Abstract
Recurrent gene fusions, typically associated with hematological malignancies and rare bone and soft tissue tumors1, have been recently described in common solid tumors2–9. Here we employ an integrative analysis of high-throughput long and short read transcriptome sequencing of cancer cells to discover novel gene fusions. As a proof of concept we successfully utilized integrative transcriptome sequencing to “re-discover” the BCR-ABL1 10 gene fusion in a chronic myelogenous leukemia cell line and the TMPRSS2-ERG 2,3 gene fusion in a prostate cancer cell line and tissues. Additionally, we nominated, and experimentally validated, novel gene fusions resulting in chimeric transcripts in cancer cell lines and tumors. Taken together, this study establishes a robust pipeline for the discovery of novel gene chimeras using high throughput sequencing, opening up an important class of cancer-related mutations for comprehensive characterization.
Keywords: Transcriptome sequencing, Prostate cancer, Bioinformatics, Gene fusions
Characterization of specific genomic aberrations in cancers has led to the identification of several successful therapeutic targets, such as BCR-ABL1, PDGFR, ERBB2, and EGFR etc11–14, therefore a major goal in cancer research is to identify causal genetic aberrations. Gene fusions resulting from chromosomal rearrangements in cancer are believed to define the most prevalent category of ‘cancer genes’15. Typically, an aberrant juxtaposition of two genes, may encode a fusion protein (e.g., BCR-ABL1), or the regulatory elements of one gene may drive the aberrant expression of an oncogene (e.g., TMPRSS2-ERG). While gene fusions have been widely described in rare hematological malignancies and sarcomas1, the recent discovery of recurrent gene fusions in prostate2,4 and lung cancers5–9 points to their role in common solid tumors as well. Considering their prevalence and common characteristics across cancer types, gene fusions may be regarded as a distinct class of ‘mutations’, with a causal role in carcinogenesis, and being strictly confined to cancer cells, they represent ideal diagnostic markers and rational therapeutic targets.
As a proof of concept we carried out whole transcriptome sequencing of the chronic myelogenous leukemia cell line, K562, harboring the classical gene fusion, BCR-ABL1 16. Using the Illumina Genome Analyzer, we generated 66.9 million reads of 36 nucleotides in length and screened them for the presence of reads showing partial alignment to exon boundaries from two different genes. While this approach was able to detect BCR-ABL1, it was one among a set of 111 other chimeras (with at least 2 reads). Thus, in a de novo discovery mode, it would be difficult to pin-point the BCR-ABL1 fusion in the background of the other putative chimeras. However, when we used the known fusion junction of BCR-ABL1 (Genbank No. M30829) as the reference sequence, we detected 19 chimeric reads (Supplementary Fig. 1). Thus, we considered an integrative approach for chimera detection, utilizing short read sequencing technology for obtaining deep sequence data and long read technology (Roche 454 sequencing platform) to provide reference sequences for mapping candidate fusion genes.
An important concern in transcriptome sequencing was whether we could detect chimeric transcripts in the background of highly abundant house-keeping genes (i.e., would cDNA normalization be required). To address this, we compared sequences from normalized and non-normalized cDNA libraries of the prostate cancer cell line VCaP, which harbors the gene fusion TMPRSS2-ERG (Supplementary Table 1). Overall, the normalized library showed an approximately 3.6-fold reduction in the total number of chimeras nominated. Furthermore, while we expected the normalized library would enrich for the TMPRSS2-ERG gene fusion, it failed to reveal any TMPRSS2-ERG chimeras suggesting that we would not benefit from normalization in our analyses.
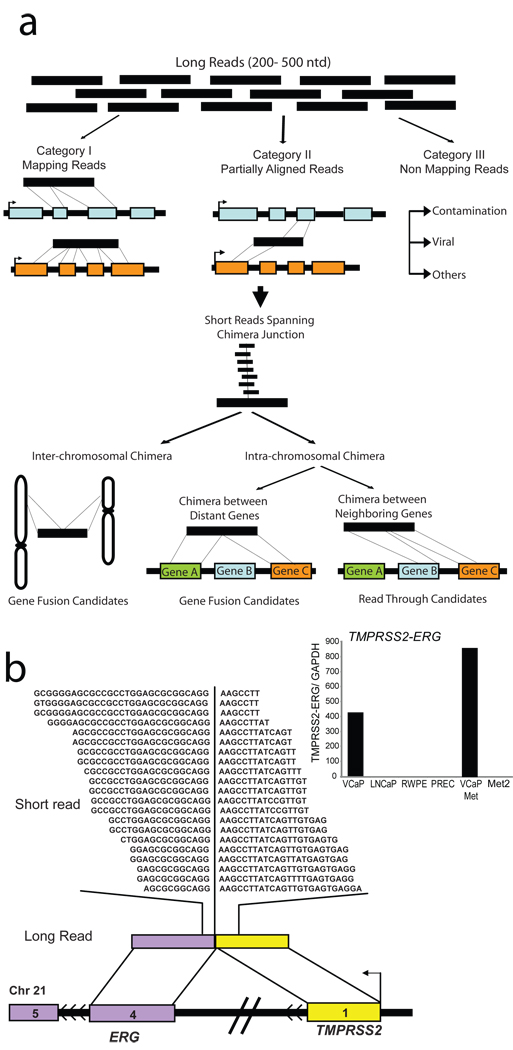
To assess the feasibility of using massively parallel transcriptome sequencing to identify novel gene fusions, we generated non-normalized cDNA libraries from the prostate cancer cell lines VCaP and LNCaP, and a benign immortalized prostate cell line RWPE. As a first step, using the Roche 454 platform, we generated 551,912 VCaP, 244,984 LNCaP, and 826,624 RWPE transcriptome sequence reads, averaging 229.4 nucleotides. These were categorized as completely aligning, partially aligning, or nonmapping to the human reference database (Fig. 1a). Sequence reads that showed partial alignments to two genes (Supplementary Methods) were nominated as first pass candidate chimeras. This yielded 428 VCaP, 247 LNCaP, and 83 RWPE candidates. Admittedly, many of these chimeric sequences could be a result of trans-splicing17 or co-transcription of adjacent genes coupled with intergenic splicing18, or simply, an artifact of the sequencing protocol. Surprisingly, among the 428 VCaP candidates, only one read spanned the TMPRSS2-ERG fusion junction using the long read sequencing platform (Supplementary Table 2).
Fig 1. Employing massively parallel sequencing to discover chimeric transcripts in cancer.
a, Schema representing our approach to employ transcriptome sequencing to identify chimeric transcripts. ‘Long read’ sequences compared with the reference database are classified as ‘Mapping’, ‘Partially Aligned’, and ‘Non-Mapping’ reads. Partially aligning reads are considered putative chimeras and are categorized as inter- or intra-chromosomal chimeras. Integration with short read sequence data is utilized for short-listing candidate chimeras and assessing the depth of coverage spanning the fusion junction. b, “Re-discovery” of TMPRSS2-ERG fusion on chromosome 21. Short reads (Illumina) are overlaid on the corresponding long read (454) represented by colored bars. Sequences spanning the fusion junction are indicated by the partition in the short reads. Chromosomal context of the fusion genes is represented by colored bars punctuated with black lines. Inset displays histogram of qRT-PCR validation of the TMPRSS2-ERG transcript.
Next, using the Illumina Genome Analyzer we obtained over 50 million short transcriptome sequence reads from VCaP, LNCaP and RWPE cDNA libraries (Supplementary Table 3). Focusing initially on VCaP cells, we identified the TMPRSS2-ERG fusion as one among 57 candidates, many of them likely false positives. To overcome the problem of false positives, lack of depth in long reads, and difficulty in mapping partially aligning short reads, we considered integrating the long and short read sequence data. Following this strategy we found the single long read chimeric sequence spanning TMPRSS2-ERG junction from VCaP transcriptome sequence, buttressed by 21 short reads (Fig. 1b), was one of only eight chimeras nominated, overall. Thus, using the integrative approach the total number of false candidates was reduced and the proportion of experimentally validated candidates increased dramatically (Supplementary Fig. 2). Extending the integrative analysis to LNCaP and RWPE sequences provided a total of fifteen chimeric transcripts, of which ten could be experimentally confirmed (Supplementary Table 4). To ensure that the integration strategy filtered out only false positives and not valid chimeras, we tested a panel of 16 long read chimera candidates that were eliminated upon integration and found that none of them confirmed a fusion transcript by qRT-PCR (Supplementary Fig. 3).
In order to systematically leverage the collective coverage provided by the two sequencing platforms, and to prioritize the candidates, we formulated a scoring function obtained by multiplying the number of chimeric reads derived from either method (Supplementary Table 4). Further, we categorized these chimeras as intra- or inter-chromosomal, based on their location on the same or different chromosomes, respectively. The latter represent bona fide gene fusions as do intra-chromosomal chimeras aligning to non-adjacent transcripts; intra-chromosomal chimeras between neighboring genes are classified as (read-throughs). Remarkably, TMPRSS2-ERG was our top ranking gene fusion sequence, second only to a read-through chimera ZNF577-ZNF649.
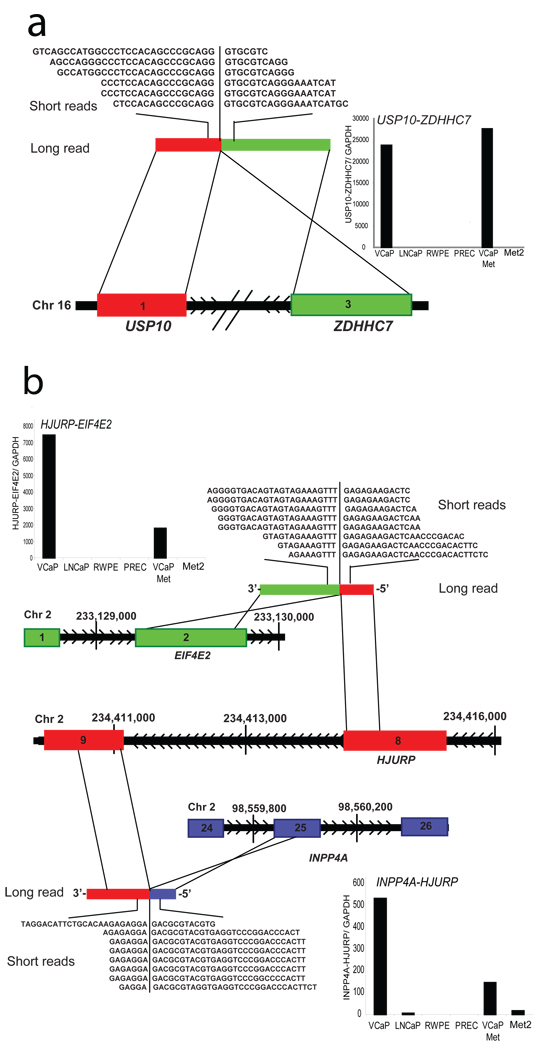
In addition to TMPRSS2-ERG we identified several new gene fusions in VCaP. One such fusion was between exon 1 of USP10, with exon 3 of ZDHHC7, both genes located on chromosome 16, approximately 200 kb apart, in opposite orientation (Fig. 2a, Supplementary Discussion). Furthermore, two separate fusions involving the gene HJURP on chromosome 2 were identified. A fusion between exon 2 of EIF4E2 with exon 8 of HJURP generated the fusion transcript EIF4E2-HJURP and a fusion between exon 9 of HJURP with exon 25 of INPP4A yielded HJURP-INPP4A (Fig. 2b, Supplementary Fig. 4).
Fig 2. Representative gene fusions characterized in the prostate cancer cell line VCaP.
a, Schematic of USP10-ZDHHC7 fusion on chromosome 16. Exon 1 of USP10 (red) is fused with exon 3 of ZDHHC7 (green), located on the same chromosome in opposite orientation. Inset displays histogram of qRT-PCR validation of USP10-ZDHHC7 transcript. b, Schematic of a complex intra-chromosomal rearrangement leading to two gene fusions involving HJURP on chromosome 2. Exon 8 of HJURP (red) is fused with exon 2 of EIF4E2 (green) to form HJURP-EIF4E2. Exon 25 of INPP4A (blue) is fused with exon 9 of HJURP (red) to form INPP4A-HJURP. Insets display histograms of qRT-PCR validation of HJURP-EIF4E2 and INPP4A-HJURP transcripts.
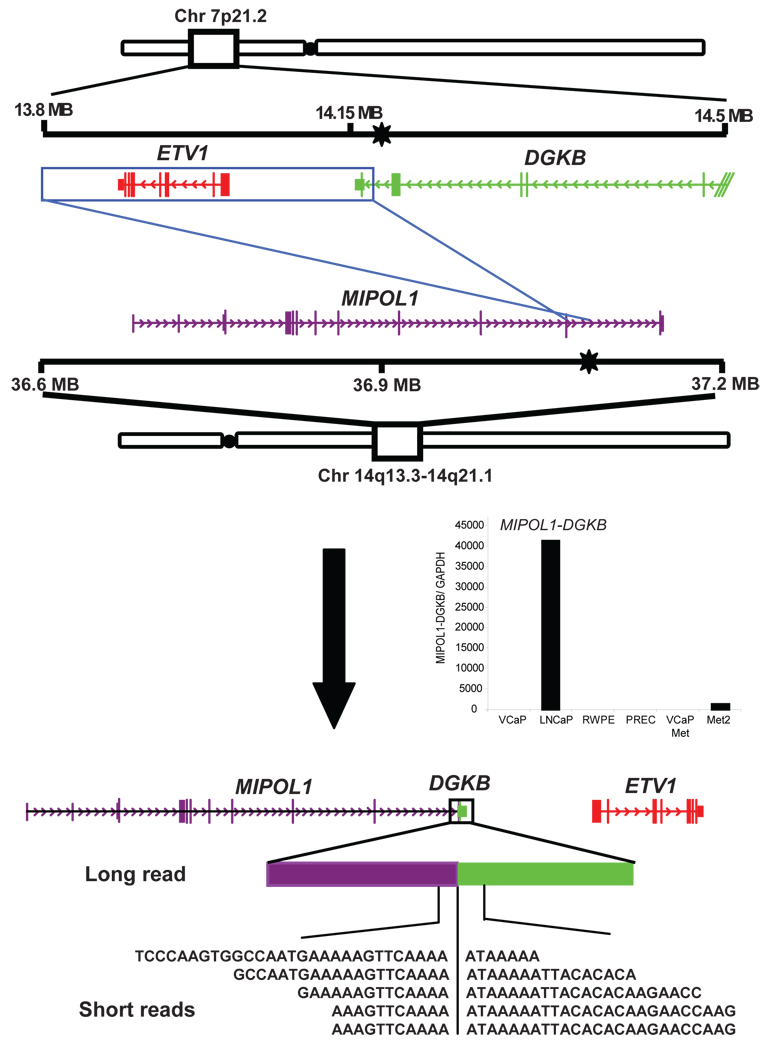
Interestingly, based on whole transcriptome sequencing, the highest ranked LNCaP gene fusion was between exon 11 of MIPOL1 on chromosome 14 with the last exon of DGKB on chromosome 7; confirmed by qRT-PCR and FISH (Fig. 3, Supplementary Fig. 5). We recently demonstrated that over-expression of ETV1, a member of the oncogenic ETS transcription factor family, plays a role in tumor progression in LNCaP cells3. The mechanism of ETV1 over-expression was attributed to a cryptic insertion of approximately 280 Kb encompassing the ETV1 gene into an intronic region of MIPOL1. Thus, while our previous study suggested that ETV1 was rearranged without evidence of an ETV1 fusion transcript, here we show the generation of a surrogate fusion of MIPOL1 to DGKB, which appears to be indicative of an ETV1 chromosomal aberration.
Fig 3. Schematic of MIPOL1-DGKB gene fusion in the prostate cancer cell line LNCaP.
MIPOL1-DGKB is an inter-chromosomal gene fusion accompanying the cryptic insertion of ETV1 locus (red) on chromosome 7 into the MIPOL1(purple) intron on chromosome 14. Previously determined genomic breakpoints (black stars) are shown in DGKB and MIPOL1. An insertion event results in the inversion of the 3’ end of DGKB and ETV1 into the MIPOL1 intron between exons 10 and 11. Inset displays histogram of qRT-PCR validation of the MIPOL1-DGKB transcript.
In addition to gene fusions, we also identified several transcript chimeras between neighboring genes, referred to as read-through events. Overall, the read-through events appear to be more broadly expressed across both malignant and benign samples whereas the gene fusions were cancer cell specific (Supplementary Fig. 6, Supplementary Discussion).
Next, we attempted to extend this methodology to tumor samples that represent the malignant cells often admixed with benign epithelia, stromal, lymphocytic, and vascular cells. Transcriptome sequencing of two TMPRSS2-ERG gene fusion positive metastatic prostate cancer tissues, VCaP-Met (from which the VCaP cell line is derived) and Met 3, and one ERG negative metastatic prostate tissue, Met 4. Interestingly, in addition to the TMPRSS2-ERG fusion sequences detected in both VCaP-Met and Met 3 tissues, three novel gene fusions were identified (Supplementary Fig. 7a). One chimeric transcript from Met 3 involves exon 9 of STRN4 with exon 2 of GPSN2 (Supplementary Fig. 7b). GPSN2 belongs to the steroid 5-alpha reductase family, the enzyme that converts testosterone to dihydrotestosterone (DHT), the key hormone that mediates androgen response in prostate tissues. DHT is known to be highly expressed in prostate cancer, and is a therapeutic target19. DHT, like its synthetic analog R1881, has been shown to induce TMPRSS2-ERG expression as well as PSA2. Additionally, we found exon 10 of RC3H2 fused to exon 20 of RGS3 in the VCaP-Met (and VCaP cells) (Supplementary Fig. 7c). Another novel gene fusion was between exon 1 of LMAN2 and exon 2 of AP3S1 (Supplementary Fig. 7d).
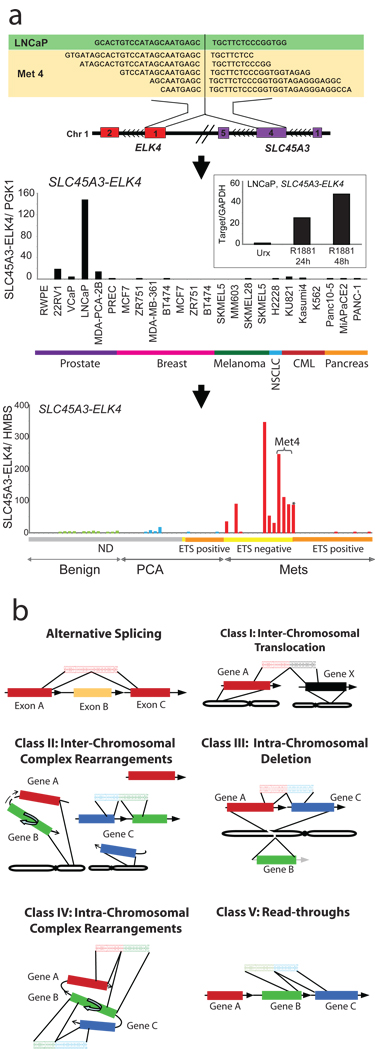
Interestingly, one read-through chimera, SLC45A3-ELK4, between the fourth exon of SLC45A3 with exon 2 of ELK4, a member of the ETS transcription factor family, was identified in metastatic prostate cancer, Met 4, and the LNCaP cell line suggesting recurrence (Fig. 4a, upper panel). Taqman qRT-PCR assay for this fusion carried out in a panel of cell lines revealed high level of expression in LNCaP cells and much lower levels in other prostate cancer cell lines including 22Rv1, VCaP, and MDA-PCA-2B. Benign prostate epithelial cells, PREC and RWPE and non-prostate cell lines including breast, melanoma, lung, CML, and pancreatic cancer cell lines were negative for this fusion (Fig. 4a, middle panel). SLC45A3 has been earlier reported to be fused to ETV1 in a prostate cancer sample3, and notably, it is a prostate specific, androgen responsive gene. Interestingly, the fusion transcript SLC45A3-ELK4 was also found to be induced by the synthetic androgen R1881 (Fig. 4a, middle panel, inset). Further, we interrogated a panel of prostate tissues for this fusion, and found it expressed in seven out of twenty metastatic prostate cancer tissues examined (Fig. 4a, lower panel). Interestingly, six of those seven positive cases have been identified as negative for ETS genes ERG, ETV1, ETV4, and ETV5 in our previous work, based on a FISH screen20. One TMPRSS2-ETV1 positive metastatic prostate cancer sample was also found to be positive for SLC45A3-ELK4 (similar to LNCaP, which is also ETV1 positive3). Unlike the previous ETS gene fusions identified, SLC45A3-ELK4 is a read-through event between adjacent genes and does not harbor detectable alterations at the DNA level by FISH (Supplementary Figure 8), array CGH (data not shown) or high-density SNP arrays (Supplementary Figure 9).As LNCaP and Met 4 harbor genomic aberrations of ETV1, and express high levels of the SLC45A3-ELK4 chimeric transcript, this suggests that ETV1 and ELK4 may cooperate to drive prostate carcinogenesis in those tumors. To our knowledge, SLC45A3-ELK4 may represent the first description of a recurrent RNA chimeric transcript specific to cancer that does not have a detectable DNA aberration. Overall, SLC45A3-ELK4 appears to be the only recurrent chimeric transcript identified in our transcriptome sequencing study, as other gene fusions tested in a panel of prostate cancer samples, appear to be restricted to the sample in which they were identified (at least in the limited number of samples we analyzed) and thus may represent rare or private mutations (Supplementary Fig. 10).
Fig. 4. Discovery of the recurrent SLC45A3-ELK4 chimera in prostate cancer and a general classification system for chimeric transcripts in cancer.
a, Upper panel, schematic of the SLC45A3-ELK4 chimera located on chromosome 1. Middle panel, qRT-PCR validation of SLC45A3-ELK4 transcript in a panel of cell lines. Inset, histogram of qRT-PCR assessment of the SLC45A3-ELK4 transcript in LNCaP cells treated with R1881. Lower panel, histogram of qRT-PCR validation in a panel of prostate tissues-benign adjacent prostate, localized prostate cancer (PCA) and metastatic prostate cancer (Mets). ETS family gene rearrangement status (by FISH) indicated by horizontal colored bars below graph. Grey not determined (ND); yellow, ETS negative; orange, ETS positive. Horizontal bracket indicates three different metastatic tissues from the same patient (Met4). Asterisk (*) denotes an ETV1 positive sample. b, Chimera classification schema (described in the text).
Next we tested if the novel gene fusions identified in this study represent acquired somatic mutations or simply, germline variations. Based on qPCR (Supplementary Fig. 11) and FISH (Supplementary Fig. 12–Supplementary Fig. 13) assessment of a representative set of fusion genes on patient matched germline tissues, we found the chimeras restricted to the cancer tissues. Further, we interrogated the 29 genes involved in our gene fusions in the Database of Genomic Variants (http://projects.tcag.ca/variation/) and found only 8 of them with previously reported copy number variations (CNVs) (Supplementary Table 5), but our matched aCGH data did not reveal any copy number variation in those genes (Supplementary Table 6), suggesting that our samples did not harbor CNVs common to the human population.
Based on the gene fusions we have characterized (Supplementary Table 7), we propose a chimera classification system (Fig. 4b). Inter-chromosomal translocation (Class I) involves fusion between two genes on different chromosomes (for example, BCR-ABL1). Inter-chromosomal complex rearrangements (Class II) where two genes from different chromosomes fuse together while a third gene follows along and becomes activated (MIPOL1-DGKB). Intra-chromosomal deletion (Class III) results when deletion of a genomic region fuses the flanking genes (TMPRSS2-ERG). Intra-chromosomal complex rearrangements (Class IV) involve a breakpoint in one gene fusing with multiple regions (HJURP-EIF4E2, and INPP4-HJURP) and Read-through chimeras (Class V) include chimeric transcripts between neighboring genes (ZNF649-ZNF577).
Overall, transcriptome sequencing was found to be a powerful tool for detecting gene fusions, exemplified by our ability to detect multiple gene fusions in cancer cell lines and tissues. One important limitation is in cases where the proximal partner contributes only the regulatory sequence to the fusion and no transcript sequence (e.g, IgH-Myc in Burkitt’s lymphoma). While it has been known that gene fusion events can play a causative role in cancer, the current study has demonstrated that a particular cancer cell line or tissue can harbor multiple gene fusions many of which are likely not recurrent. While it is unclear whether these private gene fusions play a role in malignant transformation, they could potentially cooperate with the driver mutation/gene fusions. Similar to the cataloging of point mutations associated with cancer21–27, it will be important to catalog and investigate the function of the multiple gene fusions present in a single cancer. The discovery of the chimeric transcript SLC45A3-ELK4 underscores that a refinement of next generation sequencing technologies and attendant analytical tools may well unravel the full scope of these ‘dangerous liaisons’ in carcinogenesis.
METHODS SUMMARY
Long read sequencing was conducted using 454 FLX Sequencing whereas short read sequencing was performed on the Illumina Genome Analyzer. Q-PCR for fusion candidates were performed using indicated oligonucleotide primers (Supplementary Table 8). Interphase FISH were performed in cell lines and tissues using bacterial artificial chromosome (BAC) probes (Supplementary Fig. 4a, Supplementary Fig 5a, 5c, 5e, Supplementary Fig 8, Supplementary Fig 7d, Supplementary Fig 12, Supplementary Fig 13, Supplementary Fig 14b, and 14d). Oligonucleotide comparative genomic hybridization (aCGH) was performed using Agilent arrays and copy number analysis was conducted in CGH Analytics. Affymetrix Genome-wide Human SNP Array 6.0 was processed using the Affymetrix Genotyping Console. Prostate tissues were obtained from the radical prostatectomy series at the University of Michigan and from the Rapid Autopsy Program, University of Michigan Specialized Program of Research Excellence (S.P.O.R.E.) in prostate cancer.
METHODS
Samples and cell lines
The benign immortalized prostate cell line RWPE and the prostate cancer cell line LNCaP was obtained from the American Type Culture Collection. Primary benign prostatic epithelial cells (PrEC) were obtained from Cambrex Bio Science. The prostate cancer cell line MDA-PCa 2B was provided by E. Keller. The prostate cancer cell line 22-RV1 was provided by J. Macoska. VCaP was derived from a vertebral metastasis from a patient with hormone-refractory metastatic prostate cancer28, and was provided by Ken Pienta.
Androgen stimulation experiment was carried out with LNCaP and VCaP cells grown in charcoal-stripped serum containing media for 24 h, before treatment with 1% ethanol or 1 nM of methyltrienolone (R1881, NEN Life Science Products) dissolved in ethanol, for 24 and 48 h. Total RNA was isolated with RNeasy mini kit (Qiagen) according to the manufacturer’s instructions.
Prostate tissues were obtained from the radical prostatectomy series at the University of Michigan and from the Rapid Autopsy Program29, University of Michigan Prostate Cancer Specialized Program of Research Excellence Tissue Core. All samples were collected with informed consent of the patients and prior approval of the institutional review board.
454 FLX Sequencing
PolyA+ RNA was purified from 50µg total RNA using two rounds of selection on oligo-dT containing paramagnetic beads using Dynabeads mRNA Purification Kit (Dynal Biotech, Oslo, Norway), according to the manufacturer’s instructions. 200 ng mRNA was fragmented at 82°C in Fragmentation Buffer (40 mM Tris-Acetate, 100 mM Potassium Acetate, 31.5 mM Magnesium Acetate, pH 8.1) for 2 minutes. First strand cDNA library was prepared using Superscript II (Invitrogen) according to standard protocols and directional adaptors were ligated to the cDNA ends for clonal amplification and sequencing on the Genome Sequencer FLX.
The adaptor ligation reaction was carried out in Quick Ligase Buffer (New England Biolabs, Ipswich, MA) containing 1.67 µM of the Adaptor A, 6.67 µM of the Adaptor B and 2000 units of T4 DNA Ligase (New England Biolabs, Ipswich, MA) at 37°C for 2 hours. Adapted library was recovered with 0.05% Sera-Mag30 streptavidin beads (Seradyn Inc, Indianapolis, IN) according to manufacturer’s instructions. Finally, the sscDNA library was purified twice with RNAClean (Agencourt, Beverly, MA) as per the manufacturer’s directions except the amount of beads was reduced to 1.6X the volume of the sample. The purified sscDNA library was analyzed on an RNA 6000 Pico chip on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) to confirm a size distribution between 450 to 750 nucleotides, and quantified with Quant-iT Ribogreen RNA Assay Kit (Invitrogen Corporation, Carlsbad, CA) on a Synergy HT (Bio-Tek Instruments Inc, Winooski, VT) instrument following the manufacturer’s instructions. The library was PCR amplified with 2 µM each of Primer A (5'-GCC TCC CTC GCG CCA-3') and Primer B (5'-GCC TTG CCA GCC CGC-3'), 400 µM dNTPs, 1X Advantage 2 buffer and 1 µl of Advantage 2 polymerase mix (Clontech, Mountain View, CA). The amplification reaction was performed at: 96°C for 4 min; 94°C for 30 sec, 64°C for 30 sec, repeating steps 2 and 3 for a total of 20 cycles, followed by 68°C for 3 minutes. The samples were purified using AMPure beads and diluted to a final working concentration of 200,000 molecules per µl. Emulsion beads for sequencing were generated using Sequencing emPCR Kit II and Kit III and sequencing was carried out using 600,000 beads.
Normalization by Subtraction
mRNA from the prostate cancer cell line VCaP was hybridized with the subtractor cell line LNCaP 1st-strand cDNA immobilised on magnetic beads (Dynabeads, Invitrogen), according to the manufacturers instructions. Transcripts common to both the cells were captured and removed by magnetic separation of bead-bound subtractor cDNA and the subtracted VCaP mRNA left in the supernatant was recovered by precipitation and used for generating sequencing library as described. Efficiency of normalization was assessed by qRT-PCR assay of levels of select transcripts in the sample before and after the subtraction (data not shown).
Illumina Genome Analyzer Sequencing
200ng mRNA was fragmented at 70°C for 5 min in a Fragmentation buffer (Ambion), and converted to first strand cDNA using Superscript III (Invitrogen), followed by second strand cDNA synthesis using E coli DNA pol I (Invitrogen). The double stranded cDNA library was further processed by Illumina Genomic DNA Sample Prep kit, and it involved end repair using T4 DNA polymerase, Klenow DNA polymerase, and T4 Polynucleotide kinase followed by a single <A> base addition using Klenow 3’ to 5’ exo− polymerase, and was ligated with Illumina’s adaptor oligo mix using T4 DNA ligase. Adaptor ligated library was size selected by separating on a 4% agarose gel and cutting out the library smear at 200bp (+/− 25bp). The library was PCR amplified by Phu polymerase (Stratagene), and purified by Qiaquick PCR purification kit (Qiagen). The library was quantified with Quant-iT Picogreen dsDNA Assay Kit (Invitrogen Corporation, Carlsbad, CA) on a Modulus™ Single Tube Luminometer (Turner Biosystems, Sunnyvale, CA) following the manufacturer’s instructions. 10nM library was used to prepare flowcells with approximately 30,000 clusters per lane.
Sequence datasets
Human genome build 18 (hg18) was used as a reference genome. All UCSC and Refseq transcripts were downloaded from the UCSC genome browser (http://genome.ucsc.edu/)30. Sequences of previously identified TMPRSS2-ERGa fusion transcript (Genbank accession: DQ204772) and BCR-ABL1 fusion transcript (Genbank accession: M30829) were used for reference.
Short read chimera discovery
Short reads that do not completely align to the human genome, Refseq genes, mitochondrial, ribosomal, or contaminant sequences are categorized as non-mapping. For many chimeras we expect that there will be a larger portion mapping to a fusion partner (major alignment), and smaller portion aligning to the second partner (minor alignment). Our approach is therefore divided into two phases in which we focus on first identifying the major alignment and then performing a more exhaustive approach for identifying the minor alignment. In the first phase all non-mapping reads are aligned against all exons of Refseq genes using Vmatch, a pattern matching program31. Only reads that have an alignment of 12 or more nucleotides to an exon boundary are kept as potential chimeras. In the second phase, the non-mapping portion of the remaining reads are then mapped to all possible exon boundaries using a Perl script that utilizes regular expressions to detect alignments of as few as six nucleotides. Only those short reads that show partial alignment to exon boundaries of two separate genes are categorized as chimeras. It is possible to have a chimera that has 28 nucleotides aligning to gene x and 8 nucleotides that align to gene y and z because the 8-mer does not provide enough sequence resolution to distinguish between gene y and gene z. Therefore we would categorize this as two individual chimeras. If a sequence forms more than five chimeras it is discarded because it is ambiguous. To minimize false positives, we require that a predicted gene fusion event has at least two supporting chimeras.
Long and short read integrated chimera discovery
All 454 reads are aligned against the human Refseq collection using BLAT, a rapid mRNA/DNA alignment tool32. Using a Perl script, the BLAT output files were parsed to detect potential chimeric reads. A read is categorized as completely aligning if it shows greater than 90% alignment to a known Refseq transcript. These are then discarded as they almost completely align and therefore are not characteristic of a chimera. From the remaining reads, we want to query for reads having partial alignment, with minimal overlap, to two Refseq transcripts representing putative chimeras. To accomplish this, we iterate the all possible BLAT alignments for a putative chimera, extracting only those partial alignments that have no more than a six nucleotide, or two codon, overlap. This step reduces false positive chimeras introduced by repetitive regions, large gene families, and conserved domains. Additionally, while our approach tolerates overlap between the partial alignments, it filters those having more than ten or more nucleotides between the partial alignments.
The short reads (36 nucleotides) generated from the Illumina platform are parsed by aligning them against the Refseq database and the human genome using Eland, an alignment tool for short reads. Reads that align completely or fail quality control are removed leaving only the “non-mapping” reads; a rich source for chimeras. These non-mapping short reads are subsequently aligned against all putative long read chimeras (obtained as described above) using Vmatch31, a pattern matching program. A Perl script is used to parse the Vmatch output to extract only those reads that span the fusion boundary by at least three nucleotides on each side. Following this integration, the remaining putative chimeras are categorized as inter- or intra-chromosomal chimeras based on whether the partial alignments are located on different or the same chromosomes, respectively. Those intra-chromosomal chimeras that have partial alignments to adjacent genes are believed to be the product of co-transcription of adjacent genes coupled with intergenic splicing (CoTIS)18, alternatively known as read-throughs. The remaining intra-chromosomal and all inter-chromosomal chimeras are considered candidate gene fusions.
One additional source of false positive chimeras could be an unknown transcript that is not in Refseq. Due to its absence in the Refseq database, the corresponding long read would not be able to show a complete alignment, but instead show partial hits. Subsequently, short reads spanning this transcript would naturally validate the artificially produced fusion boundary. Therefore, to remove these candidates, we aligned all of the chimeras against the human genome using BLAT. If the long read had greater than 90% alignment to one genomic location, it is considered a novel transcript rather than a chimeric read. The remaining chimeras are given a score which is calculated by multiplying the long read coverage spanning the fusion boundary against the short read coverage spanning the fusion boundary.
Coverage analysis
Transcript coverage for every gene locus was calculated from the total number of passing filter reads that mapped, via ELAND, to exons. The total count of these reads was multiplied by the read length and divided by the longest transcript isoform of the gene as determined by the sum of all exon lengths as defined in the UCSC knownGene table (Mar. 2006 assembly). Nucleotide coverage was determined by enumerating the total reads, based on ELAND mappings, at every nucleotide position within a non-redundant set of exons from all possible UCSC transcript isoforms.
Array CGH analysis
Oligonucleotide comparative genomic hybridization is a high-resolution method to detect unbalanced copy number changes at whole genome level. Competitive hybridization of differentially labeled tumor and reference DNA to oligonucleotide printed in an array format (Agilent Technologies, USA) and analysis of fluorescent intensity for each probe will detect the copy number changes in the tumor sample relative to normal reference genome. We identified genomic breakpoints at regions with a change in copy number level of at least one copy (log ratio ± 0.5) for gains and losses involving more than one probe representing each genomic interval as detected by the aberration detection method (ADM) in CGH analytics algorithm.
Real Time PCR validation
Quantitative PCR (QPCR) was performed using Power SYBR Green Mastermix (Applied Biosystems, Foster City, CA) on an Applied Biosystems Step One Plus Real Time PCR System as described3. All oligonucleotide primers were synthesized by Integrated DNA Technologies (Coralville, IA) and are listed in Table S8. GAPDH 33, primer was as described. All assays were performed in duplicate or triplicate and results were plotted as average fold change relative to GAPDH.
Quantitative PCR for SLC45A3-ELK4 was carried out by Taqman assay method using fusion specific primers and Probe #7 of Universal Probe Library (UPL), Human (Roche) as the internal oligonucleotide, according to manufacturer’s instructions. PGK1 was used as housekeeping control gene for UPL based Taqman assay (Roche), as per manufacturer’s instructions. HMBS (Applied Biosystems, Taqman assay Hs00609297_m1) was used as housekeeping gene control for Taqman assays according to standard protocols (Applied Biosystems).
Fluorescence in situ hybridization (FISH)
FISH hybridizations were performed on VCaP, LNCaP, and FFPE tumor and normal tissues. BAC clones were selected from UCSC genome browser. Following colony purification midi prep DNA was prepared using QiagenTips-100 (Qiagen, USA). DNA was labeled by nick translation labeling with biotin-16-dUTP and digoxigenin-11-dUTP (Roche, USA). Probe DNA was precipitated and dissolved in hybridization mixture containing 50% formamide, 2XSSC, 10% dextran sulphate, and 1% Denhardts solution. About 200ng of labeled probes was hybridized to normal human chromosomes to confirm the map position of each BAC clone. FISH signals were obtained using anti digoxigenin-fluorescein and alexa fluor594 conjugate for green and red colors respectively. Fluorescence images were captured using a high resolution CCD camera controlled by ISIS image processing software (Metasystems, Germany).
Affymetrix Genome-Wide Human SNP Array 6.0
1 µg each of genomic DNA samples was sent to Affymetrix service centers (Center for Molecular Medicine, Grand Rapid, MI and Vanderbilt Affymetrix Genotyping Core, Nashville, TN) for genomic level analysis of 15 samples on the Genome-Wide Human SNP Array 6.0. Copy number analysis was conducted using the Affymetrix Genotyping Console software and visualizations were generated by the Genotyping Console (GTC) browser.
Supplementary Material
Supplementary Information is linked to the online version of this paper at www.nature.com/nature.
Acknowledgements
We thank Illumina and 454 for technical support, Rohit Mehra and Javed Siddiqui for providing tissue samples, Yusong Gong, Sunita Shankar, Xiaosong Wang, and Anjana Menon for technical assistance, Jindan Yu for help with the Illumina Genome Analyzer, and Robert J. Lonigro for helpful discussions. C.A.M. was supported by an NIH Ruth L. Kirschstein post-doctoral training grant and currently derives support from the American Association of Cancer Research Amgen Fellowship in Clinical/Translational Research and the Canary Foundation and American Cancer Society Early Detection Postdoctoral Fellowship. This work was supported in part by the National Institutes of Health (to A.M.C.), Department of Defense (to A.M.C.), and the Early Detection Research Network (to A.M.C.).
References
- 1.Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nature genetics. 2004;36(4):331. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- 2.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science (New York, N.Y. 2005;310(5748):644. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 3.Tomlins SA, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448(7153):595. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 4.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nature reviews. 2008;8(7):497. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi YL, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer research. 2008;68(13):4971. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 6.Koivunen JP, et al. EML4-ALK Fusion Gene and Efficacy of an ALK Kinase Inhibitor in Lung Cancer. Clin Cancer Res. 2008;14(13):4275. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perner S, et al. EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia (New York, N.Y. 2008;10(3):298. doi: 10.1593/neo.07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rikova K, et al. Global Survey of Phosphotyrosine Signaling Identifies Oncogenic Kinases in Lung Cancer. Cell. 2007;131:14. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 9.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 10.Rowley JD. Chromosome translocations: dangerous liaisons revisited. Nature reviews. 2001;1(3):245. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- 11.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2004;350(21):2129. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 12.Slamon DJ, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. The New England journal of medicine. 2001;344(11):783. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 13.Demetri GD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. The New England journal of medicine. 2002;347(7):472. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 14.Druker BJ, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. The New England journal of medicine. 2006;355(23):2408. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 15.Futreal PA, et al. A census of human cancer genes. Nature reviews. 2004;4(3):177. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shtivelman E, Lifshitz B, Gale RP, Canaani E. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature. 1985;315(6020):550. doi: 10.1038/315550a0. [DOI] [PubMed] [Google Scholar]
- 17.Takahara T, Tasic B, Maniatis T, Akanuma H, Yanagisawa S. Delay in synthesis of the 3' splice site promotes trans-splicing of the preceding 5' splice site. Molecular cell. 2005;18(2):245. doi: 10.1016/j.molcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 18.Communi D, Suarez-Huerta N, Dussossoy D, Savi P, Boeynaems JM. Cotranscription and intergenic splicing of human P2Y11 and SSF1 genes. The Journal of biological chemistry. 2001;276(19):16561. doi: 10.1074/jbc.M009609200. [DOI] [PubMed] [Google Scholar]
- 19.Gleave M, et al. The effects of the dual 5alpha-reductase inhibitor dutasteride on localized prostate cancer--results from a 4-month pre-radical prostatectomy study. Prostate. 2006;66(15):1674. doi: 10.1002/pros.20499. [DOI] [PubMed] [Google Scholar]
- 20.Han B, et al. A fluorescence in situ hybridization screen for E26 transformation-specific aberrations: identification of DDX5-ETV4 fusion protein in prostate cancer. Cancer research. 2008;68(18):7629. doi: 10.1158/0008-5472.CAN-08-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barber TD, Vogelstein B, Kinzler KW, Velculescu VE. Somatic mutations of EGFR in colorectal cancers and glioblastomas. The New England journal of medicine. 2004;351(27):2883. doi: 10.1056/NEJM200412303512724. [DOI] [PubMed] [Google Scholar]
- 22.Cheung VG, et al. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature. 2001;409(6822):953. doi: 10.1038/35057192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephens P, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nature genetics. 2005;37(6):590. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 25.Strausberg RL, Buetow KH, Emmert-Buck MR, Klausner RD. The cancer genome anatomy project: building an annotated gene index. Trends Genet. 2000;16(3):103. doi: 10.1016/s0168-9525(99)01937-x. [DOI] [PubMed] [Google Scholar]
- 26.Weir BA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450(7171):893. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science (New York, N.Y. 2007;318(5853):1108. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 28.Korenchuk S, et al. VCaP, a cell-based model system of human prostate cancer. In vivo (Athens, Greece) 2001;15(2):163. [PubMed] [Google Scholar]
- 29.Rubin MA, et al. Rapid ("warm") autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res. 2000;6(3):1038. [PubMed] [Google Scholar]
- 30.Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32((Database issue)):D493. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abouelhoda MI, Kurtz S, Ohlebusch E. Replacing suffix trees with enhanced suffix arrays. Journal of Discrete Algorithms. 2004;2(1):53. [Google Scholar]
- 32.Kent WJ. BLAT--the BLAST-like alignment tool. Genome research. 2002;12(4):656. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3(7):34. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of this paper at www.nature.com/nature.