Abstract
Motivation
Sequences produced by automated Sanger sequencing machines frequently contain fragments of the cloning vector on their ends. Software tools currently available for identifying and removing the vector sequence require knowledge of the vector sequence, specific splice sites and any adapter sequences used in the experiment—information often omitted from public databases. Furthermore, the clipping coordinates themselves are missing or incorrectly reported. As an example, within the ~1.24 billion shotgun sequences deposited in the NCBI Trace Archive, as many as ~735 million (~60%) lack vector clipping information. Correct clipping information is essential to scientists attempting to validate, improve and even finish the increasingly large number of genomes released at a ‘draft’ quality level.
Results
We present here Figaro, a novel software tool for identifying and removing the vector from raw sequence data without prior knowledge of the vector sequence. The vector sequence is automatically inferred by analyzing the frequency of occurrence of short oligo-nucleotides using Poisson statistics. We show that Figaro achieves 99.98% sensitivity when tested on ~1.5 million shotgun reads from Drosophila pseudoobscura. We further explore the impact of accurate vector trimming on the quality of whole-genome assemblies by re-assembling two bacterial genomes from shotgun sequences deposited in the Trace Archive. Designed as a module in large computational pipelines, Figaro is fast, lightweight and flexible.
Availability
Figaro is released under an open-source license through the AMOS package (http://amos.sourceforge.net/Figaro).
1 INTRODUCTION
Even as new sequencing technologies become increasingly available (Margulies et al., 2005), Sanger sequencing remains the most widely used technique for decoding the DNA of organisms (Sanger et al., 1977). High-throughput Sanger sequencing begins by cloning a DNA fragment into a vector (usually a plasmid) that is then transfected into Escherichia coli in order to amplify the original DNA fragment. Short adapter sequences are often attached to the ends of the fragment to improve the efficiency of the cloning process (Andersson et al., 1996). The sequencing reaction is usually performed using universal sequencing primers that anneal within the vector in the vicinity of the fragment insertion site (splice site). As a result of this process (highlighted in Fig. 1), each sequence contains a small section of the vector, as well as the adapters used during cloning, in addition to the original DNA fragment. For the purpose of this article, we will refer to any such artifacts as vector sequence. These sequences must be flagged prior to further analysis of the data, in a process called vector trimming or vector clipping.
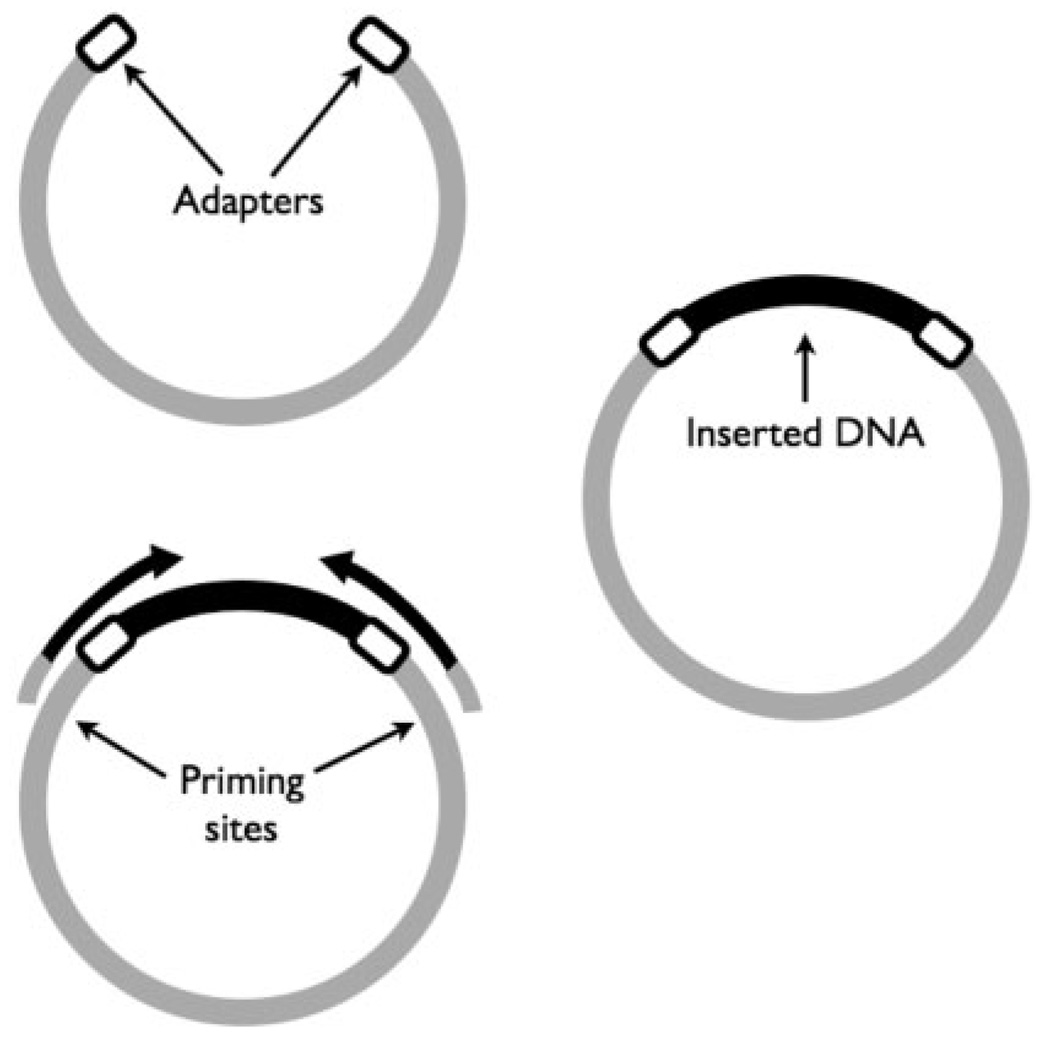
Fig. 1.
DNA from a sample (black) is cloned into a small circular piece of DNA called a vector (light gray). Short adapters (white) are used to improve efficiency of cloning the sample DNA. The molecule is then transfected into E. coli, amplified, and then sequenced from both ends starting from priming sites inside the vector.
Several software tools are available for vector removal: Lucy (Chou and Holmes, 2001), Crossmatch (www.phrap.org/phredphrapconsed.html) and VecScreen (www.ncbi.nlm.nih.gov/VecScreen). These programs compare each read to the sequence of the cloning vector, then flag sections of the read that have strong similarity to the vector (Crossmatch replaces vector sequence with Xs, Lucy provides a list of clipping coordinates in the fasta header and VecScreen provides a BLAST-like output). The alignments are performed with relaxed parameters in order to account for the higher error rates at the beginning of reads (see Fig. 2). Furthermore this approach requires three sets of information: (i) the sequence of the cloning vector; (ii) the splice site used for sequencing; and (iii) the sequence of the cloning adapters (if used—information that is often lost when the sequences are deposited in public databases). Note that the NCBI Trace Archive provides a mechanism for recording the location within the read where the vector ends (vector clip point), however this information is often missing or incorrect.
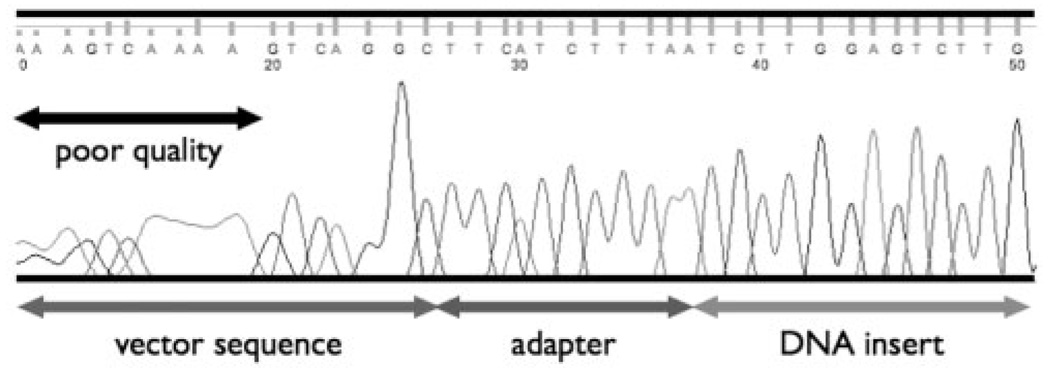
Fig. 2.
Raw output from sequencing machines contains poor quality sequence on the ends as well as vector and adapter sequence, in addition to the DNA being sequenced.
As an example, at the beginning of September, 2007, approximately 60% (735 million out of 1.24 billion) of all shotgun reads from the NCBI Trace Archive had either no vector clip information, or a vector clip point of 0 or 1, indicating the vector clipping information was not provided (clip_vector_left=0) or was arbitrarily set to the beginning of the read (clip_vector_left=1). Even when a vector coordinate is provided it is often incorrect, as described below.
We examined the shotgun reads used to assemble the Xanthomonas oryzae pv. oryzae PX099A genome, a dataset for which both vector and quality clipping coordinates had been submitted to the Trace Archive by the sequencing center. We considered all reads whose vector clip coordinate occurred at least 8 bp inside the high-quality region, then tallied the final 8 bp (8mer) of the supposed vector sequence. These 8mers should represent the end of the vector sequence; therefore, they should be virtually identical across all reads with the exception of differences caused by sequencing errors. We examined 7997 reads originating from a single sequencing library (library id 1041054961988). Furthermore, we separately examined reads sequenced with the ‘Forward’ and ‘Reverse’ trace direction in order to avoid any variability due to differences between the vector sequences flanking the splice site. The results, summarized in Table 1, highlight a much higher variability in the set of 8mers than can be explained by sequencing error alone, suggesting the vector clip points are incorrectly assigned.
Table 1.
Frequency of 8mers extracted upstream from the annotated vector clip point in shotgun reads from Xanthomonas oryzae pv. oryzae PX099A
| Trace direction | Forward | Reverse | ||
|---|---|---|---|---|
| Number of reads | 3687 | 4310 | ||
| Four most frequent 8mers and frequency |
GCGCAGCG | 40 | GCGCAGCG | 46 |
| GCCGCAGC | 29 | GTGCTGGA | 42 | |
| GATCCATT | 29 | GGCGATCG | 37 | |
| GTGCTGGA | 26 | TGGCCGAT | 35 | |
| Number of distinct 8mers |
1679 (45.5%) | 1858 (43.1%) |
We only considered reads from the library where the 5′ vector clip point was at least 8 bp to the right of the 5′ quality clip point. The reads were further binned by sequencing direction. The four most frequent 8mers are shown together with their frequency. The high level of variability indicates errors in the reported clipping coordinates.
In this article we present an algorithm for detecting and removing the vector sequence from the 5′ end of reads without prior knowledge of the vector sequences used. This algorithm can, therefore, be used to correctly identify the vector clipping points for sequences obtained from public databases. The code was implemented as a single streamlined module, named Figaro, which can be easily integrated into a high-throughput computational pipeline. The code is distributed under an open-source license through the AMOS package (http://amos.sourceforge.net).
Below we provide a detailed description of the trimming algorithm, and highlight its performance on three datasets: ~1.5 million Drosophila pseudoobscura reads; and in the de novo assembly of two bacterial genomes.
2 METHODS
For a set of shotgun reads, Figaro infers the vector sequence from the frequency of occurrence of kmers (DNA segments of length k). Under the assumption that the vector DNA flanking the inserted sequences is the same for all the sequences in a dataset, the most frequent kmers in the data likely represent vector DNA. This assumption is generally true for shotgun sequencing data, with the following exceptions: (i) different sequencing libraries may use different vectors; (ii) the vector sequences upstream and downstream from the splice site are often different (hence ‘Forward’ and ‘Reverse’ reads are prefixed by different vector DNA); and (iii) when cloning adapters are used, two different strings, corresponding to distinct adapter sequence, may prefix the reads even from a single library and orientation. To improve accuracy, the reads are partitioned by library and sequencing direction, if such information is available.
Figaro operates in two phases: (i) identification of frequent kmers likely to represent vector DNA (called vectormers throughout the text); and (ii) estimation of the vector clip point for every read, on the basis of the vectormers identified in step (i). These two components of the algorithm are described in detail below.
2.1 Detection of vectormers
The vector sequence can be recognized by identifying kmers that are more frequent at the beginning of reads than anywhere else. Intuitively, the beginning of reads represents the DNA from the vector which is shared by the majority of reads in a dataset. The remaining section of each read should be randomly sampled from the genome, leading to few commonalities between distinct reads in the dataset.
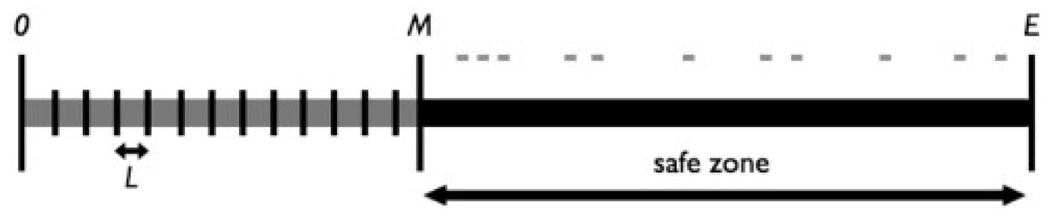
A kmer frequency table is created which records the number of occurrences of each word of length k within adjacent windows of length L over the first E bases of all reads (a kmer is assigned to the window in which it starts, thus allowing us to count kmers that cross window boundaries). We truncate all reads to a same length E in order to avoid artifacts due to the increased error rates at the ends of reads. Given a maximum vector cut length, M, we declare the safe zone of the reads to be the region from base M to E (Fig. 3). For each kmer Ki, if si is the number of occurrences of Ki in the safe zone across all reads, then we define its arrival rate αi to be:
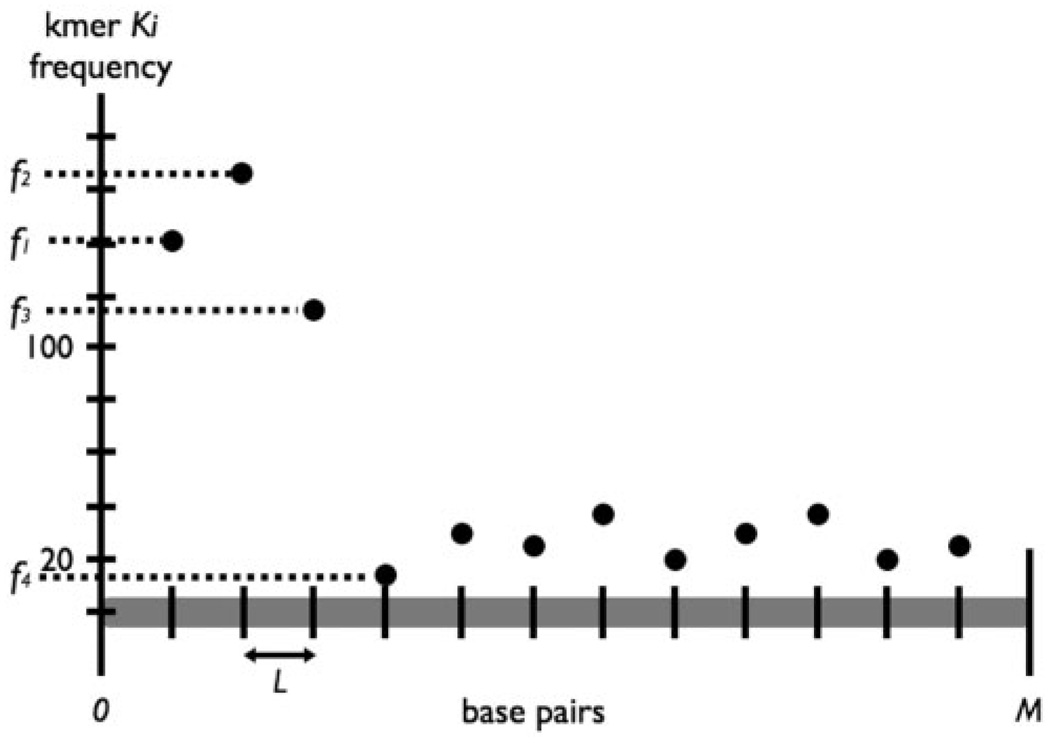
Given αi, we model the number of occurrences of Ki as a Poisson process. Letting X be the frequency of Ki in a window of length t, X follows a Poisson distribution with parameter λ=tαi. Considering fj, the frequency of occurrence of Ki within the jth window of length L (Fig. 4), we can estimate the likelihood of observing at least fj occurrences of Ki in L base pairs given αi. Mathematically,
where λ=Lαi. A kmer is declared to be a vectormer if P(X ≥ fj)<0.001 for a window within the first M base pairs of a read. By definition, we expect that 0.001*M/L of all kmers are incorrectly classified as vectormers. For example, assuming the average length of a read is 800 bp, four false vectormers are expected within any read for M = 100 and L = 20.
Fig. 3.
Within the safe zone of all reads, we consider the number of occurrences of each kmer Ki, and calculate its average arrival rate. The beginning of the read is separated into bins of length L and the frequency of each kmer within each bin is recorded.
Fig. 4.
Frequency distribution for kmer Ki across first M bases of all reads. High frequency counts at the beginning of reads indicate that Ki is a likely vectormer.
In large datasets we observed that our algorithm produced many false positives due to statistical noise and common sequencing errors. To correct for this phenomenon, we retain only the most abundant vectormers, specifically, for a user-selected threshold T, we retain the T × 100 most frequent vectormers. This simple heuristic significantly reduces overtrimming.
The implementation of Figaro uses k = 8 and L = 20. By default M = 100 and E = 500, but these parameters may be modified by the user. A reasonable setting for the threshold T is automatically computed by Figaro depending on the number of reads in the dataset, however this value can also be controlled by the user.
2.2 Vector clip estimation
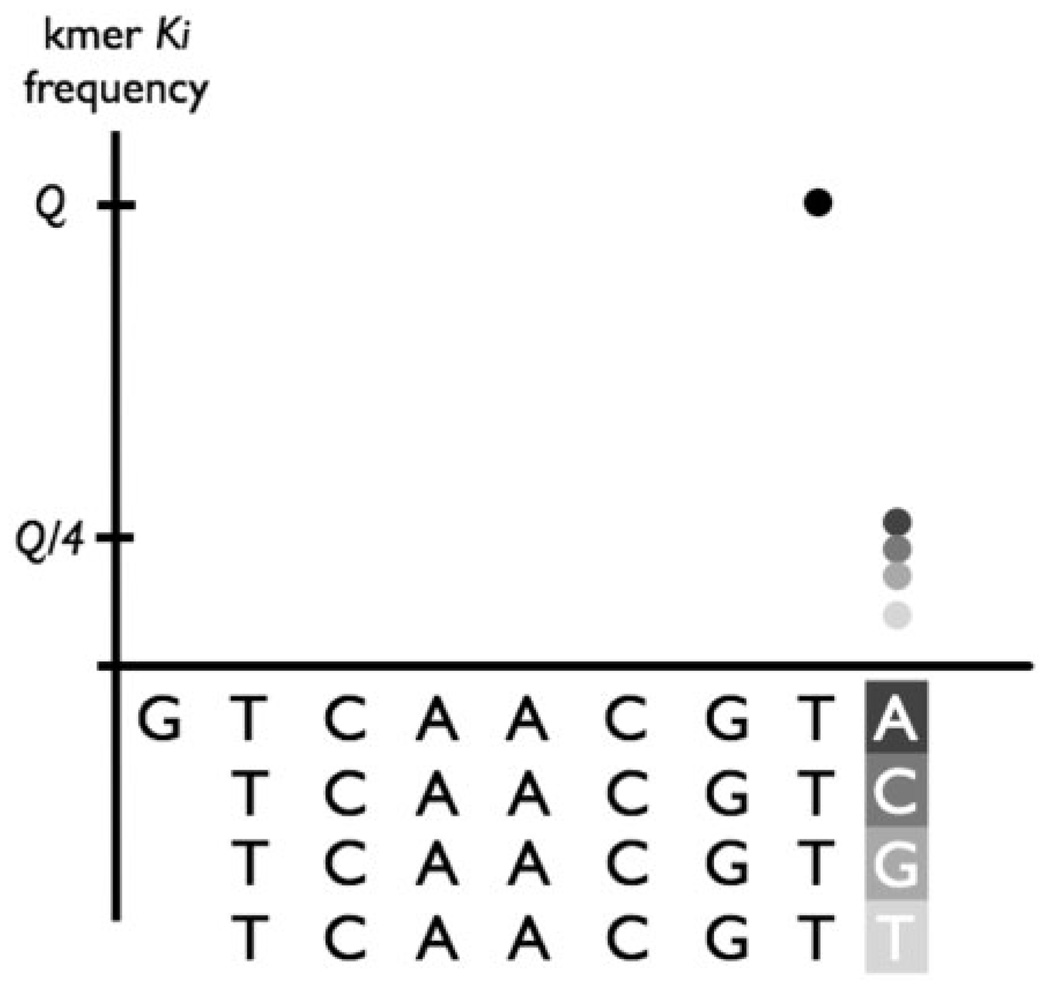
Once vectormers are computed, the algorithm first attempts to determine which vectormers are most likely to represent the ends of the vector sequences. We call these vectormers endmers. Assume a vectormer K has frequency of occurrence Q. If it is the true end of the vector, all kmers directly to the right of this vectormer (kmers whose prefix is the (k − 1) suffix of K) should have a frequency of roughly ¼ × Q (Fig. 5). The 1/4 parameter assumes equal distribution of the A, C, T and G nucleotides in the genome. To account for the non-uniform distribution of nucleotides, we first estimate the G/C content of the organism being sequenced and adjust this threshold accordingly. Suppose the calculated G/C content is δ and the A/T content is ε = 1 − δ. We declare a vectormer to be an endmer if the adjacent kmers ending in G and C both have frequency <Q × (δ/2+0.1), and if the kmers ending in A and T both have frequency <Q × (ε/2+0.1). Furthermore, to prevent many spurious endmer declarations when a large number of vectormers are allowed, we only consider the 100 most frequent vectormers as possible endmer candidates. Note that within these 100 vectormers, we only expect to find a small number of endmers (ideally four, however, sequencing errors might lead to a few more).
Fig. 5.
A conceptual example of identifying endmers (i.e. a vectormer that is likely to be the end of the vector sequence.) Note that the kmer GTCAAGCT has a frequency of Q (black dot). Frequencies of adjacent kmers ending in A, C, G and T (represented in different shades of gray) are significantly lower than Q.
Once endmers are computed, we trim every sequence using the following algorithm. The first M base pairs of each sequence are examined right to left, using a 17 bp (10 adjacent 8mers) moving window. We consider we have encountered the end of the vector, and set the clip point accordingly, once we encounter a window containing seven or more vectormers that ends in an endmer. To improve vector detection in the presence of sequencing errors, all kmers within one substitution of an endmer are also labeled as endmers.
Frequent sequencing errors can cause our algorithm to miss the end of the vector sequence (no window contains an endmer). To account for this situation, we simply select the rightmost window containing seven or more vectormers. Within this window, we identify a rightmost kmer whose distance from the end of the vector is known, then adjust the clip point accordingly. Note, that a side effect of our vectormer detection algorithm is that we can construct a de Bruijn graph (Pevzner et al., 2001) from the set of vectormers. Specifically, every vectormer represents a node in this graph, and two nodes are connected if the corresponding vectormers share a k − 1 substring (e.g. TAAAAAAA and AAAAAAAG are neighbors in this graph). Within this graph we mark the location of the endmers, and label each node with its distance (number of edges that need to be traversed) from the nearest endmer, i.e. its distance from the end of the vector. This information is used, as described above, to correctly identify the end of the vector even if an endmer cannot be detected. In the rare case where we cannot identify any vectormer whose distance to the end of the vector is known we simply use the position of the rightmost window with seven or more vectormers as the vector clip point. Note that the specific parameters of this process were set heuristically to values that performed well in our experiments. It is possible that in some cases they may need to be tuned for specific characteristics of the data being analyzed. We clearly mark these parameters at the beginning of the Figaro source code to allow their easy modification, as we have not yet identified a suitable automated procedure for estimating these parameters.
3 RESULTS
3.1 Vector trimming sensitivity and specificity
To create a test in which we know exactly where the true vector ends, we have generated a set of artificial sequences based on shotgun reads from the Chlamydophila caviae GPIC genome project (Read et al., 2003) containing variable length vector sequence on their ends. We trimmed off the first 300 bases from each of the 19 633 reads, and attached a vector sequence of random length ranging from 10 to 50 bp generated from the SmaI cloning site of the pUC18 vector (GenBank accession L09136). No vector sequence was attached to about 20% of the reads. Finally, we introduced a varying amount of error within the vector sequence to assess the performance of Figaro in the presence of sequencing errors. We ran Figaro on datasets with error rates ranging from 0% to 5%, and then compared the sensitivity and specificity of the results taking into account overtrimming and undertrimming. The same parameters were used for all trials: T = 30,M = 60 and E = 500. For each value of the parameter m, we denote a true positive (TPm) whenever the identified trimpoint is within m bases of the true trimpoint. Similarly over-trimming or undertrimming by more than m bases is denoted as a false positive (FPm) and false negative (FNm), respectively. Sensitivity and specificity are defined as follows:
Table 2 displays the sensitivity and specificity of Figaro for all trials. In the absence of errors, Figaro finds the vector sequence with 100% sensitivity, and rarely overtrims. The sensitivity and specificity remain high, even after introducing errors as high as 5% (higher than commonly encountered in practice). The fact that Figaro overtrims even in the error-less test warrants further discussion. We examined the reads that were overtrimmed by Figaro and found that the majority of these contained little or no vector (90% of these reads contained <15 bp of vector and 56% contained no vector). In such situations our algorithm is unable to identify a clear vector boundary and resorts to an aggressive trimming strategy designed to avoid undertrimming. In very few cases we found that overtrimming was due to significant homology between a section of the read and the end of the cloning vector. Note that such situations also cause overtrimming when using established, similarity-based, trimming software. Furthermore Figaro is intentionally aggressive as a small amount of overtrimming is preferable to undertrimming.
Table 2.
Sensitivity and specificity results of Figaro on simulated vector contaminant sequence with different error rates
| Error rate (%) |
SN0 (%) |
SP0 (%) |
SN3 (%) |
SP3 (%) |
SN5 (%) |
SP5 (%) |
|---|---|---|---|---|---|---|
| 0 | 100 | 99.5 | 100 | 99.7 | 100 | 99.7 |
| 1 | 99.6 | 99.3 | 99.9 | 99.7 | 99.9 | 99.7 |
| 3 | 98.0 | 98.9 | 99.0 | 99.7 | 99.1 | 99.7 |
| 5 | 96.5 | 98.0 | 98.3 | 99.6 | 98.6 | 99.6 |
For each value of the parameter m, a true positive (TPm) is counted whenever the identified trimpoint is within m bases of the true trimpoint. Similarly, overtrimming or undertrimming by more than m bases is denoted as a false positive (FPm) and false negative (FNm), respectively. We define sensitivity, SNm = (TPm/(TPm+FNm)), and specificity, SPm = (TPm/(TPm+FPm)). Introducing higher error rates reduces the program’s ability to detect the vector sequence boundary, but even with an error rate of 5%, Figaro performs well, effectively removing nearly all of the vector sequence without significantly overtrimming reads.
In order to evaluate our approach on real data, we used as a test set reads from the Drosophila pseudoobscura genome sequencing project (Richards et al., 2005). We chose these particular data because the sequencing adapters used in the project are known (Andersson et al., 1996). Searching for the two adapter sequences (16 bp each) using nucmer (Delcher et al., 2002; Kurtz et al., 2004), we collected 1 506 679 reads that matched at least 8 bp of an adapter with at least 90% identity. The 3′ end of the vector was required to match within the first 50 bp of the read, and was labeled as the true vector trimpoint. We ran Figaro with T = 30 and M = 50 (maximum vector cut length of 50 bp).
Figaro found the exact end of the vector sequences with 99.98% sensitivity and 99.15% specificity (Table 3). Without prior knowledge of the vector sequence, Figaro was able to detect and remove virtually all vector with negligible overtrimming. About 0.4% of the reads were overtrimmed by more than 3 bp and 0.01% of the reads were undertrimmed by more than 3 bp. Furthermore, the running time for this test was just short of 11 min, indicating that Figaro is efficient even for large eukaryotic projects.
Table 3.
Sensitivity and specificity results of Figaro on Drosophila pseudoobscura shotgun reads
| Max distance m |
SNm (%) |
SPm (%) |
TPm | FNm | FPm |
|---|---|---|---|---|---|
| 0 | 99.98 | 99.15 | 1 493 582 | 316 | 12 781 |
| 3 | 99.99 | 99.29 | 1 500 662 | 186 | 5831 |
| 5 | ~100 | 99.72 | 1 502 428 | 67 | 4184 |
| 10 | ~100 | 99.79 | 1 503 481 | 54 | 3144 |
Using a threshold of 30, Figaro is able to remove virtually all vector sequence and only overtrims a small proportion of reads by more than 3 bp. Note false positives and false negatives are computed only if they occur in the high-quality region of a read.
We also tested Figaro on a highly repetitive genome [maize, Zea mays (Rabinowicz and Bennetzen, 2006)]. The results on 9738 sequences from this genome were similar to those obtained for Drosophila—we achieved 100% SN1 and 99.6% SP1— indicating our method is robust in the presence of repeats.
3.2 Improving assemblies with Figaro
To illustrate how Figaro can help to improve high-throughput genomic studies, we used the Celera Assembler (Myers et al., 2000; Venter et al., 2001) to assemble the genomes of Chlamydophila caviae GPIC (Read et al., 2003) and Coxiella burnetii RSA493 (Seshadri et al., 2003), and compared these assemblies to available finished sequence. These genomes were chosen because they have been recently finished, and full quality and vector trimming information is available in the NCBI Trace Archive.
We constructed ‘Official’ assemblies using the provided vector and quality trimming points explicitly; and ‘Base quality’ assemblies using only the quality trimming information. Additional assemblies were created using the output of Figaro combined with the official quality trimming information. Figaro was run separately for each sequencing library with T = 30, M = 200 and E = 500.
Table 4 reveals that not only were the Figaro assemblies far superior to the ‘Base quality’ assemblies, but they improved upon the ‘Official’ assemblies. The Figaro assemblies of Chlamydophila caviae and Coxiella brunetii produced contigs with a higher N50 size covering more of the reference sequence than their ‘Official’ counterparts. Furthermore, our trimming did not result in any additional mis-assemblies. The C. brunetii ‘Base quality’ assembly is a particularly good example of the need for accurate vector trimming. By using Figaro the resulting assembly increased the N50 contig size nearly seven fold over the ‘Base quality’ assembly and by nearly 30% over the ‘Official’ assembly.
Table 4.
Assembly results using Figaro on two microbial genom
| Assembly Run |
Number of contigs |
Contig N50 (bp) |
% coverage | Number of errors in contigs |
|---|---|---|---|---|
| Chlamydophila caviae GPIC | ||||
| Base quality | 252 | 9466 | 93.0 | 0 |
| Official | 209 | 11 731 | 95.0 | 1 |
| T = 30 | 203 | 13 044 | 96.1 | 1 |
| Coxiella brunetii RSA493 | ||||
| Base quality | 1535 | 1232 | 77.9 | 0 |
| Official | 719 | 6713 | 94.8 | 0 |
| T = 30 | 643 | 8118 | 95.6 | 0 |
The ‘Official’ assemblies used the quality and vector trims provided with the read sets. The ‘Base quality’ assemblies only used the quality trims provided. Assemblies were performed after trimming with Figaro using T = 30, M = 200 and E = 500. Assemblies created using Figaro improve upon their ‘Official’ counterparts by increasing overall contig size without introducing more errors or losing coverage. The ‘coverage’ column denotes the percent of finished sequence covered by assembled contigs; note assembly errors are not accounted for, i.e. partial contig matches are counted toward the coverage. The ContigN50 column denotes that half the bases in the assembly are contained in contigs of the given length or greater.
4 DISCUSSION
Figaro is only intended as a tool for identifying and removing vector from the 5′ end of reads. Often, entire reads consist of vector sequence (e.g. no fragment was inserted in the vector), while in short libraries vector sequence may also occur at the 3′ end of reads. In such situations, our algorithm cannot detect the 3′ vector sequence due to the large variation in the amount of vector included in each sequence (at the 5′ end the vector ends roughly at the same location in every read), thus Figaro must be augmented with traditional vector trimming software. Further-more, since Figaro does not trim based on quality values, our software should be used in conjunction with a quality trimming program such as Lucy (Chou and Holmes, 2001). The software distribution includes several scripts that automate this process for common types of sequence data. We also provide tools for actually trimming or masking the vector sequence in the dataset.
Note that many sequencing projects use more than one library, and therefore, more than one vector. When the number of libraries is large, Figaro may incur difficulties due to the statistical nature of its algorithm. To avoid such problems, the scripts provided in the Figaro package automatically run our code on each library separately when library information is provided (e.g NCBI Trace Archive XML file).
In addition, the algorithms implemented in Figaro implicitly assume the randomness of a typical shotgun process. Therefore, Figaro cannot be used for targeted sequencing experiments where a same gene is sequenced across multiple samples. Also, in EST sequencing projects, the use of Figaro may result in the incorrect removal of the polyA tail.
The various parameters controlling the execution of our code are automatically set to reasonable default values. These values can also be controlled by the users if the default values are inappropriate for the data being processed. For example, the parameter E, marking the end of the ‘good quality’ section of a read, is usually set to 500, however its value should be increased or decreased depending on the average read length being analyzed. Similarly, our code performs best if the parameter M (the window within which Figaro searches for the vector sequence) is set to a value close to the expected length of the vector. This parameter should, therefore, be adjusted if additional information is available regarding the distance of the sequencing primers from the cloning site. Note, however, that M should be set conservatively (greater than the expected length of the vector) in order to avoid undertrimming.
Raw shotgun sequences are placed in the NCBI Trace Archive at an ever increasing rate, rapidly outpacing the availability of current assemblies for many genomes. Constructing independent assemblies from these data is complicated by the often incomplete or incorrect vector trimming information reported in the public databases. The program described in this article, Figaro, provides scientists with the means to automatically detect and remove the vector sequence from shotgun reads without prior knowledge about the sequencing protocol, thereby enabling the large-scale re-assembly of public data. Furthermore, even if the vector sequence is known, Figaro provides an efficient and effective alternative to commonly used vector removal programs.
ACKNOWLEDGMENTS
We thank Art Delcher and Aleksey Zimin for their suggestions and feedback. We are grateful to Jessica Miller and Steven Salzberg for help naming our method. M.P. was supported in part by grant R01-LM006845 from NIH and grant HU001-06-1-0015 from the Uniformed Services University of the Health Sciences administered by the Henry Jackson Foundation. J.Y. and M.R. were supported in part under NSF grant DMS 0616585 and NIH grant 1R01HG0294501. J.W. was supported in part by University of Maryland NSF VIGRE fellowship DMS0240049.
Footnotes
Conflict of Interest: none declared.
Accession numbers for finished genomes in GenBank: AE015925 and AE016828. Simulated shotgun reads from this study are available online at http://amos.sourceforge.net/Figaro.
REFERENCES
- Andersson B, et al. A ‘double adaptor’ method for improved shotgun library construction. Anal. Biochem. 1996;236:107–113. doi: 10.1006/abio.1996.0138. [DOI] [PubMed] [Google Scholar]
- Chou HH, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17:1093–1104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- Delcher AL, et al. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- Pevzner PA, et al. An Eulerian path approach to DNA fragment assembly. Proc. Natl Acad. Sci. USA. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz PD, Bennetzen JL. The maize genome as a model for efficient sequence analysis of large plant genomes. Curr. Opin. Plant Biol. 2006;9:149–156. doi: 10.1016/j.pbi.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Read TD, et al. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 2003;31:2134–2147. doi: 10.1093/nar/gkg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, et al. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, et al. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R, et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl Acad. Sci. USA. 2003;100:5455–5460. doi: 10.1073/pnas.0931379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]