Summary
Ubiquitin and ubiquitin-like proteins (UBLs) are directed to targets by cascades of E1, E2, and E3 enzymes. The largest ubiquitin E3 subclass consists of cullin-RING ligases (CRLs), which contain one each of several cullins (CUL1, 2, 3, 4, or 5) and RING proteins (RBX1 or 2). CRLs are activated by ligation of the UBL NEDD8 to a conserved cullin Lys. How is cullin NEDD8ylation specificity established? Here we report that like UBE2M (aka UBC12), the previously uncharacterized E2 UBE2F is a NEDD8 conjugating enzyme in vitro and in vivo. Biochemical and structural analyses indicate how plasticity of hydrophobic E1–E2 interactions and E1 conformational flexibility allow one E1 to charge multiple E2s. The E2s have distinct functions, with UBE2M/RBX1 and UBE2F/RBX2 displaying different target cullin specificities. Together, these studies reveal the molecular basis for and functional importance of hierarchical expansion of the NEDD8 conjugation system in establishing selective CRL activation.
Keywords: Cullin, Cul1, Cul5, Rbx1, Rbx2, Cullin-RING ligase, NEDD8, E2, UBE2M, UBE2F, Ubiquitin
Introduction
Cullin RING ligases (CRLs) comprise the largest subfamily of E3 ubiquitin ligases. In humans, six cullins (CUL1, 2, 3, 4A, 4B, and 5), two RBX-family RING proteins (RBX1 and 2), and hundreds of substrate receptors assemble into distinct CRLs that mediate ubiquitination of thousands of targets to regulate a vast array of cellular processes (Cardozo and Pagano, 2004; Petroski and Deshaies, 2005; Willems et al., 2004). CRL function is regulated by attachment of the ubiquitin-like protein (UBL) NEDD8 to a conserved Lys in a cullin’s C-terminal domain (Pan et al., 2004). NEDD8 both enhances intrinsic CRL ubiquitination activity (Amir et al., 2002; Kawakami et al., 2001; Morimoto et al., 2000; Podust et al., 2000; Read et al., 2000; Wu et al., 2000), and prevents CRL binding to the inhibitor CAND1 (Liu et al., 2002; Zheng et al., 2002). At present, the NEDD8 cascade is known to contain a single E1 (NAE1-UBA3), which activates NEDD8 and ultimately catalyzes transfer of NEDD8’s C-terminus to the catalytic cysteine of the only characterized NEDD8 E2, UBE2M (also known as UBC12) (Gong and Yeh, 1999; Liakopoulos et al., 1998; Osaka et al., 1998). The resulting UBE2M~NEDD8 thioester conjugate serves as the direct source of NEDD8 to be covalently attached to a cullin’s acceptor Lys. In vivo and also in cell lysates, additional factors can also influence a cullin’s NEDD8ylation state. As examples, Dcn1 (aka SCCRO) enhances cullin NEDD8ylation (Kim et al., 2008; Kurz et al., 2008; Kurz et al., 2005) and the COP9 Signalosome (CSN) removes ligated NEDD8 from cullins (Lyapina et al., 2001).
Despite the importance of CRL activation by NEDD8, it remains unknown whether there is any specificity in cullin NEDD8ylation, or how such specificity could be established. Clues to selectivity might come from analogy to the ubiquitin (Ub) pathway. A fundamental feature of the Ub pathway is that substrate ubiquitination selectivity is generated by the hierarchical organization of the cascade (Dye and Schulman, 2007; Haas and Siepmann, 1997; Kerscher et al., 2006). In vertebrates, the Ub pathway comprises two E1s (UBA1 and UBA6), tens of E2s and hundreds of E3s promoting Ub transfer to thousands of targets (Chiu et al., 2007; Hershko et al., 2000; Jin et al., 2007; Pelzer et al., 2007; Pickart and Rose, 1985). Within this hierarchy, E1–E2 and E2–E3-substrate interactions are highly specific to ensure precise temporal and spatial regulation of targets by modification with appropriate Ub linkages, which in turn mediate particular downstream functions (Harper and Schulman, 2006; Hicke et al., 2005; Hurley et al., 2006; Pickart and Fushman, 2004; Ravid and Hochstrasser, 2008).
One pivotal feature of a hierarchical E1–E2–E3 cascade is that an E1 needs to charge multiple E2s with Ub, such that E3s can then direct the distinct charged Ub~E2s to downstream targets. Previous studies revealed that an E1’s C-terminal ubiquitin-fold domain plays a crucial role in E2 recruitment, and that UBL transfer from E1 to E2 involves rotation of this domain about a flexible linker (Huang et al., 2007; Huang et al., 2005; Lee and Schindelin, 2008; Lois and Lima, 2005; Walden et al., 2003). Nonetheless, it remains poorly understood how any E1 recognizes multiple E2s.
In contrast to the Ub pathway, the NEDD8 conjugation cascade has been reported to contain only a single E2. Given the important and diverse CRL functions in cellular regulation, we wondered whether the NEDD8 cascade is equipped with additional levels of selectivity for cullin NEDD8ylation. Here we describe that like the Ub cascade, the NEDD8 cascade is expanded at the E2 level. We identify UBE2F as a NEDD8 E2. Biochemical studies and a structure of NEDD8 E1’s ubiquitin-fold domain in complex with UBE2F’s core domain reveal the basis for how an E1 recognizes distinct E2s. We further show that UBE2M-RBX1 and UBE2F-RBX2 pairs specifically regulate NEDD8ylation of Cullins 1–4 and CUL5, respectively, in an E2-RING-dependent manner. Thus, hierarchical expansion of the NEDD8 cascade at the E2-RING level confers selective CRL activation.
Results and Discussion
UBE2F is an E2 for NEDD8 in vitro and in vivo
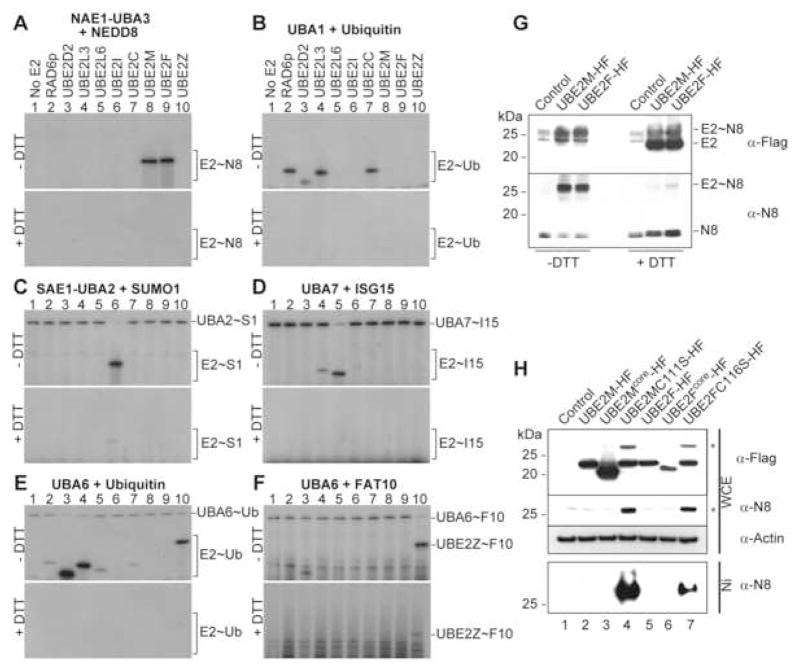
In characterizing a previously undescribed E2, UBE2F, we discovered that it is a specific NEDD8 conjugating enzyme in vitro and in vivo. First, using purified components, we tested the ability of UBE2F and several well-characterized E2s to be charged by E1~UBL (~ covalent bond) pathways as follows: NAE1-UBA3~NEDD8 (Fig. 1A); UBA1~Ub (Fig. 1B); SAE1-UBA2~SUMO1 (Fig. 1C); UBA7~ISG15 (Fig. 1D); UBA6~Ub (Fig. 1E); UBA6~FAT10 (Fig. 1F). Control E2s were charged only by their known cognate E1~UBLs (Chiu et al., 2007; Johnson and Blobel, 1997; Johnson et al., 1997; McGrath et al., 1991; Osaka et al., 1998; Pelzer et al., 2007{Jin, 2007 #249; Yuan and Krug, 2001; Zhao et al., 2004). Furthermore, a DTT-sensitive band corresponding to a UBE2F~UBL thioester product was detected only in the presence of NAE1-UBA3 and NEDD8, suggesting that like UBE2M, UBE2F is a NEDD8 conjugating enzyme in vitro (Fig. 1A, lanes 8, 9).
Figure 1. UBE2F forms a NEDD8 thioester conjugate in vitro and in vivo.
Autoradiograms showing 32P-UBL thioesters after E2 charging by E1~32P-UBLs (top) and reduction with 100 mM DTT (bottom): A, NAE1-UBA3~NEDD8; B, UBA1~Ub; C, SAE1-UBA2~SUMO1; D, UBA7~ISG15; E, UBA6~Ub; F, UBA6~ FAT10. G, Immunoblot showing 100 mM DTT reduction of E2~NEDD8 complex detected with antibodies against E2 (α-Flag) and NEDD8 (α-N8) from NIH 3T3 cells infected with indicated retroviruses. H, Immunoblot as in G (top) from whole cell extracts (WCE) of NIH 3T3 cells infected with indicated retroviruses and lysed in urea buffer, and from nickel-agarose pull-downs (His). * expected MW of E2~NEDD8 oxy-ester complex.
We next examined in vivo conjugate formation in NIH 3T3 cells between UBE2F with a C-terminal His-Flag tag (UBE2F-HF) and NEDD8. Because thioester bonds are not stable with standard cell lysis procedures, we examined formation of a covalent UBE2F~NEDD8 complex in two ways. First, we used a recently described acid lysis method that preserves thioester conjugates (Jin et al., 2007), and saw a slow migrating band, reactive with anti-Flag and -NEDD8 antibodies, which is sensitive to reduction. This indicates that like UBE2M-HF~NEDD8, a UBE2F-HF~NEDD8 thioester product was formed in cells (Fig. 1G). As a second approach, we generated a UBE2F catalytic Cys to Ser mutant, enabling purification of a stable oxy-ester complex (Wada et al., 2000). Bands corresponding to E2~NEDD8 complexes reacted with antibodies to NEDD8 were observed in cell extracts expressing UBE2M C111S-HF and UBE2F C116S-HF, and after nickel agarose pull-down (Fig. 1H lanes 4 and 7). Together, the results indicate that like UBE2M, UBE2F is an E2 for NEDD8 in vitro and in vivo.
UBE2F interacts with NEDD8 E1 in a bipartite manner
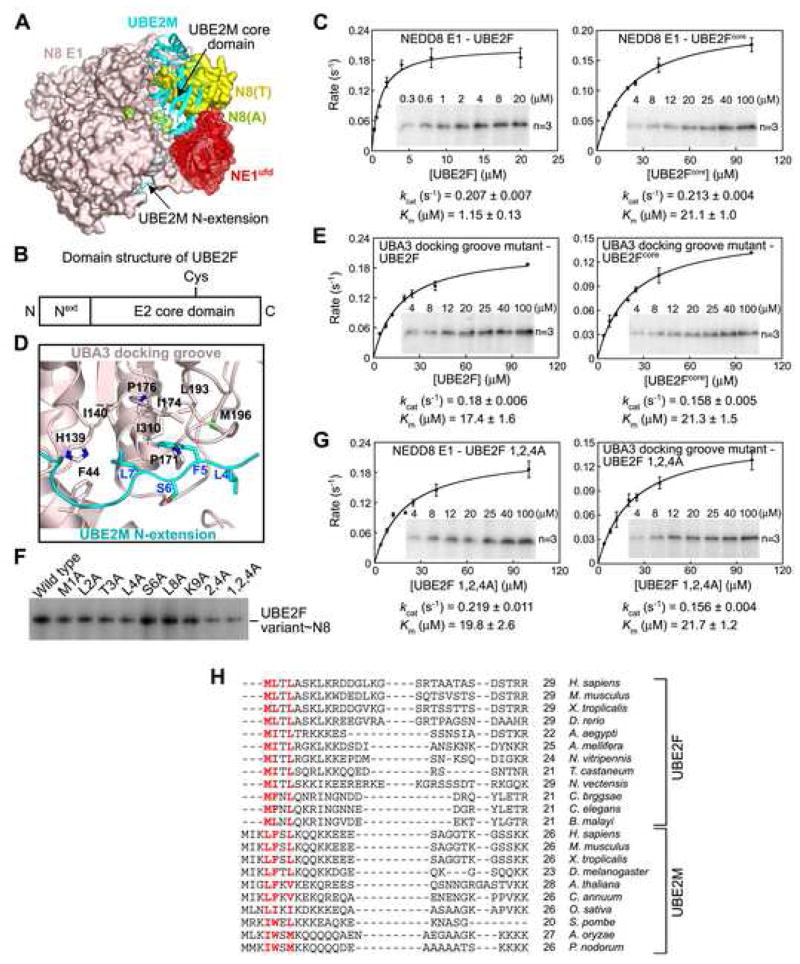
How does one E1 (NAE1-UBA3, i.e. NEDD8’s E1) interact with multiple E2s (UBE2M and UBE2F)? To address this question, we examined UBE2F’s E1-binding domains based on previous studies of bipartite interactions between NEDD8’s E1 and UBE2M. UBE2M’s N-terminal extension binds UBA3’s hydrophobic groove; also, UBE2M’s catalytic core domain interacts with NEDD8 E1’s ubiquitin-fold domain (ufd) (Fig. 2A) (Huang et al., 2007; Huang et al., 2004; Huang et al., 2005). This latter interaction mode is thought to be common among UBLs cascades (Durfee et al., 2008; Huang et al., 2007; Huang et al., 2005; Jin et al., 2007; Lee and Schindelin, 2008; Lois and Lima, 2005; Walden et al., 2003). UBE2F’s sequence also includes the two domains, an N-terminal extension and E2 core (Fig. 2B). We tested their binding to NEDD8’s E1 by analyzing kinetics of NEDD8 transfer. In comparison to wild-type UBE2F, a mutant (UBE2Fcore) consisting only of the core domain showed ~18-fold higher Km (Fig. 2C). Thus, both UBE2F’s N-extension and core domains bind NEDD8’s E1.
Figure 2. Bipartite UBE2F interaction with UBA3: a conserved Φ-Φ-X-Φ UBA3 in NEDD8 E2 N-extensions.
A, Structure of NAE1 UBA3~NEDD8(T)-NEDD8(A)-MgATP-UBE2M(C111A) (Huang et al., 2007) depicting interactions between NEDD8’s E1 (NAE1-UBA3, light pink, with ufd in red) and UBE2M (cyan). The thioester-bound NEDD8 (N8(T)) is yellow and adenylation site NEDD8 (N8(A)) is lime. UBE2M’s N-extension and core domains are indicated. B, Domain structure of UBE2F. C, Michaelis-Menten curves for 32P-NEDD8 thioester conjugate formation by UBE2F and UBE2Fcore. D, Detailed interactions between UBE2M’s N-extension (blue) and UBA3’s docking groove (black) (Huang et al., 2004). E, Michaelis-Menten curves for UBA3 docking groove mutant-catalyzed 32P-NEDD8 thioester conjugate formation by UBE2F and UBE2Fcore. F, Autoradiogram of 32P-NEDD8 thioester conjugates formed by the indicated Ala mutant of UBE2F. G, Michaelis-Menten curves for 32P-NEDD8 thioester conjugate formation by UBE2F and NAE1-UBA3 and mutants. H, Structure-based sequence alignment of UBE2F and UBE2M’s N-terminal extension from various species (Tables S2, S3).
Kinetic analyses (C, E, G) show standard errors, insets are representative autoradiograms indicating E2 concentrations and the number of independent replicates, and kinetic constants (kcat and Km values) are indicated.
Defining a Φ-Φ-X-Φ UBA3-binding motif common among NEDD8 E2 N-extensions
To define NAE1-UBA3-UBE2F interactions, we performed mutational analysis. Similar Km values were measured for UBE2Fcore with wild-type NAE1-UBA3, for wild-type UBE2F and a previously described NAE1-UBA3 docking groove mutant (UBA3 F44, H139, I140, P171, I174, P176, L193, M196, I310A (Huang et al., 2004)), and for UBE2Fcore with the docking groove mutant (Fig. 2C–E). Thus, UBA3’s docking groove binds UBE2F’s N-terminal extension. Further Ala mutational analysis of UBE2F’s N-terminal sequence showed a similar Km value for a Met1Ala, Leu2Ala, Leu4Ala triple mutant (Fig. 2F, G). This suggests that UBE2F‘s N-terminal Met1-Leu2-X-Leu4 sequence mediates the bulk of the N-extension’s interaction with UBA3’s docking groove. Comparing UBA3-binding sequences from UBE2F and UBE2M (Leu4-Phe5-X-Leu7 (Huang et al., 2004)) across species revealed a conserved Φ-Φ-X-Φ motif (Φ-hydrophobic; X - any residue) (Fig. 2H, Tables S1–S3).
Structural basis for hierarchical expansion of a UBL cascade at the E2 level
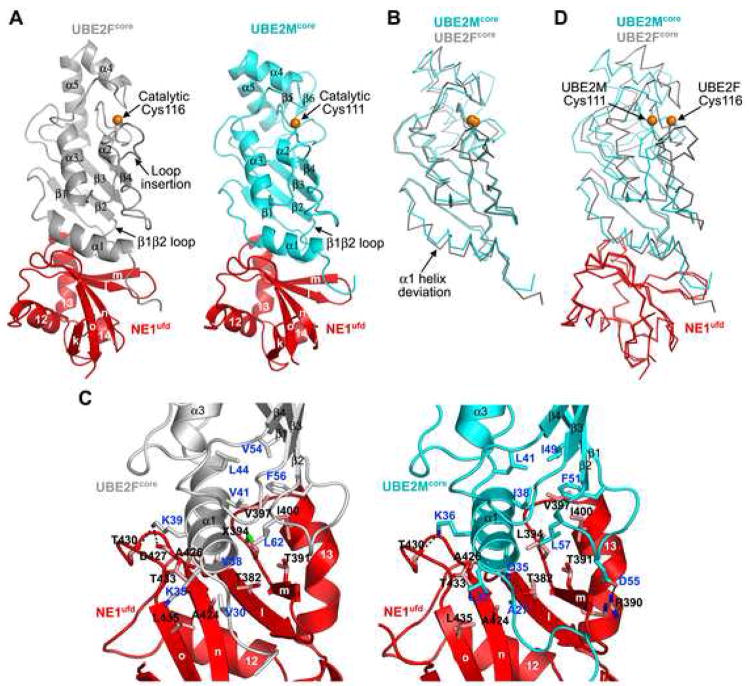
A common feature of ubiquitin and NEDD8 E1s is a critical role for their C-terminal ubiquitin-fold domains (ufds) in E2 binding. Consistent with this notion, we found that the isolated NEDD8 E1 ufd (NE1ufd) formed a complex with UBE2Fcore (Fig. S1), and determined the complex crystal structure at 2.5 Å resolution (Fig. 3A left, S2, Table S4). Overall, NE1ufd-UBE2Fcore adopts a compact, globular ovoid structure similar to NE1ufd-UBE2Mcore (PDB 1Y8X; Fig. 3A) (Huang et al., 2005), with UBE2Fcore displaying a canonical E2 catalytic core domain fold. Superimposing UBE2Fcore with prior UBE2Mcore structures (Huang et al., 2007; Huang et al., 2005) revealed distinct features of UBE2Fcore including: (1) an offset orientation and a half-turn extension for UBE2Fcore’s N-terminal αa1 helix; (2) a loop insertion immediately following the catalytic Cys116; and (3) an extended C-terminal helix more reminiscent of ubiquitin E2 structures than the 2-stranded β-sheet at the C-terminus of UBE2M (Fig. 3B, S3).
Figure 3. Structural basis for NEDD8 E1 ufd’s binding to varying E2 sequences.
A, Overall structure of NE1ufd-UBE2Fcore (left) compared to NE1ufd-UBE2Mcore (Huang et al., 2005) (right). NE1ufd is red, UBE2Fcore grey, UBE2Mcore cyan, and E2 catalytic cysteines orange spheres. B, UBE2Fcore and UBE2Mcore structural superposition. C, Close-ups of NE1ufd-UBE2Fcore (left) and NE1ufd-UBE2Mcore (right) interfaces. Oxygens are red, nitrogens blue, ionic interactions dashed. E2 residues are labeled in blue, and NE1ufd’s in black. X – Leu394SeMet. D, Structural superposition of the NE1ufd portions of NE1ufd-UBE2Fcore and NE1ufd-UBE2Mcore.
Interestingly, structural comparison shows how NEDD8’s E1 binds distinct UBE2F and UBE2M α 1-helix and β 1 β2-loop sequences. For both E2s, interactions are centered around two hydrophobic clusters (Fig. 3C). One cluster involves NE1ufd’s Ala424, Ala426, Thr433, and Leu435, and UBE2Fcore’s Val30 and the hydrophobic portion of Lys35 from the α1-helix. These correspond to UBE2Mcore’s Ala27 and Leu32, respectively. The second cluster involves NE1ufd’s Thr382, Thr391, SeMet394 (in our structure, Leu394 in wild type NE1ufd), Val397, and Ile400 packing against UBE2Fcore’s Val38, Val41, and Leu44 from the α1-helix, and Val54, Phe56, and Leu62 from the β 1 β 2-loop. Although Leu44, Phe56 and Leu62 are conserved between UBE2Fcore and UBE2Mcore, other residues differ significantly. For example, UBE2Fcore’s Val38 makes hydrophobic interactions, whereas the corresponding UBE2Mcore Gln35 forms a hydrogen bond with NEDD8 E1’s Thr382. Unlike the NE1ufd-UBE2Mcore complex, which is stabilized by numerous ionic interactions, only one stabilizes NE1ufd-UBE2Fcore. These structural variations may account for the different Km values for UBE2Fcore (21.1 ± 1.0 μM) and UBE2Mcore (6.07 ± 0.36μM; (Huang et al., 2004)) during NEDD8 transfer from E1 to E2.
General implications for hierarchical expansion of UBL cascades
Our data provide general insights into how an E1 can recognize multiple E2s. First, NEDD8’s E1 recognizes cognate E2s via two surfaces. The ufd-E2core interaction is common among UBL pathways. The second interaction between E2 N-extensions and UBA3’s docking groove both selects for the NEDD8 pathway, and prevents mis-association with a non-cognate E1 (Huang et al., 2008). Notably, a second, unique E2-binding site was recently identified to play a role in specificity for the yeast Ub E1 Uba1p (Huang et al., 2008; Lee and Schindelin, 2008). Thus, multiple E1–E2 interaction surfaces may be a means to increase affinity in a manner that is adaptable to multiple cognate E2 sequences, while selecting against mischarging the wrong E2. Second, both E2 binding sites focus on hydrophobic interactions. Pliability of hydrophobic interactions (Lim and Sauer, 1989) likely accommodates a range of sequences and structures (Fig. 3). Indeed, the corresponding ufd surface from the yeast Ub E1 Uba1p is also hydrophobic, which may facilitate binding to numerous Ub E2s (Table S5). Third, in addition to local structural malleability, E1 conformational flexibility at a more global level is also likely to be important for binding to multiple E2s. This is highlighted by superimposing the NE1ufd portion of NE1ufd-UBE2Fcore with previous structures of NE1ufd-UBE2Mcore and NAE1-UBA3~NEDD8(T)-NEDD8(A)-MgATP-UBE2M (C111A) (Huang et al., 2007; Huang et al., 2005). Due to different UBE2F and UBE2M N-terminal α 1-helix orientations, the corresponding positions of the two E2s distal from the E1-interface diverge by as much as 6Å (Fig. 3D). In particular, the relative locations for their catalytic Cys thiols differ by ~3.5Å. Given these differences, how might a single E1 be able to transfer a UBL to E2s with different E1-to-E2 Cys geometries and distances? It seems likely that the ability of UBA3’s ufd to rotate around a flexible hinge (Huang et al., 2007; Huang et al., 2005) may be important for accommodating multiple E2s. Indeed, ufd rotation has also been observed for the Ub E1 Uba1p (Lee and Schindelin, 2008).
Different global consequences of knocking down Ube2m and Ube2f levels in cells
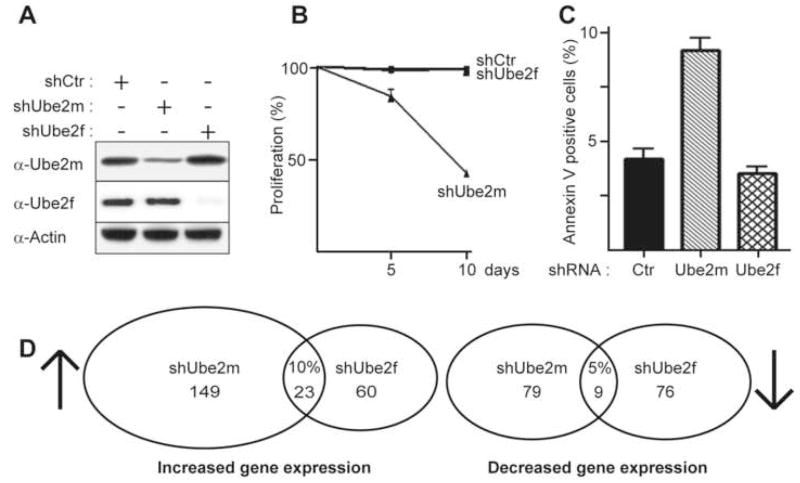
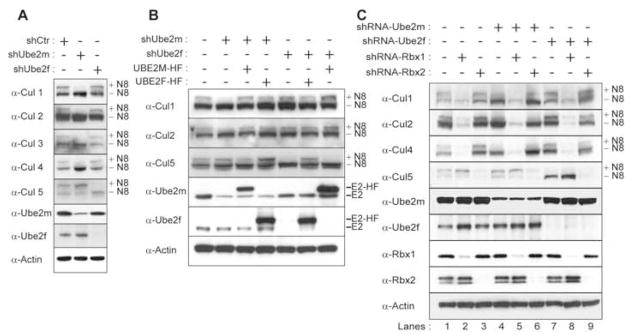
To begin to address the cellular roles of the multiple NEDD8 E2s, we examined some global properties of UBE2F and UBE2M. First, we compared expression patterns of UBE2M and UBE2F in various tissues and cell lines, and observed similar mRNA and protein expression trends, although UBE2F was generally expressed at lower levels (Fig. S4). Next, we knocked down Ube2f or Ube2m (hereafter, human and mouse genes are designated by capital and small letters, respectively) in NIH 3T3 cells by retroviral shRNA expression (Fig. 4A) and examined effects on proliferation and apoptosis. Ube2m knockdown led to increased cell and nuclear size (Fig. S5). Moreover, FACS profiles revealed accumulation of cells in G2/M phase and increased aneuploidy, and these cells also displayed budding nuclei characteristic of apoptosis (Fig. S5). Accordingly, Ube2m knockdown led to decreased cell numbers and increased Annexin V staining (Fig. 4B, C). By contrast, cells appeared normal when Ube2f was knocked down (Fig. 4B, C).
Figure 4. Effects of Ube2f and Ube2m knockdown on global cellular properties.
A, Immunoblots of Ube2f and Ube2m proteins after infecting NIH 3T3 cells with retroviruses expressing shRNAs for control (Ctr), Ube2m or Ube2f. B, Average % of cells in culture (3 independent experiments) relative to control 5 and 10 days after infecting NIH 3T3 cells with the indicated retroviruses. C, Average % of Annexin V positive cells (3 independent experiments) detected by flow cytometry 8 days after infecting NIH 3T3 cells with the indicated retroviruses. Error bars – standard error. D, Venn diagram displaying the results of transcriptome analysis from 3 independent experiments. Total numbers of up- and down-regulated genes for NIH 3T3 cells treated with shRNA against Ube2m or Ube2f versus control, displaying % overlap.
To examine whether knocking down Ube2f and Ube2m leads to distinct molecular consequences in cells, we performed transcriptome analysis with Affymetrix gene chip microarrays. In both cases, different gene expression profiles were observed in comparison to controls (Tables S6–S9). Furthermore, knockdown of Ube2f and Ube2m led to distinct patterns of gene expression, with only 10% and 5% overlap for the up- and down-regulated genes, respectively (Fig. 4D, S6). Thus, although the molecular pathways leading to the transcriptome differences remain to be elucidated, the differential effects of knocking down the two NEDD8 E2s compared to controls, and compared to each other, likely reflects distinct functions for Ube2m and Ube2f.
Ube2f and Ube2m mediate distinct patterns of Cullin NEDD8ylation
To identify direct downstream functions of UBE2F, we examined effects of Ube2f and Ube2m knockdown on NEDD8ylation of the best-characterized targets, members of the cullin family. In NIH 3T3 cells, knocking down Ube2m led to decreased Nedd8ylation of endogenous Cullins 1–4, and a marginal increase in Nedd8-modified Cul5 (Fig. 5A, S7). By contrast, knocking down Ube2f virtually eliminated the presence of Nedd8ylated Cul5, and had no effect on the levels of Nedd8-modified Cullins 1–4 (Fig. 5A). Similar results were obtained using early passage primary mouse embryonic fibroblasts (MEFs) (data no shown) suggesting that the effects were not exclusive to NIH 3T3 cells. Knocking down Ube2f had no effect on Ube2m levels and vice-versa, so endogenous Ube2f and Ube2m were unable to complement the effects of knocking down the other NEDD8 E2.
Figure 5. Distinct E2-Rbx pairs regulate cullin NEDD8ylation specificity in vivo-N8 indicates unNedd8ylated and +N8 indicates Nedd8ylated cullin.
A, Immunoblots of urea lysates from NIH 3T3 cells infected with retroviruses expressing shRNA against control (Ctr), Ube2m or Ube2f, probed with the indicated antisera. B, Immunoblots of urea lysates from retrovirally infected NIH 3T3 cells as in A, in the absence or presence of retroviruses expressing human UBE2M-HF or human UBE2F-HF, probed with the indicated antisera. C, Immunoblots of urea lysates from retrovirally infected NIH 3T3 cells as in A, in the absence or presence of retroviruses expressing shRNA against Rbx1 or Rbx2, probed with the indicated antisera.
Next, we asked whether overexpressing UBE2F could rescue defects caused by Ube2m knockdown,and vice-versa. Human UBE2F and UBE2M, which have 97% and 100% identical protein sequences as their mouse counterparts, are encoded by distinct DNA sequences that escape targeting by shRNAs against the mouse genes. In controls, CMV-mediated UBE2F-HF or UBE2M-HF expression rescued defects from Ube2f or Ube2m knockdowns, respectively (Fig. 5B). UBE2M-HF overexpression did not rescue the Ube2f knockdown-mediated decrease in Nedd8ylated Cul5. However, over-expressing UBE2F-HF partially rescued the Ube2m shRNA-dependent decrease in cullin Nedd8ylation (Fig. 5B) and increase in aneuploidy (Fig. S8). Thus, in vivo, the two NEDD8 E2s have distinct roles in cullin NEDD8ylation, although when overexpressed, UBE2F displayed a broader range of substrates.
NEDD8ylation selectivity in vivo is dictated combinatorially by E2-RBX-Cullin interplay
Cullins interact with RING box proteins (RBX) to form CRLs, and Rbx1 plays a crucial role in CUL1 NEDD8ylation (Gray et al., 2002; Kamura et al., 1999a; Morimoto et al., 2003). In higher eukaryotes, there are two RBX proteins, RBX1 and RBX2, with distinct cullin association preferences. RBX1 preferentially interacts with CULs 1–4, and under some circumstances can bind CUL5 (Kamura et al., 2001; Kamura et al., 1999b; Mahrour et al., 2008; Ohta et al., 1999; Querido et al., 2001; Seol et al., 1999; Skowyra et al., 1999; Yu et al., 2003). By contrast, RBX2 associates exclusively with CUL5 (Kamura et al., 2004; Reynolds et al., 2008). Thus, we tested whether the differential effects of UBE2M and UBE2F on cullin NEDD8ylation might depend on RBX1 and RBX2. Upon knocking down Rbx1 levels in NIH 3T3 cells, the total protein amounts for Cullins 1–4 decreased, while the level of Nedd8ylated Cul5 increased slightly compared to control. By contrast, Rbx2 knockdown only decreased the total levels of Cul5 protein (Fig. 5C). Thus, the cellular levels of distinct cullins are related to the levels of their distinct Rbx partners.
Next we examined the effects of Rbx1 or Rbx2 knockdown in combination with either Ube2m or Ube2f shRNA in NIH 3T3 cells. Rbx1 and Ube2f double knockdown reduced the levels of Nedd8ylated Cul5, while Rbx1 and Ube2m double knockdown had no defect for Cul5 Nedd8ylation. On the other hand, Rbx2 and Ube2m double knockdown reduced the levels of Nedd8ylated Cullins 1–4, while Rbx2 and Ube2f double knockdown had no effect on Nedd8ylation of Cullins 1–4 (Fig. 5C). Together, these data suggested that in vivo, NEDD8ylation specificity is established combinatorially: Ube2f pairs with Rbx2 to control Cul5 Nedd8ylation, and Ube2m functions through Rbx1 to mediate Nedd8ylation of Cullins 1–4.
RBX2 is specific for UBE2F in vitro
In order to address the extent to which NEDD8ylation specificity is established by inherent E2-RBX-CUL properties, we turned to in vitro assays, using components purified from E. coli. As the conserved target Lys is located in the cullin C-terminal domain (CTD), we took advantage of a system to generate highly pure native or non-native complexes between RBX proteins and cullin CTDs by bacterial coexpression (H. Walden, M.S. Lee, D. Duda, and L. Borg, unpublished results). Indeed, RBX1-CUL1CTD and RBX2-CUL5CTD exhibited similar activities as their full-length counterparts in assays testing 32P-NEDD8 transfer from UBE2M or UBE2F to CUL1 and CUL5 (Fig. S9). To extend our analysis, we purified all 10 possible RBX-CULCTD pairs (RBX 1–2 x CUL 1–5, Fig. S10), and performed pulse-chase assays. First, equivalent amounts of UBE2F~32P-NEDD8 and UBE2M~32P-NEDD8 were generated in pulse reactions. After the pulse was quenched, RBX-CULCTDs were added, and 32P-NEDD8 transfer from E2 to a given CULCTD was monitored. RBX2-associated CULCTDs showed substantially greater NEDD8ylation by UBE2F, whereas RBX1-associated CULCTDs were NEDD8ylated by both UBE2F and UBE2M (Fig 6A, B). Also, UBE2M is specific for RBX1, whereas UBE2F NEDD8ylates both RBX1- and RBX2-associated CULCTDs. The cullins themselves further influence NEDD8ylation. UBE2M preferred CUL1CTD over CUL2CTD and CUL5CTD. UBE2F preferred CUL5CTD and CUL1 CTD over CUL2CTD (Fig. 6A,B). Both NEDD8 E2s exhibited lower activity toward CUL3CTD and CUL4CTD, irrespective of their RBX partner. Taken together, the results indicate that NEDD8ylation specificity is established combinatorially.
Figure 6. Combinatorial roles of RBXs and E2s establish cullin NEDD8ylation specificity in vitro.
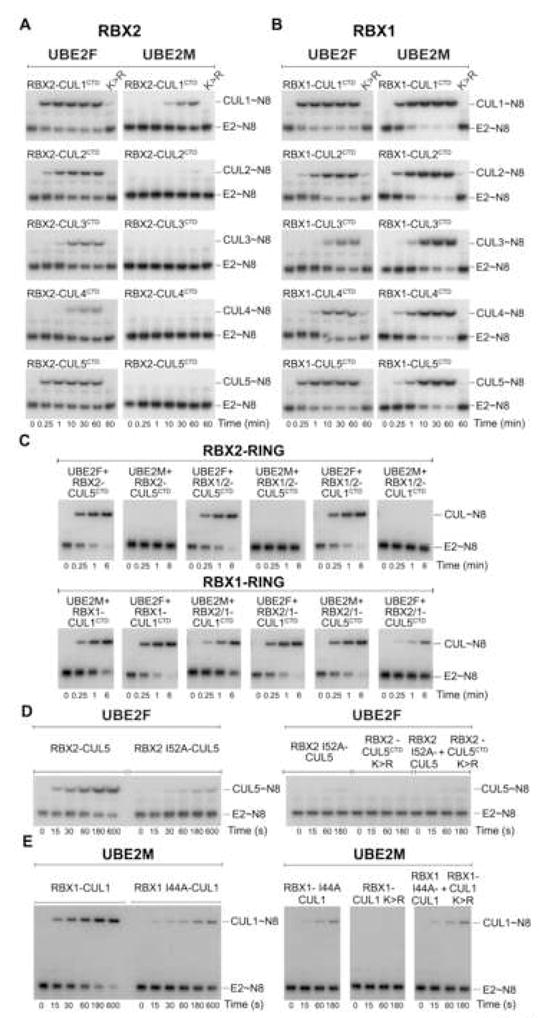
Autoradiograms of chase reactions monitoring A, RBX1- or B, RBX2-mediated 32P-NEDD8 transfer from UBE2F (left) and UBE2M (right) to the indicated CULCTD. C, Autoradiograms for chase reactions monitoring 32P-NEDD8 transfer from UBE2F or UBE2M to the indicated CULCTD mediated by RBX chimeras harboring the indicated N-terminus/RBX2 RING (top panel), and the indicated N-terminus/RBX1 RING (bottom panel). D, UBE2F reactions as in A and B, except with RBX2(Ile52Ala)-CUL5 (left) and wild-type RBX2 in complex with non-NEDD8ylatable CUL5CTD(K724R) as indicated (right). E, UBE2M reactions as in A and B, except with RBX2(Ile44Ala)-CUL1 (left) and wild-type RBX1 in complex with non-NEDD8ylatable CUL1CTD(K720R) as indicated (right).
To further define RBX-NEDD8 E2 specificity, we examined CULCTD NEDD8ylation with chimeric RBXs harboring an N-terminal CULCTD-binding strand from one RBX, and a C-terminal RING from the other. An RBX’s NEDD8 E2 specificity correlates with the identity of the RING domain (Fig. 6C). Thus, the RBX2 RING displays intrinsic specificity for UBE2F, and UBE2M displays intrinsic specificity for RBX1’s RING.
An RBX RING’s E2-binding surface is critical for cullin NEDD8ylation
In Ub cascades, RING domains recruit E2s via hydrophobic surfaces (Brzovic et al., 2003; Dominguez et al., 2004; Zheng et al., 2000). Indeed, an Ala mutation in place of a critical conserved BRCA1 RING surface Ile decreased E2 binding without perturbing the RING fold (Brzovic et al., 2003) (Fig. S11). Thus, we tested a role for the corresponding Ile in the RBX2 (Ile52) and RBX1 (Ile44) RINGs, and found that their mutation to Ala reduced cullin NEDD8ylation (Fig. 6D, E). We next asked whether a wild-type RBX RING domain could restore NEDD8ylation activity in trans. For these experiments, we generated RBX-CUL mutants with wild-type RING domains, but which cannot be NEDD8ylated due to Arg substitutions replacing the acceptor Lys. Adding these in trans did not rescue defects caused by E2-binding surface RBX2 Ile52Ala or RBX1 Ile44Ala mutations (Fig. 6D,E). Future structural studies of RBX2-UBE2F and RBX1-UBE2M will be required to understand the detailed basis for NEDD8 E2 specificity.
UBE2F-RBX2-CUL5-BC/Cul5box – a metazoan-specific CRL pathway complete with activation mechanism
Phylogenetic analyses identify UBE2F only in metazoans (Fig. S12), appearing late in evolution relative to UBE2M. Interestingly, the evolutionary histories of UBE2F and UBE2M mirror their CRL partners: RBX2 and CUL5 also appear restricted to metazoa, whereas UBE2M, RBX1 and CUL1 are present in all eukaryotes. Thus, our results help to further define the distinct RBX2-CUL5 pathway as also including UBE2F.
At present, it is not known why RBX2-CUL5 would have evolved as a separate branch of the CRL/NEDD8 pathway. CUL5 assembles into CRLs via the ElonginB/C heterodimeric adaptor complex, which recruits substrate-receptors harboring BC(SOCS)/Cul5-boxes. Although potential physiological roles of RBX2-CUL5 are only beginning to become apparent, known functions are associated with signaling pathways consistent with emergence of the pathway in metazoans. Some BC(SOCS)/Cul5-box substrate receptors regulate cytokine signaling (Kile et al., 2002), and CUL5-based CRLs have been implicated in regulating neuronal migration (Feng et al., 2007), chondrocyte differentiation (Dentice et al., 2005), and myogenesis (Nastasi et al., 2004).
The importance of CUL5-based CRLs is highlighted by findings that viral pathogens hijack these ligases. As examples, HIV-1 Vif promotes degradation of the host antiviral factor APOBEC3G (Yu et al., 2003), and human adenoviral E4orf6 and E1B55K together promote degradation of the tumor suppressor protein p53 (Querido et al., 2001), both via CUL5-based CRLs. NEDD8 activation was shown to be critical for these CUL5 degradation pathways through the use of a non-NEDD8ylatable mutant CUL5, or ts41 cells harboring a temperature-sensitive NEDD8 E1. However, CUL5 can bind RBX1, and Vif and E4orf6/E1B55K-based ligases have been found with CUL5-RBX1 (Querido et al., 2001; Yu et al., 2003). Thus, determining which CUL5 pathways depend on RBX2, RBX1, UBE2F, UBE2M, or other as yet unknown cullin NEDD8ylation specificity factors may provide opportunities for therapeutic intervention.
Implications for NEDD8ylation of other targets
In addition to cullins, MDM2, p53, p73, breast-cancer associated protein 3, epidermal growth factor receptor, and ribosomal subunits have been reported as NEDD8 targets (Gao et al., 2006; Jones et al., 2008; Oved et al., 2006; Watson et al., 2006; Xirodimas et al., 2004; Xirodimas et al., 2008). Little is known about when or how NEDD8 is conjugated to non-cullin targets. Given that many ubiquitin E2s work with a range of RING proteins, it is plausible that NEDD8 E2s may function with other E3s. Indeed, we found that UBE2M and UBE2F can interact with multiple RINGs in vitro (Fig. 6, S13), albeit to substantially different extents. Although Ube2f appears to play a more restricted role than Ube2m (Fig. S14), the discovery of UBE2F as a NEDD8 conjugating enzyme may facilitate identifying NEDD8ylation pathways for non-cullin targets.
Implications of hierarchical expansion of the NEDD8 cascade for regulating CRL activation
Our results provide a new view of the cullin NEDD8ylation machinery. Rather than a single NEDD8ylation cascade for all target conjugation, we find that NEDD8ylation is a specific process, with discrete molecular pathways dictating modification of different cullin targets (Fig. 7). Indeed, RBX2 is specific for UBE2F, and UBE2M is specific for RBX1. It appears that RBX proteins establish a primary layer of selectivity as classic RING-type E3s for the NEDD8 pathway, by recruiting both a particular cullin target and a specific NEDD8 E2.
Figure 7. Hierarchical E2-RING expansion of the NEDD8 cascade.
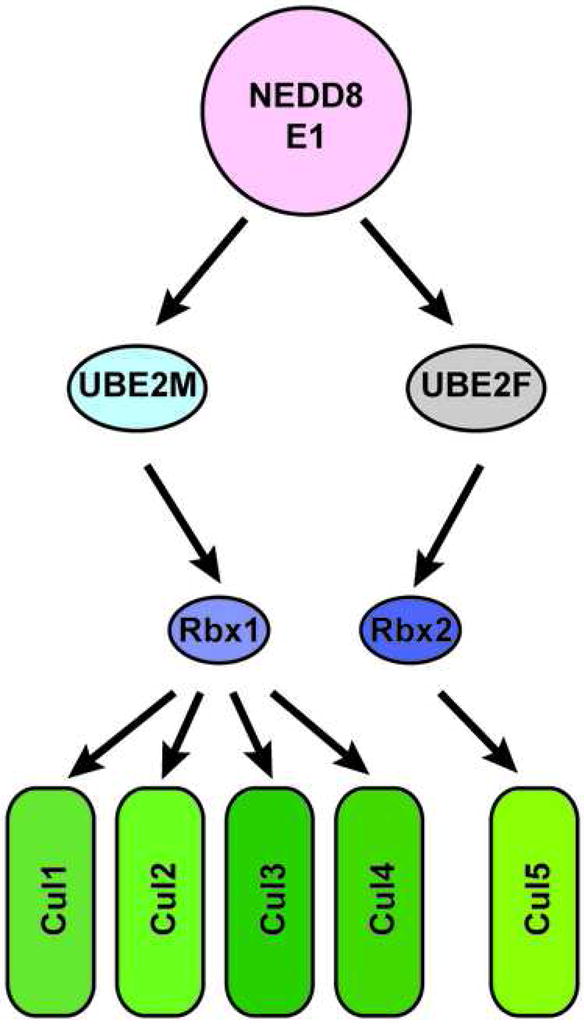
Schematic view of distinct molecular pathways dictating specificity of cullin NEDD8ylation.
Selectivity may also be influenced by differences in protein levels, as cellular concentrations of UBE2F are generally lower than of UBE2M (Fig. S4). E1-E2 transthiolation efficiency also may affect flux through the different pathways: consistent with findings for the Ub pathway that distinct E2s differ in their charging efficiencies by ~4-orders of magnitude (Haas et al., 1988; Huang et al., 2008), kcat/Km for NEDD8 transfer to UBE2F (Fig. 2) is ~7-fold lower than for UBE2M (Huang et al., 2004). Taken together, it seems that under normal conditions the larger cellular pool of UBE2M~NEDD8 would preferentially NEDD8ylate RBX1-associated cullins, whereas RBX2 could only work with the more limited amounts of UBE2F~NEDD8.
Our identification of cullin-specific enzymes implies that target NEDD8ylation is regulated. Indeed, we find UBE2F activity restricted toward Rbx2-Cul5 in vivo, even though it can cooperate with both RBX1 and RBX2 upon elevated expression in cells (Fig. 5B) and in vitro (Fig. 6). Also, different cullins display a range of in vitro NEDD8ylation propensities (Fig. 6). Thus, it seems that in vivo, additional mechanisms regulate NEDD8ylation of specific cullins.
A complete view of NEDD8ylation will require understanding how UBE2F and UBE2M concentrations are established, and integrating the roles of other factors such as the NEDD8ylation enhancer DCN1, the inhibitor CAND1, and the deNEDD8ylating CSN (Kurz et al., 2008; Kurz et al., 2005; Lyapina et al., 2001). Also, some organisms have additional RBXs and NEDD8s. For example, D. melanogaster has three RBXs, each with distinct cullin preferences (Noureddine et al., 2002; Reynolds et al., 2008), and A. thaliana has three NEDD8s (Rao-Naik et al., 1998). Thus, we anticipate that many exciting future studies will reveal how these, and perhaps other as yet unknown factors work together to NEDD8ylate specific CRLs under distinct physiological settings to control important ubiquitination pathways.
Experimental Procedures
General methods
Detailed experimental procedures are provided in Supplemental Information. Briefly, constructs, purified proteins, and crystals were prepared by adapting methods described previously (Duda et al., 2008; Huang et al., 2004; Huang et al., 2005; Huang et al., 2008). The structure was determined by molecular replacement using PDB 1Y8X as the search model for PHASER (Storoni et al., 2004). The model was built in O (Jones et al., 1991), refined to 2.5 Å with REFMAC (1994). For mammalian cell expression, UBE2M, UBE2F and variants with a C-terminal His-Flag tag, and NEDD8 with an N-terminal HA-tag were cloned into an MSCV-IRES-GFP vector (Hawley et al., 1994). Retroviral shRNAs were prepared using the MSCV-LMP shRNA vector (Open Biosystems) carrying a GFP gene. Biochemical assays were performed using methods similar to those described previously (Eletr et al., 2005; Huang et al., 2004; Huang et al., 2008).
Supplementary Material
Acknowledgments
We are grateful to B. Dye for much assistance, H. Walden and M.S. Lee for help with CULCTD-RBX constructs, D. King and V. Pagala for mass spectrometry, J.W. Harper for cDNAs and helpful discussions, Y. Sun for anti-Rbx2 antibodies, W. DenBesten for the Rbx1 shRNA construct, D. Campana for NK cells, C. Ross for computational support, R. Ashmun for FACS, and D. Miller and S. Bozeman for technical and administrative support. This work was supported in part by ALSAC (American Syrian Lebanese Associated Charities), an American Cancer Society fellowship to DMD, grants from the NIH (R01GM069530 and R01GM077053 to BAS, 5P01CA0719075 to MFR., P30CA021765 to St. Jude), a Beckman Young Investigator Award to BAS, and the Howard Hughes Medical Institute. NECAT beamlines of the Advanced Photon Source (APS), are supported by award RR-15301 from the NCRR at the NIH. The APS is supported by the U.S. DOE, Office of Basic Energy Sciences, under contract No. W-31-109-ENG-38. BAS is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Accession numbers
Coordinates/structure factors for the NE1ufd-UBE2Fcore crystal structure and microarray data have RCSB and GEO accession codes of 3FN1 and GSE14088, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- The CCP4 suite: programs for protein crystallography. Acta crystallographica. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Amir RE, Iwai K, Ciechanover A. The NEDD8 pathway is essential for SCF(beta -TrCP)-mediated ubiquitination and processing of the NF-kappa B precursor p105. The Journal of biological chemistry. 2002;277:23253–23259. doi: 10.1074/jbc.M200967200. [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D, 3rd, Fukuda M, Ohta T, Klevit R. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nature reviews. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Chiu YH, Sun Q, Chen ZJ. E1-L2 activates both ubiquitin and FAT10. Molecular cell. 2007;27:1014–1023. doi: 10.1016/j.molcel.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Dentice M, Bandyopadhyay A, Gereben B, Callebaut I, Christoffolete MA, Kim BW, Nissim S, Mornon JP, Zavacki AM, Zeold A, et al. The Hedgehog-inducible ubiquitin ligase subunit WSB-1 modulates thyroid hormone activation and PTHrP secretion in the developing growth plate. Nature cell biology. 2005;7:698–705. doi: 10.1038/ncb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez C, Bonvin AM, Winkler GS, van Schaik FM, Timmers HT, Boelens R. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure. 2004;12:633–644. doi: 10.1016/j.str.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee LA, Kelley ML, Huibregtse JM. The Basis for Selective E1-E2 Interactions in the ISG15 Conjugation System. The Journal of biological chemistry. 2008;283:23895–23902. doi: 10.1074/jbc.M804069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- Feng L, Allen NS, Simo S, Cooper JA. Cullin 5 regulates Dab1 protein levels and neuron positioning during cortical development. Genes & development. 2007;21:2717–2730. doi: 10.1101/gad.1604207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Cheng J, Shi T, Yeh ET. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFkappaB-dependent transcription. Nature cell biology. 2006;8:1171–1177. doi: 10.1038/ncb1483. [DOI] [PubMed] [Google Scholar]
- Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. The Journal of biological chemistry. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- Gray WM, Hellmann H, Dharmasiri S, Estelle M. Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell. 2002;14:2137–2144. doi: 10.1105/tpc.003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AL, Bright PM, Jackson VE. Functional diversity among putative E2 isozymes in the mechanism of ubiquitin-histone ligation. The Journal of biological chemistry. 1988;263:13268–13275. [PubMed] [Google Scholar]
- Haas AL, Siepmann TJ. Pathways of ubiquitin conjugation. Faseb J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- Harper JW, Schulman BA. Structural complexity in ubiquitin recognition. Cell. 2006;124:1133–1136. doi: 10.1016/j.cell.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Hawley RG, Lieu FH, Fong AZ, Hawley TS. Versatile retroviral vectors for potential use in gene therapy. Gene therapy. 1994;1:136–138. [PubMed] [Google Scholar]
- Hershko A, Ciechanover A, Varshavsky A. Basic Medical Research Award. The ubiquitin system. Nature medicine. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nature reviews. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Huang DT, Hunt HW, Zhuang M, Ohi MD, Holton JM, Schulman BA. Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature. 2007;445:394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Miller DW, Mathew R, Cassell R, Holton JM, Roussel MF, Schulman BA. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat Struct Mol Biol. 2004;11:927–935. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8’s E1. Molecular cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Huang DT, Zhuang M, Ayrault O, Schulman BA. Identification of conjugation specificity determinants unmasks vestigial preference for ubiquitin within the NEDD8 E2. Nat Struct Mol Biol. 2008;15:280–287. doi: 10.1038/nsmb.1387. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. The Biochemical journal. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 2007;447:1135–1138. doi: 10.1038/nature05902. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. The Journal of biological chemistry. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. The EMBO journal. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J, Wu K, Yang Y, Guerrero C, Nillegoda N, Pan ZQ, Huang L. A Targeted Proteomic Analysis of the Ubiquitin-Like Modifier Nedd8 and Associated Proteins. J Proteome Res. 2008;7:1274–1287. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kamura T, Burian D, Yan Q, Schmidt SL, Lane WS, Querido E, Branton PE, Shilatifard A, Conaway RC, Conaway JW. Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. The Journal of biological chemistry. 2001;276:29748–29753. doi: 10.1074/jbc.M103093200. [DOI] [PubMed] [Google Scholar]
- Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes & development. 1999a;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Jr, Elledge SJ, Conaway RC, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science (New York, NY) 1999b;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes & development. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Chiba T, Suzuki T, Iwai K, Yamanaka K, Minato N, Suzuki H, Shimbara N, Hidaka Y, Osaka F, et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. The EMBO journal. 2001;20:4003–4012. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Kile BT, Schulman BA, Alexander WS, Nicola NA, Martin HM, Hilton DJ. The SOCS box: a tale of destruction and degradation. Trends in biochemical sciences. 2002;27:235–241. doi: 10.1016/s0968-0004(02)02085-6. [DOI] [PubMed] [Google Scholar]
- Kim AY, Bommelje CC, Lee BE, Yonekawa Y, Choi L, Morris LG, Huang G, Kaufman A, Ryan RJ, Hao B, et al. SCCRO (DCUN1D1) Is an Essential Component of the E3 Complex for Neddylation. The Journal of biological chemistry. 2008;283:33211–33220. doi: 10.1074/jbc.M804440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, Peter M, Sicheri F. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Molecular cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Kurz T, Ozlu N, Rudolf F, O’Rourke SM, Luke B, Hofmann K, Hyman AA, Bowerman B, Peter M. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435:1257–1261. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- Lee I, Schindelin H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 2008;134:268–278. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. The EMBO journal. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WA, Sauer RT. Alternative packing arrangements in the hydrophobic core of lambda repressor. Nature. 1989;339:31–36. doi: 10.1038/339031a0. [DOI] [PubMed] [Google Scholar]
- Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Molecular cell. 2002;10:1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. The EMBO journal. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science (New York, NY) 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- Mahrour N, Redwine WB, Florens L, Swanson SK, Martin-Brown S, Bradford WD, Staehling-Hampton K, Washburn MP, Conaway RC, Conaway JW. Characterization of cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to elongin BC-based ubiquitin ligases. The Journal of biological chemistry. 2008;283:8005–8013. doi: 10.1074/jbc.M706987200. [DOI] [PubMed] [Google Scholar]
- McGrath JP, Jentsch S, Varshavsky A. UBA 1: an essential yeast gene encoding ubiquitin-activating enzyme. The EMBO journal. 1991;10:227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto M, Nishida T, Honda R, Yasuda H. Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF(skp2) toward p27(kip1) Biochem Biophys Res Commun. 2000;270:1093–1096. doi: 10.1006/bbrc.2000.2576. [DOI] [PubMed] [Google Scholar]
- Morimoto M, Nishida T, Nagayama Y, Yasuda H. Nedd8-modification of Cul1 is promoted by Roc1 as a Nedd8-E3 ligase and regulates its stability. Biochem Biophys Res Commun. 2003;301:392–398. doi: 10.1016/s0006-291x(02)03051-6. [DOI] [PubMed] [Google Scholar]
- Nastasi T, Bongiovanni A, Campos Y, Mann L, Toy JN, Bostrom J, Rottier R, Hahn C, Conaway JW, Harris AJ, et al. Ozz-E3, a muscle-specific ubiquitin ligase, regulates beta-catenin degradation during myogenesis. Developmental cell. 2004;6:269–282. doi: 10.1016/s1534-5807(04)00020-6. [DOI] [PubMed] [Google Scholar]
- Noureddine MA, Donaldson TD, Thacker SA, Duronio RJ. Drosophila Roc1a encodes a RING-H2 protein with a unique function in processing the Hh signal transducer Ci by the SCF E3 ubiquitin ligase. Developmental cell. 2002;2:757–770. doi: 10.1016/s1534-5807(02)00164-8. [DOI] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Molecular cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. A new NEDD8-ligating system for cullin-4A. Genes & development. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oved S, Mosesson Y, Zwang Y, Santonico E, Shtiegman K, Marmor MD, Kochupurakkal BS, Katz M, Lavi S, Cesareni G, et al. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. The Journal of biological chemistry. 2006;281:21640–21651. doi: 10.1074/jbc.M513034200. [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- Pelzer C, Kassner I, Matentzoglu K, Singh RK, Wollscheid HP, Scheffner M, Schmidtke G, Groettrup M. UBE1L2, a novel E1 enzyme specific for ubiquitin. The Journal of biological chemistry. 2007;282:23010–23014. doi: 10.1074/jbc.C700111200. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nature reviews. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Rose IA. Functional heterogeneity of ubiquitin carrier proteins. Progress in clinical and biological research. 1985;180:215. [PubMed] [Google Scholar]
- Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, Pierce JW, Lightcap ES, Chau V. A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, Kaelin WG, Conaway RC, Conaway JW, Branton PE. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes & development. 2001;15:3104–3117. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao-Naik C, delaCruz W, Laplaza JM, Tan S, Callis J, Fisher AJ. The rub family of ubiquitin-like proteins. Crystal structure of Arabidopsis rub1 and expression of multiple rubs in Arabidopsis. The Journal of biological chemistry. 1998;273:34976–34982. doi: 10.1074/jbc.273.52.34976. [DOI] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nature. 2008 doi: 10.1038/nrm2468. reviews. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS, Chau V, et al. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Molecular and cellular biology. 2000;20:2326–2333. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds PJ, Simms JR, Duronio RJ. Identifying determinants of cullin binding specificity among the three functionally different Drosophila melanogaster Roc proteins via domain swapping. PLoS ONE. 2008;3:e2918. doi: 10.1371/journal.pone.0002918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes & development. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science (New York, NY) 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta crystallographica. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Wada H, Yeh ET, Kamitani T. A dominant-negative UBC12 mutant sequesters NEDD8 and inhibits NEDD8 conjugation in vivo. The Journal of biological chemistry. 2000;275:17008–17015. doi: 10.1074/jbc.275.22.17008. [DOI] [PubMed] [Google Scholar]
- Walden H, Podgorski MS, Schulman BA. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. 2003;422:330–334. doi: 10.1038/nature01456. [DOI] [PubMed] [Google Scholar]
- Watson IR, Blanch A, Lin DC, Ohh M, Irwin MS. Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. The Journal of biological chemistry. 2006;281:34096–34103. doi: 10.1074/jbc.M603654200. [DOI] [PubMed] [Google Scholar]
- Willems AR, Schwab M, Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochimica et biophysica acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Wu K, Chen A, Pan ZQ. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. The Journal of biological chemistry. 2000;275:32317–32324. doi: 10.1074/jbc.M004847200. [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Sundqvist A, Nakamura A, Shen L, Botting C, Hay RT. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 2008 doi: 10.1038/embor.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science (New York, NY) 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Yuan W, Krug RM. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. The EMBO journal. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Beaudenon SL, Kelley ML, Waddell MB, Yuan W, Schulman BA, Huibregtse JM, Krug RM. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7578–7582. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Molecular cell. 2002;10:1519–1526. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.