INTRODUCTION
Section in situ hybridization (SISH) is a high-resolution tool used to analyze gene expression patterns. This protocol utilizes the Tecan Freedom EVO150 platform to perform high-throughput SISH on paraffin sections to detect mRNA with a digoxigenin (DIG)-labeled probe. The slide is mounted and imaged before performing immunohistochemistry (IHC) on the same section. The dual reaction enables a marker of protein expression to be localized on the same section as the mRNA and facilitates more accurate annotation of the gene expression.
RELATED INFORMATION
This protocol was adapted from Wilkinson and Nieto (1993) and protocols supplied by J. Yu from A. McMahon’s laboratory. The Tecan script was supplied by Tecan and G. Eichele (Yaylaoglu et al. 2005) but has been modified for paraffin sections by B. Rumballe. A protocol for synthesis of riboprobe is described in In Situ Hybridization of Whole-Mount Mouse Embryos with RNA Probes: Preparation of Embryos and Probes (Lufkin 2007).
MATERIALS
Reagents
Acetylation solution
Antibodies for immunohistochemistry, primary and secondary
Anti-DIG-alkaline phosphatase (AP) (Fab fragments; Roche 11093274910)
BM Purple (Roche Applied Science)
DAB substrate kit with hydrogen peroxide (Vector Laboratories SK-4100)
Ethanol (100%, 95%, 80%, 70%, 60%, 30%)
Hybridization buffer for SISH (preheated to 65°C for Step 19)
IHC blocking buffer
Levamisole in NTMT (2 mM)
MBST
MBST blocking buffer
Mounting medium (aqueous)
Mouse tissues (7-µm paraffin sections on SuperFrost Plus slides [VWR])
NaCl (0.85% [w/v] in RO-H2O)
NaCl/ethanol (0.85% [w/v] NaCl in 70% ethanol)
NTMT for SISH
Paraformaldehyde for SISH (4% w/v) (cold for Step 4)
Phosphate-buffered saline (PBS) for SISH
Proteinase K in PBS for SISH (10 µg/mL)
Riboprobe (DIG-labeled, 500–800 bp) (see In Situ Hybridization of Whole-Mount Mouse Embryos with RNA Probes: Preparation of Embryos and Probes (Lufkin 2007).
RNase A in TNE (2 µg/mL) (optional; see Step 27)
RO-H2O (H2O that has been purified by reverse osmosis [18 megaohm])
SSC (pH 5) (5X, 2X, 0.2X)
SSC (pH 5) (1X) with 50% formamide
TNE for SISH
Vectastain Elite ABC Kit (Vector Laboratories)
Vector M.O.M. Basic immunodetection kit (for use with mouse antibodies; Vector Laboratories)
Xylene
Equipment
Centrifuge
Chambers (humidified, covered with aluminium foil)
Coverslips (22 mm × 50 mm)
dotSlide imaging system incorporating an Olympus BX51 microscope, motorized stage and workstation, charge-coupled device [CCD] digital camera, SL50 automated slide loader, dotSlide software, and OlyVIA software (Olympus)
Freedom EVO150 robotic platform (Tecan)
Fume hood
- GenePaint system (Tecan) incorporating the following components:
- PC with Gemini pipetting software (version 4.2 or higher)
- Lauda Ecoline immersion circulator (E-200)
- Lauda Integral thermostat (T2200/E)
- Mounting Plate
- Tecan reagent troughs (12 × 100 mL)
- Tecan reagent troughs (4 × 400 mL)
- Tecan reagent troughs (2 × 1000 mL)
- Tecan Freedom EVO150 GenePaint Te-Flow module
- Te-Flow module chamber racks (2)
- Te-Flow module flow-through chambers (96)
- Te-Flow module spacers
- Temperature-controlled carrier for 32 microcentrifuge tubes
- Temperature-controlled carrier for 4 × 400 mL glass troughs
- Temperature Measuring Instrument PT100 temperature sensor
- Waste Pump (Heidolph PD 5201 peristaltic pump and SP quick [tubing wall thickness 2.5mm] pump head)
Ice
Incubator preset to 25°C, 65°C, 80°C
Microscope (dissecting)
Pen (Mini PAP wax; Invitrogen)
Photoshop (CS2 or better, Adobe)
Sterile glass slide staining dishes and stainless steel racks
Sterile plastic tubes (2mL, 15mL, 50mL)
METHOD
All solutions and containers must be free of RNase. Prepare all solutions in H2O that has been purified by reverse osmosis (18 megaohm) and use sterile plastic vials or glassware that has been baked for 2 h at 180°C.
Negative (no riboprobe/no anti-DIG-AP, etc.) and positive controls should always be used in both the SISH and the IHC. Incorporate three positive controls of low-, medium-, and high-expressing genes to assess the sensitivity of each SISH run. Include a control for each tissue type being tested.
Manual Dewaxing of Paraffin Sections
In the fume hood, assemble baked glass staining jars and label with appropriate solution names. Prepare 300 mL of each solution. Use slide staining dishes and racks to take the paraffin sections through the following dewaxing process at room temperature.
-
1.
Immerse the slides in xylene for 10 min. Repeat.
The xylene can be used for as many as 4 runs if it has been stored in RNase-free bottles.
-
2.
Rehydrate the sections by soaking the slides in the following ethanol series: 100%, 95%, 80%, 60%, 30%, for 1 min at each concentration.
-
3.
Wash in PBS for SISH for 5 min.
-
4.
Fix in cold, freshly made 4% paraformaldehyde for SISH for 10 min.
-
5.
Wash the slides twice in PBS for SISH for 5 min each. Transfer to fresh PBS for SISH.
Chamber Assembly
-
6.
Assemble the slides into Te-Flow flow-through chambers immersed in PBS for SISH. Ensure that no bubbles are present on the slides.
-
7.
Place the chambers into degassed Te-Flow chamber racks on the Tecan Freedom EVO150 platform equipped with the GenePaint system as indicated by the Gemini script. Ensure that reagents and troughs are in the correct locations on the platform.
Automated Pretreatment of Paraffin Sections
-
8.
Begin the Tecan Gemini script with a maintenance wash program to flush the system: Incubate tips and tubing in 70% ethanol for 5 min and flush thoroughly with RO-H2O.
Dispense reagents in 300 µL volumes unless otherwise stated.
-
9.
Incubate the slides in PBS for SISH for 5 min at 25°C.
-
10.
Incubate the slides twice with 10 µg/mL proteinase K in PBS for SISH for 5–10 min each.
Add the proteinase K to the buffer as the script commences. Concentration and incubation time depend on the type and developmental stage of the tissue sections, and may need to be adjusted. Try a total of 10 min for embryonic tissue and 20 min for adult tissue.
-
11.
Incubate the slides three times in PBS for SISH for 3 min each.
-
12.
Fix the slides in 4% paraformaldehyde for SISH for 5 min.
-
13.
Wash three times with PBS for SISH three times for 3 min each.
-
14.
Incubate five times in acetylation solution for 2 min each.
The script is designed to pause at this step to allow freshly prepared acetylation solution to be added to the platform.
-
15.
Incubate three times in PBS for SISH for 5 min each.
-
16.
Incubate in 0.85% NaCl for 3 min.
-
17.
Wash with NaCl/ethanol for 5 min.
-
18.
Wash with 95% ethanol for 5 min.
-
19.
Incubate twice with preheated (65°C) hybridization buffer for SISH. During this time, increase the temperature of the chamber rack to 68°C.
The slides can incubate for as long as necessary until the chamber racks are at temperature and/or the DIG-labeled riboprobes are ready to be dispensed, generally 10–20 min.
Hybridization
-
20.
Mix the DIG-labeled riboprobe with hybridization buffer for SISH to a concentration of 0.5 µg/mL. Heat the probe for 5 min at 80°C, put it on ice for 5 min, centrifuge briefly and place it on the Tecan platform.
-
21.
Use the Tecan script to dispense a 250-µL aliquot of diluted probe to each chamber. Incubate for 4–6 h at 68°C.
-
22.
Add a second 250-µL aliquot of riboprobe to the chamber and continue to incubate for another 4–6 h at 68°C.
Stringency Washes
-
23.
Preheat the stringency wash solutions (5X SSC [pH 5], 1X SSC [pH 5] with 50% formamide, 2X SSC [pH 5], and 0.2X SSC [pH 5]) in the incubator at 65°C.
-
24.
Dispense the wash solutions into 400-mL glass troughs and place into the temperature-controlled carrier on the platform. Maintain the solutions at 65°C using a script-controlled circulation bath connected to the chamber.
-
25.
Incubate the slides in 5X SSC (pH 5) for 5 min.
-
26.
Incubate the slides three times in 1X SSC (pH 5) with 50% formamide for 10 min each.
The script pauses after this step to ensure that you are ready to continue.
-
27.
(Optional) Perform the following RNase A treatment protocol:
Treat with RNase if you are trying to improve the signal-to-background ratio.
Incubate twice in TNE for SISH for 5 min each. During this time, the script ramps down the chamber rack temperature to 37°C.
-
Manually dispense 300 µL RNase A (2 µg/mL) in TNE. Incubate the slides twice for 7 min each at 37°C.
The script pauses at this step to allow manual dispensation of the RNase A. Manually dispensing the RNase A decreases the risk of contamination in the robot.
Incubate the slides twice with TNE for SISH for 5 min each at 37°C.
Increase chamber rack temperature to 65 °C.
-
28.
Incubate twice in 2X SSC (pH 5) for 10 min each at 65°C.
-
29.
Incubate four times in 0.2X SSC (pH 5) for 10 min each at 65°C. After the second wash, the script decreases the temperature of the chamber rack to 25°C.
Riboprobe Detection
-
30.
Wash the slides three times with MBST for 5 min each at 25°C.
-
31.
Incubate the slides three times with MBST blocking buffer for 20 min each.
-
32.
Prepare anti-DIG-AP diluted 1/4000 in MBST blocking buffer. Incubate the slides three times with this solution for 2 h each at 4°C.
Alternatively, incubate twice with anti-DIG-AP diluted 1/1000 for 1 h each at 25°C.
The script pauses here until you are ready to begin color development.
Color Development
-
33.
Wash the slides three times in MBST for 5 min each.
-
34.
Incubate the slides twice for 5 min with freshly prepared 2 mM Levamisole in NTMT.
-
35.
Disassemble the hybridization chambers while they are immersed in fresh NTMT for SISH. Dry the edges and place into the humidified chamber ready for color development.
-
36.
Aliquot 200 µL of BM Purple directly onto each slide. Allow the color to develop until staining is sufficient.
See Troubleshooting.
-
37.
Wash the slides with PBS for SISH for 5 min.
-
38.
Fix the slides in 4% paraformaldehyde for 10 min.
-
39.
Wash the slides twice with PBS for SISH for 5 min each.
-
40.
Mount the slides with aqueous mounting medium and 22 mm × 50 mm coverslips.
Image Capture
-
41.
Load the slides onto the SL50 automated slide loader to be batch scanned and imaged with the automated dotSlide system. Analyze using the dotSlide OlyVIA software and Adobe Photoshop (CS2 or better).
Immunohistochemistry over mRNA SISH
Dewaxing of slides and antigen retrieval is not necessary if tissue has already been through the SISH process. The general protocol for use with non-mouse antibodies is described in Steps 44–51. For detection of mouse primary monoclonal antibodies on mouse tissue, use the M.O.M. kit (Steps 52 and 53).
Negative (no primary antibody/no secondary antibody, etc.) and positive controls should always be used in the IHC.
-
42.
Soak the coverslips off the mounted slides by immersing in H2O or PBS for SISH for up to 24 h.
-
43.
Remove excess PBS for SISH. Dry around each section and circle with a mini PAP wax pen to reduce the volume of reagents needed for antibody application and detection.
Application of Non-mouse Antibodies
-
44.
Incubate the sections with 50–100 µL of IHC blocking buffer for 1 h at room temperature in a humidified chamber.
-
45.
Remove the IHC blocking buffer.
-
46.
Prepare primary antibody diluted in IHC blocking buffer. Incubate the sections with 50–100 µL of the diluted antibody for either 1 h at room temperature or overnight at 4°C in a humidified chamber.
-
47.
Remove the primary antibody solution.
-
48.
Wash the sections with PBS for SISH for 2 min. Wash again with PBS for SISH for 3 min.
-
49.
Prepare diluted secondary antibody. Add 100 µL of diluted secondary antibody to the sections.
We usually dilute the secondary antibody 1/200 in PBS for SISH.
-
50.
Incubate for 60 min at room temperature or overnight at 4°C in a humidified chamber.
-
51.
Prepare Vectastain Elite ABC Reagent according to the manufacturer’s instructions and allow it to stand for 30 min before use in Step 55. Proceed to Step 54.
Application of Mouse Monoclonal Antibodies
-
52.
Perform blocking and antibody incubation steps according to the manufacturer’s instructions that accompany the M.O.M. Kit.
-
53.
Prepare Vectastain Elite ABC Reagent according to the manufacturer’s instructions and allow it to stand for 30 min before use in Step 55. Continue with Step 54.
Detection of Antibodies
After incubation with secondary antibody, continue protocol here.
-
54.
Wash once with PBS for SISH for 2 min. Wash again with PBS for SISH for 3 min.
-
55.
Incubate sections for 30 min with the Vectastain Elite ABC Reagent that was prepared in Steps 51 or 53.
-
56.
Wash sections twice with PBS for SISH for 3 min each.
-
57.
Prepare DAB peroxidase substrate solution according to manufacturer’s instructions just before use.
-
58.
Incubate sections in peroxidase substrate solution for 30–60 sec; monitor color development continuously under a dissecting microscope.
See Troubleshooting.
-
59.
Rinse sections briefly in H2O after the appropriate color has developed.
-
60.
Wash sections once in PBS for SISH for 5 min.
-
61.
Fix sections in 4% paraformaldehyde for 10 min.
-
62.
Rinse sections twice with PBS for SISH for 5 min each.
-
63.
Mount the slides with aqueous mounting medium and 22 mm × 50 mm coverslips.
Image Capture
-
64.
Load the slides onto the SL50 automated slide loader to be batch scanned and imaged with the automated dotSlide system. Analyze using the dotSlide OlyVIA software and Adobe Photoshop (CS2 or better).
TROUBLESHOOTING
Problem: Background signal is detected for SISH.
[Step 36]
Solution: There are many possible ways to decrease SISH background, including the following:
Increase the dilution of anti-DIG-AP.
To increase the specificity of riboprobe binding, increase the hybridization temperature.
Use RNase A after hybridization if not already doing so.
Be sure that the 4% paraformaldehyde is prepared fresh on the day of use.
Centrifuge the BM Purple before use to remove any precipitate.
Remake the riboprobe and ensure the purification step is performed.
Problem: SISH signal is low.
[Step 36]
Solution: Consider the following:
Allow the slides to develop with BM Purple for >24 h. We include a low signal gene (Shh) to assess each run’s sensitivity. If no signal has developed after 24 h, allow the sample to incubate for longer (maximum 120 h), but ensure that the slides do not dry out.
Decrease the temperature of hybridization and the subsequent stringency washes by 5°C.
Increase the length of the probe.
Increase the thickness of the tissue sample.
Problem: Background signal for IHC is too high.
[Step 58]
Solution: To decrease the IHC signal, consider the following:
Increase the dilution of primary antibody.
Ensure that antibodies and all other reagents are within their expiration dates.
Repeat the antibody-binding step with a freshly thawed aliquot of primary antibody.
To ensure that the tissue has no endogenous peroxidases, include a hydrogen peroxidase step before blocking (e.g., incubate sections with 3% hydrogen peroxide for 5 min).
Problem: There is no signal for IHC.
[Step 58]
Solution: To increase the IHC signal, consider the following:
Decrease the dilution of primary antibody.
Incubate with the primary antibody overnight at 4°C.
Ensure that antibodies and all other reagents are within their expiration dates.
Repeat the antibody-binding step with a freshly thawed aliquot of primary antibody.
Sometimes immunoreactivity decreases after SISH, so consider testing the primary antibody on pre-SISH tissue.
DISCUSSION
Analysis of the developing gene expression patterns in murine brain (Gong et al. 2003; Lein et al. 2007), central nervous system (Gong et al. 2003), and whole embryo (Visel et al. 2004) is being augmented by large-scale SISH screens. Many of these projects involve automated SISH on cryosections, which tend to give a greater sensitivity of signal than paraffin-embedded tissue but result in a loss of morphological resolution. We adapted a high-throughput robotic approach for use with paraffin sections to take advantage of the superior morphology achieved. This method is applicable to any tissue, but is most relevant to tissues with a high level of morphological complexity and cellular diversity. We have applied this method to tissues of the urogenital tract. Although the sensitivity is decreased, from an anatomical point of view the quality of the cellular resolution overcomes this disadvantage. Annotation of the mRNA signal can be defined to a single cell. We have had particular success with this method using paraffin sections of embryonic (15.5 days postcoitum [dpc], 17.5 dpc) and adult kidney, gonads and bladder. Other complex tissue types with several distinct cell populations, either in murine or human tissue, may also benefit from the paraffin embedded approach.
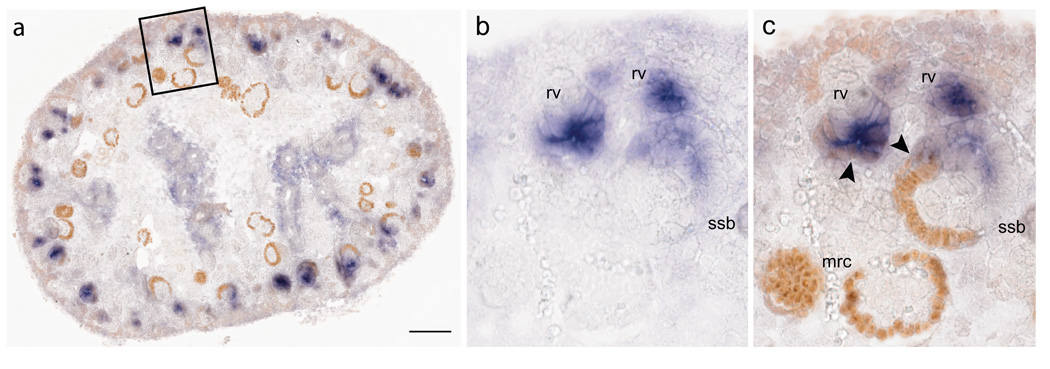
Labeling of a specific cell or cell population within a tissue section based on production of a marker protein can assist in verifying the identity of neighbouring cells or cell populations expressing a particular gene. While this can be instructive even when antibodies are applied to adjacent sections, this tool is markedly improved when performed in a dual mRNA in situ hybridization/IHC reaction. We found that in many instances the antibodies produced a stronger signal and less background on tissue that had previously been through an SISH experiment. This is possibly due to greater antigen retrieval. The ability to remove a coverslip after imaging a SISH slide ensures that high-resolution images can be taken of the gene expression and the same resolution image recaptured with the additional marker of protein expression (see Fig. 1). This can be particularly useful if the antibody co-localizes to the same structure as the gene and/or if the antibody masks the signal of a low-expressing gene.
Figure 1.
Example of dual SISH/IHC on 15.5 dpc kidney. (a) Sagittal section through kidney indicating region enlarged in b and c. Wnt4 mRNA (blue); Wt1 protein (orange). (b) SISH of Wnt4 mRNA. Expression detected in renal vesicle (rv) and S-shaped body (ssb). (c) IHC with anti-Wt1 antibody over the top of the same Wnt4 mRNA SISH. Wt1 protein expression co-expressed with Wnt4 in the proximal end of the renal vesicle (arrow). Wt1 protein localized to the proximal segment of the S-shaped body, in the visceral and parietal epithelium. This highlighted the restriction of Wnt4 expression to the medial segment of the S-shaped body. There was a slight co-expression of Wnt4 and Wt1 in the border of the medial and proximal segments of the ssb (arrowhead). Wt1 was also detected in cap mesenchyme, nephrogenic zone interstitium and maturing renal corpuscles (mrc). Scale bar = 100 µm.
Automation has provided the means to analyze hundreds of gene expression patterns, and the ability to overlay protein expression on the same SISH slide enables a more thorough annotation of the activity of the gene at a single-cell resolution. In an effort to accurately catalog this expression data, members of the GenitoUrinary Development Molcular Anatomy Project (GUDMAP) consortium have created a high-resolution anatomical ontology to provide a common language to describe the components of the developing murine urogenital tract (Little et al. 2007). The annotated results are in a publicly available database (http://www.gudmap.org), and examples of SISH with and without IHC can be found there. The continued output of SISH/IHC expression data will increase our knowledge of the molecular basis of urogenital development and facilitate our understanding of the defects and diseases affecting this organ system.
ACKNOWLEDGMENTS
We acknowledge Jing Yu and Andy McMahon for technical advice regarding the SISH protocol and Gregor Eichele and his Genepaint.org team at the Max-Planck-Institute of Biophysical Chemistry for technical advice regarding the Tecan GenePaint platform. We thank Han Sheng Chiu and Emmanuelle Lesieur for technical assistance. This work was supported by the National Institute of Diabetes, Digestion and Kidney Diseases, National Institutes of Health, USA (DK070136-02).
REFERENCES
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Little MH, Brennan J, Georgas K, Davies JA, Davidson DR, Baldock RA, Beverdam A, Bertram JF, Capel B, Chiu HS, et al. A high-resolution anatomical ontology of the developing murine genitourinary tract. Gene Expr. Patterns. 2007;7:680–699. doi: 10.1016/j.modgep.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lufkin T. In situ hybridization of whole-mount mouse embryos with RNA probes: Preparation of embryos and probes. CSH Protocols. 2007 doi: 10.1101/pdb.prot4822. [DOI] [PubMed] [Google Scholar]
- Visel A, Thaller C, Eichele G. GenePaint.org: An atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- Yaylaoglu MB, Titmus A, Visel A, Alvarez-Bolado G, Thaller C, Eichele G. Comprehensive expression atlas of fibroblast growth factors and their receptors generated by a novel robotic in situ hybridization platform. Dev. Dyn. 2005;234:371–386. doi: 10.1002/dvdy.20441. [DOI] [PubMed] [Google Scholar]