Abstract
The DNA interstrand cross-link (ICL) resulting from the C4′-oxidized abasic site (C4-AP) is a unique clustered lesion comprised of a cross-link adjacent to a nick. The ICL is a substrate for the UvrABC nucleotide excision repair system. The strand containing the nick is preferentially incised, but the nick influences the cleavage sites. Moreover, in approximately 15% of the molecules the strand opposite the nick is incised, resulting in a more toxic double strand break. This is the first example in which an interstrand cross-link is converted by nucleotide excision misrepair into a more deleterious double strand break.
DNA double-strand breaks (DSBs) are considered to be the most dangerous form of DNA damage. Even one DSB is sufficient to kill a cell (1). Much of this danger arises from the intrinsic difficulty in repairing a severed DNA molecule. Erroneous rejoining of DNA DSBs leads to genomic instability, which can lead to tumorgenesis (2). DNA DSBs are formed by endogenous reactive oxygen species and mechanical chromosomal stress. DSBs also result from exogenous sources such as ionizing radiation and the chemotherapeutic agent bleomycin due to the accumulation of single-strand breaks (SSBs) in close proximity to one another (4,5). DNA interstrand crosslinks (ICLs) are also highly deleterious DNA lesions. ICLs are absolute blocks to replication and transcription and may also give rise to DSBs during repair. Studies in cells and lysates indicate that DSBs are produced from psoralen cross-links during repair (6,7). However, the protein(s) responsible have not been identified.
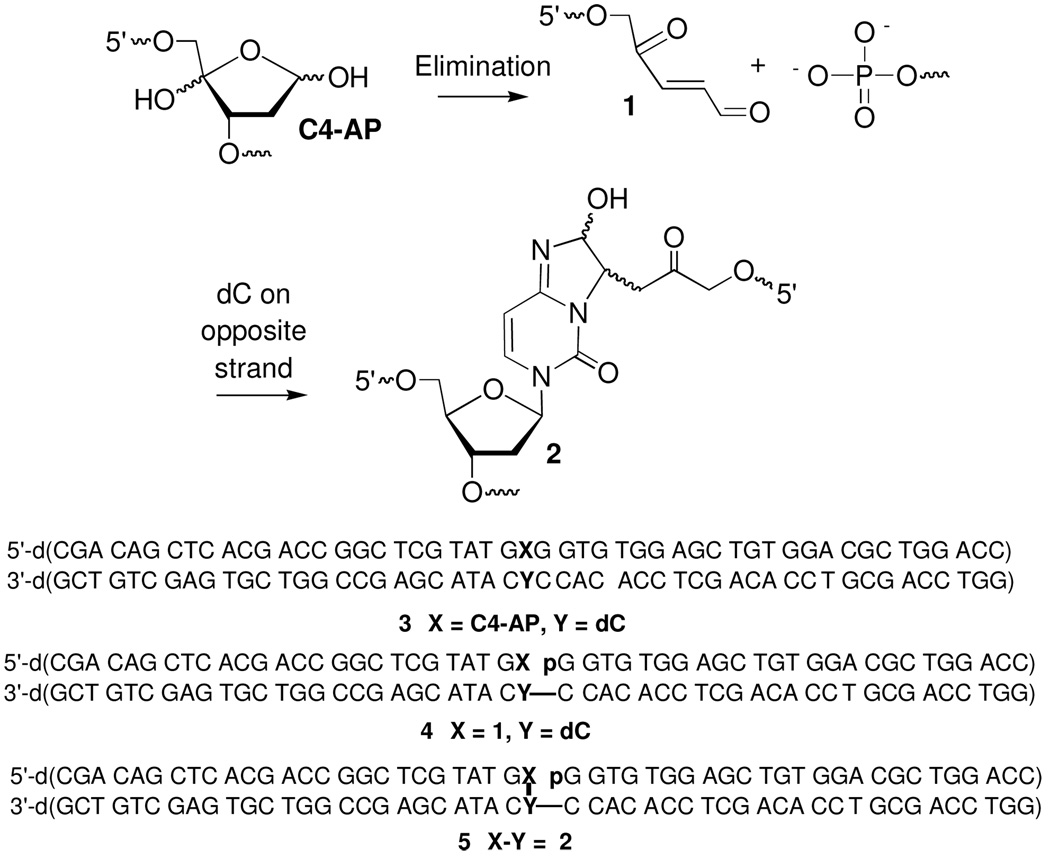
The C4′-oxidized abase site (C4-AP) is produced by a variety of DNA damaging agents (8). It is most often associated with bleomycin reactions where it accounts for as much as 40% of the products (9). We recently reported that C4-AP forms stable ICLs in DNA following its elimination to 1 (Scheme 1) (10). Elimination is catalyzed by an opposing 2′-deoxyadenosine and by DNA lyases such as endonuclease III (Nth). The unsaturated aldehyde (1) reacts with dA or dC in the opposite strand to form ICLs (e.g. 2) (10, 11). DNA damage of this type is referred to as a complex or clustered lesion. Clustered lesions are defined by the presence of two or more damage sites within ~1.5 helical turns (12). Although it has been suggested that clustered lesions can be converted to DSBs during repair (13, 14), we provide the first direct evidence that an ICL (2) is converted to a DSB during nucleotide excision repair (NER).
Scheme 1.
Formation of interstrand cross-links.
In bacteria, the three-component endonuclease UvrABC recognizes and incises damage including cross-links in distorted DNA (15). The UvrA2B complex is responsible for locating DNA damage. Once detected, UvrC is recruited and incises the damaged strand on the 5′- and 3′-side of the lesion. For example, UvrABC preferentially incises the strand covalently linked to the furan ring of psoralen-DNA interstrand cross-links. Strand scission occurs primarily at the 9th and 3rd phosphodiester bonds relative to the 5′- and 3′-phosphates of the ICLs respectively (16). UvrABC also incises the complementary DNA strand bonded to the pyrone of psoralen, albeit to a lesser extent. However, the protein complex incises only a single strand in any one cross-linked molecule, thus avoiding DSB formation.
The C4-AP ICL (2) presents an unusual situation to UvrABC because incising the strand opposite the nick would result in a DSB. In order to examine NER of 2 by UvrABC, we used a 51 base-pair substrate containing a single ICL (5). The substrate was prepared by treating 3 with Nth for synthetic expediency, to generate 4, which rapidly yields the cross-linked product (Scheme 1) (17). The ICL was purified by denaturing polyacrylamide gel electrophoresis (PAGE). Hydroxyl radical cleavage showed that the ICL results from reaction with the dC opposite the initial C4-AP. Following purification, 5 was reassembled by hybridizing the cross-linked product with the 5′-phosphorylated fragment lost during purification. Complete hybridization was verified by non-denaturing PAGE and by digesting 5 with the restriction endonuclease AluI (17). The substrate remains completely hybridized under the UvrABC assay conditions. However, some of the cross-link is decomposed at the site of cross-linking (4.5 ± 2.1%) during isolation and hybridization to form the ternary complex (17). (See blue arrows in Figures 1A–C.) An additional 1–2% of decomposition products are produced during the reaction of 5 with UvrABC at 55 °C.
Figure 1.
Incision by UvrABC on substrates 5a (A), 5b (B), 5c (C), and 5d (D). Lanes 1–3 are identical in A–D. Lane 1: G+A sequencing, lane 2: G sequencing, and lane 3: UvrABC reaction. The decomposition products formed prior to incision are indicated by the blue arrow. The inset (A) highlights the minor products of 5a, which are a result of incision on the unlabeled strand. (E) Histogram of incision sites. Relative intensities are shown by red arrows. (F) Cartoon depiction of anomalously slow migrating incision products 6a and 6b formed from cleavage of nicked strand in 5b and 5c, respectively. Radiolabeled position is indicated (*)
In separate experiments, ICL that was labeled at one of its termini (5a–d) was incubated with UvrABC. The nuclease preferentially incised the 5′-side of the ICL on the nicked strand (5a, Figure 1A, 1E, Figure 2), cutting at the 10th and 11th phosphodiester bonds relative to the cross-link (G16, G17). The yield of these incisions progressed steadily and was 48.3 ± 4.2 % after 3 h. Incision was observed only when all 3 proteins of UvrABC were present (17). Incisions 3′-from the ICL in the same strand (5d, Figure 1D, 1E) reached only a fraction of this level (7.7 ± 1 %) after 3 h. Only a fraction of these occurred at the 3rd phosphate from the lesion (T30). The majority was further downstream from the ICL (G36–T38). This is unusual because UvrABC typically makes incisions on both sides of the lesion during a single binding event (15). We propose that the presence of the nick on the 3′-side of the lesion is responsible for the reduced cleavage. DNA substrates containing a nick 3′ to a damage site are incised on the 5′-side of the lesion 3–5 times more efficiently than a substrate without the nick (18, 19). This is most likely due to the recognition of the nicked substrates as pre-incised lesions by the UvrA2B recognition complex approaching from the 5′-side of the damage. In addition, recognition of the preexisting nick shifts the incisions on 5a further from the lesion in the 5′-direction compared to other types of damage, including a psoralen ICL. From the protein's perspective, this enables it to maintain the typical spacing between incision sites.
Figure 2.
Incision product formation as a function of time. If not specified in the legend incision on the labeled strand is recorded.
Importantly, the strand opposite the nick was also incised albeit to a lesser extent (Figure 1B, 1C, 1E, Figure 2), indicating that NER of the C4-AP ICL results in double strand breaks. Cleavage of this strand on the 3′- (6.7 ± 1.3 %) and 5′-sides (6.7 ± 1.1 %) of the ICL were equal after 3 h. The UvrB-DNA complex is stabilized by the presence of a 3′-nick, which may explain the preferential incision of the nicked strand (20). The presence of the nick on the opposing strand affected the incision sites. The majority of cleavage on the 3′-side of the cross-link occurred at the 11th and 12th phosphodiester bonds from the ICL (G88, G89, Figure 1B, 1E), whereas incisions on the 5′-side of the ICL were produced at the 8th and 11th phosphodiester bonds 5′ from the damage (T69, C70, Figure 1C, 1E). Time course experiments (5b–c) show that this strand is incised on either side of the cross-link at the same rate (Figure 2). This observation suggests the 5′- and 3′-incisions result from a single binding event and is in agreement with the dual incision mechanism of UvrABC (16).
Incisions on both strands are detectable in 5a–c in a given experiment even though only one strand is radiolabeled. Products that migrate more slowly than the non-crosslinked DNA strand represent incision in the unlabeled strand. For instance, minor products from incision of 5a on the opposite, unnicked strand are observed just below the ICL (Figure 1A, inset). As expected, a large amount of incision on the opposite strand was observed when 5b (50.0 ± 5.9%) and 5c (48.7 ± 2.9%) were treated with UvrABC (Figure 1B, 1C). The yields of these products were in good agreement with major incisions detected from cleavage of 5a. Unlike 5a only a minor amount of these products migrated faster than the ICL. The majority of the labeled products that were presumed to be 6a and 6b (Figures 1B, 1C, 1F) migrated more slowly than the ICL despite its longer length. Because of its anomalous migration, one of these products (6a) was characterized further. This product is insensitive to protease digestion, suggesting it is not a result of bound protein. The identity of the anomalously migrating product was verified using a combination of hydroxyl radical cleavage of isolated 6a and comparison to an independently synthesized model compound (17).
This behavior was observed in another ICL derived from C4-AP (data not shown). However, given the importance of DSB formation resulting from misrepair of ICL 5, we considered other explanations for incision of the strand opposite the nick. The decomposition product in 5b,c (Figure 1B, 1C, blue arrow) was not a substrate for UvrABC as it persisted throughout the reaction and was unaffected by the enzymes. Furthermore, the time course showed that there was no incubation period for producing incisions on the strand opposite the nick (Figure 2), also arguing against any decomposition product from 5 as a substrate. The minor products from 5a (Figure 1A, inset) resulting from incisions on the unlabeled strand would not be present if incision of 5b,c were from decomposed ICL, because these products would be far shorter and migrate faster than material that is not cross-linked. In addition, the rate at which these products are formed coincides well with incisions for 5b,c (Figure 2). Based upon these observations and those noted above, we propose that NER of the cross-link product produced from C4-AP (5) results in a double-strand break in approximately 15% of the ICLs.
Unlike previously studied ICLs (21), incision of 5 requires absolute strand specificity by UvrABC in order to avoid generating double-strand breaks. Although the protein complex preferentially incises the nicked strand, it is unable to achieve this goal. To our knowledge, this is the first direct evidence for double-strand break formation associated with repair of an ICL by UvrABC. This observation could be biologically significant because the C4′-oxidized abasic lesion is produced by many therapeutically relevant DNA damaging agents, including the bleomycin and enediyne families of anti-cancer agents and ionizing radiation. In addition, there is evidence that the cross-links that are formed from this lesion are present in cells (11). Hence, the formation of double-strand breaks during nucleotide excision repair of these cross-links is a possible source of the cytotoxicity of the agents that produce C4-AP.
Supplementary Material
Acknowledgment
MMG is grateful for support from the National Institute of General Medical Sciences (GM-063028). JTS thanks JHU for a Sonneborn fellowship. BVH was supported by Intramural funds NIEHS, NIH.
Footnotes
Supporting Information. Experimental procedures, characterization of 6a, control experiments, and hydroxyl radical cleavage gels. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Rich T, Allen RL, Wyllie AH. Nature. 2000;407:777–783. doi: 10.1038/35037717. [DOI] [PubMed] [Google Scholar]
- 2.Khanna KK, Jackson SP. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 3.Vamvakas S, Vock EH, Lutz W. Crit. Rev. Toxicol. 1997;27:155–174. doi: 10.3109/10408449709021617. [DOI] [PubMed] [Google Scholar]
- 4.Jeggo PA. Mut. Res. 1990;239:1–16. doi: 10.1016/0165-1110(90)90028-a. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Ghorai MK, Kenney G, Stubbe J. Nucl. Acids Res. 2008;36:3781–2790. doi: 10.1093/nar/gkn302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dardalhon M, Averbeck D. Mut. Res. 1995;336:49–60. doi: 10.1016/0921-8777(94)00037-7. [DOI] [PubMed] [Google Scholar]
- 7.Bessho T. J. Biol. Chem. 2003;278:5250–5254. doi: 10.1074/jbc.M212323200. [DOI] [PubMed] [Google Scholar]
- 8.Dedon PC. Chem. Res. Toxicol. 2008;21:206–219. doi: 10.1021/tx700283c. [DOI] [PubMed] [Google Scholar]
- 9.Rabow LE, Stubbe J, Kozarich JW. J. Am. Chem. Soc. 1990;112:3196–3203. [Google Scholar]
- 10.Sczepanski JT, Jacobs AC, Greenberg MM. J. Am. Chem. Soc. 2008;130:9646–9647. doi: 10.1021/ja8030642. [DOI] [PubMed] [Google Scholar]
- 11.Regulus P, Duruox B, Bayle P, Favier A, Cadet J, Ravanat J. Proc. Natl. Acad. Sci. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomax ME, Gulston MK, O’Neill P. Radiat. Prot. Dosimetry. 2002;99:63–68. doi: 10.1093/oxfordjournals.rpd.a006840. [DOI] [PubMed] [Google Scholar]
- 13.Gulston MK, de Lara C, Jenner T, Davis E, O’Neill P. Nucleic Acids Res. 2004;32:1602–1609. doi: 10.1093/nar/gkh306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blaisdell JO, Harrison L, Wallace SS. Radiat. Prot. Dosimetry. 2001;97:25–31. doi: 10.1093/oxfordjournals.rpd.a006634. [DOI] [PubMed] [Google Scholar]
- 15.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Chem. Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 16.Van Houten B, Gamper H, Hearst JE, Sancar A. J. Biol. Chem. 1986;261:14135–14141. [PubMed] [Google Scholar]
- 17.See Supporting Information.
- 18.Zou Y, Walker R, Bassett H, Geacintov NE, Van Houten B. J. Biol. Chem. 1997;272:4820–4827. doi: 10.1074/jbc.272.8.4820. [DOI] [PubMed] [Google Scholar]
- 19.Snowden A, Wah Kow Y, Van Houten B. Biochemistry. 1990;29:7251–7259. doi: 10.1021/bi00483a013. [DOI] [PubMed] [Google Scholar]
- 20.Moolenaar GF, Monaco V, van der Marel G, van Boom JH, Visse R, Goosen N. J. Biol. Chem. 2000;275:8038–8043. doi: 10.1074/jbc.275.11.8038. [DOI] [PubMed] [Google Scholar]
- 21.Noll DM, Mason TM, Miller PS. Chem. Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.