636–640. doi: 10.1093/embo-reports/kvf132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kikonyogo A, Bouamr F, Vana ML, Xiang Y, Aiyar A, Carter C, Leis J. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc Natl Acad Sci USA. 2001;98:11199–11204. doi: 10.1073/pnas.201268998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pitha-Rowe IF, Pitha PM. Viral defense, carcinogenesis and ISG15: novel roles for an old ISG. Cytokine Growth Factor Rev. 2007;18:409–417. doi: 10.1016/j.cytogfr.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sen GC, Sarkar SN. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr Top Microbiol Immunol. 2007;316:233–250. doi: 10.1007/978-3-540-71329-6_12. [DOI] [PubMed] [Google Scholar]
- 62.Kim JH, Luo JK, Zhang DE. The level of hepatitis B virus replication is not affected by protein ISG15 modification but is reduced by inhibition of UBP43 (USP18) expression. J Immunol. 2008;181:6467–6472. doi: 10.4049/jimmunol.181.9.6467. [DOI] [PubMed] [Google Scholar]
- 63.Osiak A, Utermohlen O, Niendorf S, Horak I, Knobeloch KP. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol Cell Biol. 2005;25:6338–6345. doi: 10.1128/MCB.25.15.6338-6345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moore EC, Barber J, Tripp RA. Respiratory syncytial virus (RSV) attachment and nonstructural proteins modify the type I interferon response associated with suppressor of cytokine signaling (SOCS) proteins and IFN-stimulated gene-15 (ISG15) Virol J. 2008;5:116. doi: 10.1186/1743-422X-5-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, Wolff T, Osiak A, Levine B, Schmidt RE, Garcia-Sastre A, Leib DA, Pekosz A, Knobeloch KP, Horak I, Virgin HW., 4th IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci USA. 2007;104:1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giannakopoulos NV, Arutyunova E, Lai C, Lenschow DJ, Haas AL, Virgin HW. ISG15 Arg151 and the ISG15-conjugating enzyme UbE1L are important for innate immune control of Sindbis virus. J Virol. 2009;83:1602–1610. doi: 10.1128/JVI.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pitha PM, Staal SP, Bolognesi DP, Denny TP, Rowe WP. Effect of interferon on murine leukemia virus infection. II. Synthesis of viral components in exogenous infection. Virology. 1977;79:1–13. doi: 10.1016/0042-6822(77)90329-4. [DOI] [PubMed] [Google Scholar]
- 68.Pitha PM, Rowe WP, Oxman MN. Effect of interferon on exogenous, endogenous, and chroniv murine leukemia virus infection. Virology. 1976;70:324–338. doi: 10.1016/0042-6822(76)90275-0. [DOI] [PubMed] [Google Scholar]
- 69.Kunzi MS, Pitha PM. Interferon targeted genes in host defense. Autoimmunity. 2003;36:457–461. doi: 10.1080/08916930310001605855. [DOI] [PubMed] [Google Scholar]
- 70.Kunzi MS, Pitha PM. Role of interferon-stimulated gene ISG-15 in the interferon-omega-mediated inhibition of human immunodeficiency virus replication. J Interferon Cytokine Res. 1996;16:919–927. doi: 10.1089/jir.1996.16.919. [DOI] [PubMed] [Google Scholar]
- 71.Okumura A, Lu G, Pitha-Rowe I, Pitha PM. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci USA. 2006;103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan W, Krug RM. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. Embo J. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takeuchi T, Yokosawa H. ISG15 modification of Ubc13 suppresses its ubiquitin-conjugating activity. Biochem Biophys Res Commun. 2005;336:9–13. doi: 10.1016/j.bbrc.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 74.Takeuchi T, Yokosawa H. Detection and analysis of protein ISGylation. Methods Mol Biol. 2008;446:139–149. doi: 10.1007/978-1-60327-084-7_10. [DOI] [PubMed] [Google Scholar]
- 75.Okumura A, Pitha PM, Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci USA. 2008;105:3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Malakhova OA, Zhang DE. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem. 2008;283:8783–8787. doi: 10.1074/jbc.C800030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martin-Serrano J, Perez-Caballero D, Bieniasz PD. Context-dependent effects of L domains and ubiquitination on viral budding. J Virol. 2004;78:5554–5563. doi: 10.1128/JVI.78.11.5554-5563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lai C, Struckhoff JJ, Schneider J, Martinez-Sobrido L, Wolff T, Garcia-Sastre A, Zhang DE, Lenschow DJ. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J Virol. 2009;83:1147–1151. doi: 10.1128/JVI.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt JA, Rowland RR, Schmaljohn CS, Lenschow DJ, Snijder EJ, Garcia-Sastre A, Virgin HW., 4th Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guerra S, Caceres A, Knobeloch KP, Horak I, Esteban M. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 2008;4:e1000096. doi: 10.1371/journal.ppat.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]