Abstract
Protein S is a major spore coat protein of Myxococcus xanthus, consisting of two homologous domains, the N-terminal domain (NTD) and the C-terminal domain, both of which contain a Ca2+-binding site. Protein S tightly binds to myxospores in a Ca2+-dependent manner. Here, we constructed a novel expression vector, pCold-PST, encoding two tandem repeat NTDs (PrS2). By using this vector, a number of human proteins that were expressed at low levels or in insoluble forms by a pET vector were expressed not only at high levels but also in soluble forms. We also demonstrated that an Escherichia coli protein tagged with PrS2 fully retained its function, indicating that it is folded independently from the tag. This technology not only allows simple, one-step protein purification using myxospores, but can also be used for the identification of proteins interacting with a protein of interest and will prove immensely useful for structural studies of proteins which are difficult to produce or are insoluble.
A major bottleneck in structural biology is that many human proteins are expressed very poorly or are expressed well but not in soluble forms. These difficulties may arise due to the improper folding of human proteins in Escherichia coli cells, where they are quickly digested by proteases or accumulated as inclusion bodies. A number of expression systems have been developed to overcome this problem, for example, by coexpressing human proteins with molecular chaperones (12), expressing them as fusions with maltose-binding protein (4), glutathione S-transferase (16), small ubiquitin-related modifier (SUMO) (10), and others (2), or expressing them at low temperatures using cold shock vectors (15).
In the present study, we attempted to develop an expression vector for human proteins or other proteins which are difficult to express at high levels or in soluble forms in E. coli by using other expression systems. For this strategy, we took advantage of protein S, a major Ca2+-binding spore coat protein from Myxococcus xanthus which binds to the M. xanthus spore (myxospore) surface in the presence of Ca2+, even in 1 M NaCl (8). Protein S can be readily dissociated from myxospores in its soluble form with EDTA. Previously, we also observed that when OmpR, an E. coli transcription factor, is fused to the N-terminal domain (NTD) of protein S, not only does the expression of the OmpR protein improve but also its solubility dramatically increases, by more than 20-fold (9). Importantly, the protein S-OmpR fusion protein exhibits DNA binding almost identical to that of OmpR alone (19).
To create this novel expression vector, we decided to use the pCold vector, which was created in our laboratory (15), as a template. The pCold vector contains the promoter of the cold shock-inducible gene for the major cold shock protein CspA and the gene's 134-base 5′ untranslated region, allowing a very high level of expression of a cloned gene upon cold shock (15). This vector also contains a lac operator downstream of the cspA promoter so that the expression of a cloned gene can be tightly regulated not only by temperature but also by lac inducers such as isopropyl-β-d-1-thiogalactopyranoside (IPTG). We combined the novel features of both protein S and CspA to create the new vector pCold-PST. Using this vector, we demonstrate here that the expression and solubility levels of a number of human proteins are significantly improved when these proteins are expressed as fusions with the NTDs of protein S. Importantly, the fusion proteins can be purified from the cell lysate in one step using myxospores in the presence of Ca2+. This new technology can also be used for the analysis of protein-protein interactions and for the structural study of proteins which are difficult to express or are insoluble.
MATERIALS AND METHODS
Plasmid construction.
The pCold-PST vector was constructed by cloning the gene which encodes two tandem repeat NTDs (the PrS2 tag) into the pCold IV vector (TaKaRa Bio) with an additional tobacco etch virus (TEV) protease cleavage site, a multiple-cloning site (MCS), and a sequence encoding a hexahistidine (His6) tag. The plasmid pPR010, which contains the fusion gene encoding the PrS2-OmpR protein (5), was used as a template to obtain PrS2 by PCR. PCR was carried out with a 5′ PCR primer containing an NcoI site and a 3′ PCR primer containing a KpnI site to clone the entire PrS2 coding region. The appropriate PCR product was digested with NcoI and KpnI and then cloned into the pCold IV vector digested with NcoI and KpnI to construct pCold-PrS2. Note that in this pCold IV vector construct, the sequence of the NdeI site of an MCS was mutated to create an NcoI site. Since there is another NdeI site between the first and second NTD sequences, the sequence CAT in the NdeI site was deleted by site-directed mutagenesis, resulting in pCold-PrS2 (ΔNdeI). Two oligonucleotides (5′-CGGGGTACCGAAAACCTGTATTTTCAGGGAGCTGCAGCTCATATGGTCGACCTCGAGGGATCCCGTGGTGAAATCCATCACCATCACCATCACTAATCTAGAATTATT-3′ and 5′-AATAATTCTAGATTAGTGATGGTGATGGTGATGGATTTCACCACGGGATCCCTCGAGGTCGACCATATGAGCTGCAGCTCCCTGAAAATACAGGTTTTCGGTACCCCG-3′) were annealed, and the resultant DNA fragment contains a KpnI site at the 5′ end, a TEV protease digestion site, an MCS (for NdeI, SalI, XhoI, and BamHI), and a His6 tag sequence followed by a stop codon, TAA, and an XbaI site at the 3′ end (see Fig. 1b). This DNA fragment was digested with KpnI and XbaI and inserted into pCold-PrS2 (ΔNdeI) digested with KpnI and XbaI. The final construct was named pCold-PSTTEV.
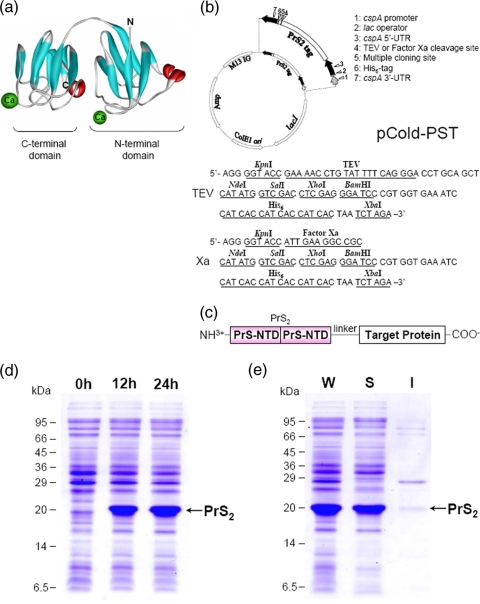
FIG. 1.
NMR structure of protein S and map of the pCold-PST vector. (a) NMR structure of M. xanthus protein S with Ca2+ ions bound (3). (b) Structure and schematic map of pCold-PSTTEV or pCold-PSTXa. (c) Schematic presentation of a PrS2 fusion protein. Two tandem repeats of the NTD of protein S (amino acids 1 to 92; PrS2) are fused at the N-terminal end of a protein of interest, and a short linker which contains a protease cleavage site is added between the PrS2 tag and the target protein. (d) Expression of PrS2. Aliquots of a culture growing at 15°C were removed at 0, 12, and 24 h after induction with 1 mM IPTG and analyzed by SDS-PAGE. (e) Solubility of PrS2 as determined using a culture aliquot obtained 24 h after induction. M13 IG, intergenic region of M13 bacteriophage; UTR, untranslated region; W, whole-cell lysate; S, soluble fraction; I, insoluble fraction of PrS2.
To construct pCold-PSTXa, a fragment was amplified by PCR with 5′-ACGGTACCATTGAAGGCCGCCATATGGTCGACCTCGAGGGATCCGT-3′ and 5′-ACGGATCCCTCGAGGTCGACCATATGGCGGCCTTCAATGGTACCGT-3′ (underlining indicates the factor Xa cleavage site). The fragment was then inserted into pCR 2.1-TOPO (Invitrogen) after the addition of an adenine residue to the 3′ end and subcloned into pCold-PST after treatment with KpnI and NdeI.
The pCold His6 tag vector was constructed by replacing the PrS2 tag sequence with the His6 tag sequence by using NcoI and NdeI. After the replacement, the NcoI site was destroyed by site-directed mutagenesis.
Expression of fusion proteins.
E. coli BL21 cells were transformed with pCold-PST and pCold His6 tag vectors harboring different genes of interest. The transformed cells were grown at 37°C in M9-Casamino Acid medium to an optical density at 600 nm of 0.8. The cultures were shifted to 15°C or room temperature, and IPTG (1 mM) was added to induce protein expression. The cells were harvested at 14 h after induction and disrupted by sonication in 200 μl (per 1 ml culture) of buffer (50 mM Tris-HCl [pH 8.0], 50 mM KCl, and 5% glycerol) containing 1 mM phenylmethylsulfonyl fluoride. The cell debris and insoluble proteins were removed by centrifugation at 9,000 × g for 20 min, and the supernatant was further centrifuged at 1.8 × 106 × g for 10 min to remove the membrane fraction.
Complex formation by EnvZc and OmpR or PrS2-OmpR.
A C-terminal fragment of EnvZ (EnvZc; 4 μM) and PrS2-OmpR (4 μM) or OmpR (4 μM) (20) were mixed and incubated in buffer A (50 mM Tris-HCl [pH 8.0], 50 mM KCl, 5 mM CaCl2, 5% glycerol) at room temperature for 5 min.
Phosphorylation of OmpR and PrS2-OmpR.
Purified 32P-labeled phosphorylated EnvZc ([32P]EnvZc-P; 2 μM) was mixed with OmpR (4 μM), PrS2-OmpR (4 μM), or the mixture of OmpR (2 μM) and PrS2-OmpR (2 μM) in buffer A. The final reaction mixtures were incubated at room temperature. Aliquots were removed at 20, 40, 60, and 120 s, and the reaction was stopped by adding 5× sodium dodecyl sulfate (SDS) loading buffer. The samples were subjected to SDS-17.5% polyacrylamide gel electrophoresis (SDS-17.5% PAGE).
Dephosphorylation of OmpR and PrS2-OmpR.
Purified 32P-labeled EnvZc-P (2 μM) was mixed with OmpR (4 μM), PrS2-OmpR (4 μM), or the mixture of OmpR (2 μM) and PrS2-OmpR (2 μM), and the reaction mixture was incubated in buffer A at room temperature for 2 min to generate phosphorylated OmpR (OmpR-P) or PrS2-OmpR-P. After the addition of ADP at the final concentration of 1 mM, aliquots were removed at 20, 40, 60, and 120 s and the reaction was stopped by adding 5× SDS loading buffer. The samples were subjected to SDS-17.5% PAGE.
Preparation of myxospores.
Myxospores were prepared as described previously (8). Harvested myxospores in buffer (10 mM Tris-HCl [pH 7.6], 25 mM EDTA [pH 8.0]) were heated at 70°C three times for 10 min each time to remove protein S (8) and protein C (11) bound to myxospores. Myxospores were then autoclaved at 121°C for 20 min to eliminate inherent protease activity and to inactivate the myxospores for germination. Note that treated myxospores can no longer germinate, even under optimum conditions (8 mM MgSO4, 1 mM CaCl2, and 0.2% Casitone) (13), while they retain their ability to bind to protein S or PrS2-tagged proteins.
One-step purification.
To remove Ca2+-dependent proteins, CaCl2 (final concentration, 1 mM) was added to the soluble fraction of fusion proteins and the mixture was centrifuged at 1.8 × 106 × g for 10 min. The supernatants were mixed with 1.0 × 107 myxospores in buffer B (10 mM Tris-HCl [pH 7.6], 50 mM NaCl, and 1 mM CaCl2) and incubated for 30 min at 4°C. The myxospores were collected at 3,000 × g and washed first with buffer C (10 mM Tris-HCl [pH 7.6], 1 M NaCl, and 10 mM CaCl2) two times and further washed with buffer B and analyzed by SDS-PAGE.
Factor Xa cleavage on myxospores.
The soluble fraction of PrS2-HR2898 was collected and bound to myxospores (1.0 × 107 spores). PrS2-HR2898 bound to myxospores was treated with 0.2 μg of factor Xa (BioLabs) with buffer B to cleave PrS2 fusion proteins. After incubation at room temperature for 30 min, the mixture was separated into the supernatant and myxospores by centrifugation at 3,000 × g for 5 min, and the supernatant was further centrifuged at 9,000 × g for 20 min and analyzed by SDS-PAGE.
Pulldown assay with protein S tag (PST).
Der, an essential GTPase that is required for ribosome assembly in E. coli (7), was overexpressed in E. coli by a derivative of the pET28 plasmid carrying a T7 promoter. The soluble fraction of Der and a PrS2-YihI fusion protein lacking the 45 N-terminal residues of YihI (PrS2-Δ45 YihI) (J. Hwang and M. Inouye, unpublished results) or purified PrS2 were mixed together and incubated at 4°C for 30 min. Myxospores (1.5 × 107 cells) were added to the mixture with 1 mM CaCl2, and the mixture was incubated at 4°C for 30 min to allow the binding of the myxospores to the Der-PrS2-Δ45 YihI complex. Unbound proteins were washed out with buffer (10 mM Tris-HCl [pH 7.6] and 1 mM CaCl2) two times and analyzed by SDS-PAGE.
RESULTS
Construction of pCold-PST vector using the NTD of protein S.
Protein S, consisting of 173 residues, is composed of two independent Ca2+-binding domains, the 92-residue NTD and the 81-residue C-terminal domain (Fig. 1a) (3). It is a heat-stable protein and, particularly, the NTD is more stable (melting temperature, 68°C) than the C-terminal domain (melting temperature, 48°C) in the presence of Ca2+ (18). The fusion of a single NTD to the N-terminal end of the OmpR protein results in dramatic improvement in the solubility, as well as the expression, of OmpR (9). However, a single NTD is not capable of binding to myxospores in the presence of Ca2+ (data not shown). Therefore, in order to retain the spore-binding capability together with the heat stability, we constructed a pCold vector that produces a fusion protein containing PrS2 at the N-terminal end of a target protein (Fig. 1b and c). This vector, termed pCold-PST, was constructed using the pCold IV vector (15). In the pCold-PST vector, a short linker which encodes a TEV protease cleavage site (ENLYFQG) or a factor Xa cleavage site (IEGR) (for pCold-PSTTEV or pCold-PSTXa, respectively) is followed by an MCS and a His6 tag sequence (Fig. 1b). The transcription of the cloned gene can be terminated by the 3′ untranslated region from the cspA gene.
To test the efficiency of this vector for allowing high-level expression of a cloned gene, the induction of PrS2 alone was first examined as an example. As shown in Fig. 1d, when BL21 cells transformed with pCold-PSTTEV were incubated at 15°C in the presence of 1 mM IPTG, dramatic induction of PrS2 was observed after 12 h and the amount of PrS2 was further increased to account for more than 20% of the total cellular protein synthesis at 24 h postinduction. The PrS2 thus produced was highly soluble, as indicated by the ultracentrifugation profile (Fig. 1e).
Application of pCold-PSTTEV for human proteins.
We chose seven human proteins that are associated with cancer as target proteins. The expression of these proteins with the use of a pET vector has been attempted previously by Northeast Structural Genomics (NESG). However, their expression was not satisfactory for structural study by both nuclear magnetic resonance (NMR) and X-ray crystallography analyses, as some of these proteins (HR79, HR2930, and HR3073) were expressed very poorly, one (HR3111) was not expressed at all, and the others (HR2898, HR2921, and HR2929) were expressed well but not in soluble forms (Table 1) (1). The function(s) of each protein is described in Table 1, footnote a.
TABLE 1.
Expression levels and solubilities of human proteins with or without a PrS2 tag
| NESG identification no.a | No. of residues | pET
|
pCold His6 tag
|
pCold-PST
|
|||
|---|---|---|---|---|---|---|---|
| Expressionb | Solubilityc | Expressionb | Solubilityc | Expressionb | Solubilityc | ||
| HR79 | 168 | + | + | 0 | NAd | ++ | + |
| HR2898 | 165 | ++++ | + | ++ | + | ++++ | ++++ |
| HR2921 | 134 | +++ | 0 | + | 0 | + | + |
| HR2929 | 182 | ++++ | + | ++++ | +++ | +++++ | +++++ |
| HR2930 | 175 | + | + | 0 | NA | ++ | + |
| HR3073 | 184 | ++ | 0 | ++ | 0 | +++ | +++ |
| HR3111 | 119 | 0 | NA | 0 | NA | +++ | +++ |
HR79, human Ras-like transcription suppressor; HR2898, human growth arrest and DNA damage-inducible protein GADD45-alpha, involved in apoptosis, DNA repair, nuclear localization, and regulation of cyclin-dependent kinase activity, has limited sequence similarity to ribosomal protein L30; HR2921, human DNA-binding inhibitor ID-2, required for G1 progression; HR2929, human HSP20 heat shock protein beta 2, protein chaperone, enzyme activator; HR2930, Bcl2-related protein A1 (human Bcl-2-related protein, hematopoiesis-specific early response protein, Glasgow rearranged sequence protein); HR3073, human Bcl2-modifying protein, involved in apoptosis, myosin-binding protein; HR3111, human DNA-binding inhibitor ID-3, transcriptional corepressor (http://nmr.cabm.rutgers.edu:9090/PLIMS/targetViewerQuickSearchContent.jsf; 1).
Expression levels (where 100% indicates total protein expression by E. coli) are semiquantitatively indicated from 0 (not detected) and + (>5%) to +++++ (∼50%).
Protein solubility is also indicated from 0 to +++++ as described in footnote b.
NA, not applicable.
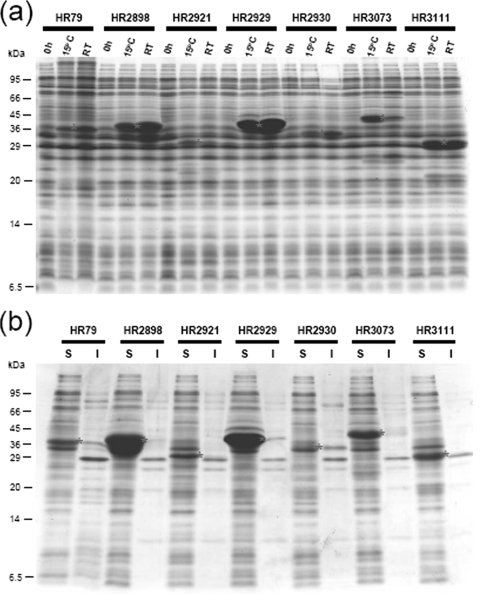
The genes for these proteins were obtained from NESG and cloned into pCold-PSTTEV. BL21 cells were transformed with the resulting plasmids, and the expression was induced for 14 h either at 15°C or at room temperature in the presence of 1 mM IPTG; whole cells were analyzed by SDS gel electrophoresis (Fig. 2a). Note that previously we found that although the pCold vector is designed for the production of proteins at 15°C, the CspA promoter is also active at 37°C and some proteins can be produced in high yields at room temperature with the pCold vector (data not shown). Notably, HR3111, which was not expressed by a pET vector, was well expressed at both 15°C and room temperature, and the other three proteins that were expressed poorly by a pET vector, HR79, HR2930, and HR3073, were also expressed reasonably well by the pCold-PSTTEV vector at both temperatures. Interestingly, HR79 and HR2930 were expressed better at room temperature, while HR3073 was expressed better at 15°C. The level of expression of HR2921 at 15°C was the lowest observed for the seven proteins tested. Two other proteins, HR2898 and HR2929, were expressed at very high levels, particularly at room temperature. Most significantly, all fusion proteins except the HR2930 fusion protein were soluble (Fig. 2b). Highly expressed HR2898 and HR2929 proteins were completely soluble, and those which were previously not soluble at all (HR2921 and HR3073) also became fully soluble. HR79 was mostly soluble (Fig. 2b). We also attempted to compare the expression and solubility levels with those obtained by using pCold His6 tag. Some of the proteins (HR2898, HR2921, HR2929, and HR3073) were expressed with the His6 tag; however, the others (HR79, HR2930, and HR3111) were not expressed at all. Furthermore, only one (HR2929) of the four proteins expressed was soluble (Table 1).
FIG. 2.
Improvement of expression levels and solubilities of human proteins by the PrS2 tag. (a) Expression of PrS2-tagged human proteins. The PrS2 tag is fused to human proteins that were expressed at low levels or in insoluble forms by a pET system or pCold His6 tag. Each PrS2-tagged protein was expressed at 15°C and room temperature (RT) for 14 h, and cell pellets were examined by SDS-PAGE. The sample at 0 h (before protein induction) was used as a negative control. (b) The solubilities of PrS2-tagged proteins for which expression patterns are shown in panel a were examined. S, soluble fraction; I, insoluble fraction. Asterisks indicate the corresponding proteins in panels a and b.
One-step purification of PrS2 fusion proteins by using myxospores.
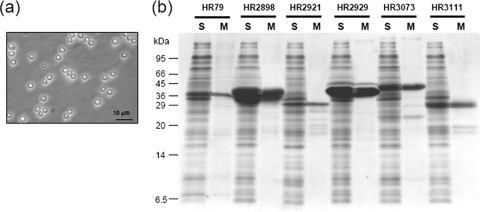
Next, we attempted to purify the soluble PrS2 fusion proteins (all but PrS2-HR2930) directly from cell lysates (after ultracentrifugation to remove the membrane fraction and insoluble material) in one step with the use of myxospores. Myxospores were purified from 10-day-old fruiting bodies of M. xanthus. The myxospores were extensively washed with 20 mM EDTA to completely remove protein S (8) and protein C (11). The washed myxospores were then completely inactivated by autoclaving at 121°C for 20 min to prevent germination and also to completely inactivate associated proteases (13). The myxospores thus prepared are homogeneous particles of approximately 1.5 μm in diameter (Fig. 3a).
FIG. 3.
One-step purification of human proteins with the use of myxospores. (a) Phase-contrast light microscope picture of myxospores. (b) One-step purification of human proteins by myxospores. The human proteins for which expression patterns are shown in Fig. 2 were subjected to one-step purification from cell lysates. S, soluble fraction; M, myxospore fraction. Asterisks indicate the full-sized human proteins fused with PrS2.
Myxospores were added to the cell lysate in the presence of 1 mM CaCl2. The mixture was incubated at 4°C for 1 h. Myxospores were then collected by centrifugation and washed three times. The final myxospore pellets were analyzed by SDS-PAGE (Fig. 3b). Most proteins were almost pure, with little background contamination, and were obtained at high yields. As for HR2898, although the yield of the protein was very high, its recovery was less efficient than that of other proteins such as HR2929. Further optimization of the recovery step for this protein may be necessary. Except for the fusion protein expressed at very high levels (the HR2929 fusion protein), the proteins exhibited a number of minor bands at smaller molecular weights than the full-sized bands. These minor bands are due possibly to proteolytic digestion occurring in E. coli cells. These proteins can be further purified by using the respective C-terminally His6-tagged proteins expressed by the pCold-PST vector as depicted in Fig. 1b.
Effect of PrS2 on the function of a fusion protein.
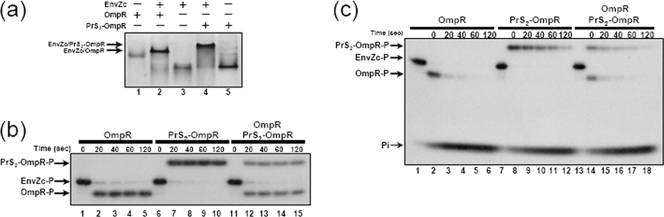
Previously, we have demonstrated that PrS2-OmpR fully retains the ability of OmpR to bind to the specific DNA sequence upstream of the ompF and ompC promoters (19). We further examined if PrS2 interferes with other functions of OmpR by analyzing the abilities of PrS2-OmpR to form a complex with EnvZ, a histidine kinase for OmpR, and to serve as an enzymatic substrate for EnvZ, which functions as both a kinase for OmpR and a phosphatase for OmpR-P. Figure 4a shows that PrS2-OmpR (lane 4), similar to OmpR alone (lane 2), is able to form a complex (shown by an arrow) with EnvZc (14) as seen by native PAGE analysis. Note that both complexes migrate more slowly than either OmpR (lane 1), PrS2-OmpR (lane 5), or EnvZc (lane 3).
FIG. 4.
Comparison of wild-type OmpR and PrS2-OmpR. (a) Complex formation between EnvZc and PrS2-OmpR. EnvZc and PrS2-OmpR (lane 4) or OmpR (lane 2) were mixed and incubated in the reaction buffer at room temperature for 5 min. The samples were subjected to 10% native PAGE. +, present; −, absent. (b) Phosphotransfer from EnvZc-P to PrS2-OmpR. Purified 32P-labeled EnvZc was mixed with OmpR, PrS2-OmpR, or the mixture of OmpR and PrS2-OmpR in the reaction buffer. The final reaction mixtures were incubated at room temperature. Aliquots were removed at 20, 40, 60, and 120 s, and the reaction was stopped with 5× SDS loading buffer. (c) Dephosphorylation of PrS2-OmpR-P by EnvZc. First, purified 32P-labeled EnvZc-P was mixed with OmpR, PrS2-OmpR, or the mixture of OmpR and PrS2-OmpR, and the final mixture was incubated in the reaction buffer at room temperature for 2 min to generate OmpR-P or PrS2-OmpR-P. After the addition of ADP (1 mM), aliquots were removed at 20, 40, 60, and 120 s and the reaction was stopped with 5× SDS loading buffer.
The phosphotransfer reaction between EnvZc-P and PrS2-OmpR (Fig. 4b, lanes 6 to 10) was as efficient as that with OmpR alone (Fig. 4b, lanes 1 to 5). The equal reactivities of PrS2-OmpR and OmpR were evident when PrS2-OmpR and OmpR were combined in the phosphotransfer reaction mixture (Fig. 4b, lanes 11 to 15). Similarly, the phosphatase reaction of EnvZc was examined using OmpR-P (Fig. 4c, lanes 1 to 6), PrS2-OmpR-P (Fig. 4c, lanes 7 to 12), and the mixture of both proteins (Fig. 4c, lanes 13 to 18). Again, PrS2-OmpR-P serves as an efficient substrate for the phosphate reaction, although the half-life of PrS2-OmpR-P appears to be several times longer than that of OmpR-P. These results indicate that OmpR fused to PrS2 is folded independently from the PrS2 domain to almost fully retain its biological function. It appears that these two independent domains have very little interaction so that no severe interference by PrS2 with the function of OmpR is observed.
Other applications of PST technology.
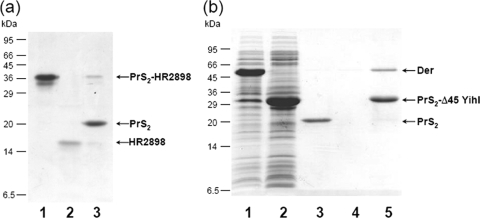
We next attempted to cleave the PrS2 tag from a myxospore-bound fusion protein by using TEV protease or factor Xa. For this purpose, pCold-PSTXa-HR2898 was constructed. Note that, surprisingly, PrS2 has a TEV protease cleavage site which cannot be predicted from the TEV protease consensus cleavage sequence. The PrS2-HR2898 protein was purified from the cell lysate in one step (Fig. 5a, lane 1). The fusion protein bound to the myxospores was then treated with factor Xa. After incubation for 30 min at room temperature in the presence of 1 mM CaCl2, the myxospores were precipitated by centrifugation. HR2898 cleaved from the fusion protein was released into a solution (Fig. 5a, lane 2), while the PrS2 tag was detected in the myxospore fraction, together with a small amount of uncleaved PrS2-HR2898 (Fig. 5a, lane 3). Importantly, although HR2898 was insoluble when expressed by a pET vector (Table 1), this protein stayed soluble even after the PrS2 tag was removed. These results indicate that a target protein can be isolated from the PrS2 fusion protein in one step.
FIG. 5.
Applications of PST technologies. (a) Cleavage of PrS2-tagged protein by factor Xa. PrS2-HR2898 bound to myxospores was treated with factor Xa on the myxospores at room temperature for 30 min. After the reaction, the mixture was centrifuged at 3,000 × g for 5 min and the supernatant was further centrifuged at 9,000 × g for 20 min and analyzed by SDS-17% PAGE. Lanes: 1, PrS2-HR2898 purified by one-step purification; 2, supernatant; and 3, myxospore fraction after treatment with factor Xa. (b) Results from a pulldown assay using PST technology. The interaction between PrS2-Δ45 YihI and Der was examined using the PrS2 tag and myxospores. The soluble fraction of a cell lysate containing Der was mixed with a lysate containing PrS2-Δ45 YihI (lane 5) or purified PrS2 (lane 3), and the mixture was incubated at 4°C for 30 min. Myxospores were added, and the mixture was further incubated at 4°C for 30 min, washed with buffer (10 mM Tris-HCl [pH 7.6] and 1 mM CaCl2) two times, and checked by SDS-17% PAGE. Lanes: 1, whole-cell lysate containing Der; 2, whole-cell lysate containing PrS2-Δ45 YihI; 3, Der, PrS2, and myxospores; 4, Der and myxospores; and 5, Der, PrS2-Δ45 YihI, and myxospores.
Another interesting application of PST technology is a pulldown assay to identify a protein(s) interacting with a target protein. For this analysis, Der, an essential GTPase that is required for ribosome assembly in E. coli (7) and interacts with YihI (or Δ45 YihI) (Hwang and Inouye, unpublished), was used as a model system. Δ45 YihI was fused with PrS2, and the fusion protein was used as bait. A cell lysate from 1 ml of a culture expressing Der (Fig. 5b, lane 1) was mixed with a cell lysate from 1 ml of a culture expressing PrS2-Δ45 YihI (Fig. 5b, lane 2). Myxospores were added to the mixture of cell lysates. The myxospores retained not only PrS2-Δ45 YihI but also Der (Fig. 5b, lane 5) by virtue of the interaction between Δ45 YihI and Der. Myxospores added to the cell lysate containing only Der protein did not retain any protein (Fig. 5b, lane 4). When myxospores were added to the cell lysate containing Der alone and purified PrS2, only PrS2, but not the Der protein, was precipitated (Fig. 5b, lane 3). Thus, using this small-scale pulldown assay, we were able to demonstrate the interaction between Der and Δ45 YihI.
DISCUSSION
To simplify protein purification, a number of tagging strategies have been developed (17). Among them, the His tag technology has been a major breakthrough, as a His-tagged protein can be easily trapped by nickel-nitrilotriacetic acid resin and the trapped protein can be eluted by imidazole (6). However, quite often a His-tagged fusion protein is eluted with high background levels of contaminating proteins. Other protein tags, glutathione S-transferase (16) and maltose-binding protein (4), also have problems with high background levels of contamination. In contrast to the use of these tags, PST technology with the PrS2 tag and myxospores yields a very low background. Most importantly, however, cold shock induction of PrS2 fusion proteins significantly enhances the expression as well as the solubility of the target proteins. Furthermore, the PrS2 fusion proteins can be readily purified from cell lysates in one step with the use of myxospores. Myxospores appear to be an excellent tool for protein purification, as they are very stable and biologically inert material once they are autoclaved. They are very homogeneous and can be well dispersed in a solution but readily precipitated by low-speed centrifugation. We found that some human proteins (HR2921, HR3073, and HR3111) can be obtained in soluble form only with the PrS2 tag. Therefore, the functional and structural study of these proteins may be carried out only with the use of the corresponding PrS2 fusion proteins. It may be possible to determine their NMR structures if NMR signals from the PrS2 domain can be silenced by the addition of myxospores. This approach is currently in progress in our laboratory.
Acknowledgments
We thank Gaetano T. Montelione for helpful discussions and Sangita Phadtare for critical reading of the manuscript.
This work was partially supported by research grants from the National Institute of General Medical Sciences Protein Structure Initiative (U54-GM074958, to G. T. Montelione), the NIH (grant no. 1R01GM085449, to M.I.), and TaKaRa Bio Inc. (to M.I.).
Footnotes
Published ahead of print on 19 June 2009.
REFERENCES
- 1.Acton, T. B., K. C. Gunsalus, R. Xiao, L. C. Ma, J. Aramini, M. C. Baran, Y. W. Chiang, T. Climent, B. Cooper, N. G. Denissova, S. M. Douglas, J. K. Everett, C. K. Ho, D. Macapagal, P. K. Rajan, R. Shastry, L. Y. Shih, G. V. Swapna, M. Wilson, M. Wu, M. Gerstein, M. Inouye, J. F. Hunt, and G. T. Montelione. 2005. Robotic cloning and protein production platform of the Northeast Structural Genomics Consortium. Methods Enzymol. 394:210-243. [DOI] [PubMed] [Google Scholar]
- 2.Arnau, J., C. Lauritzen, G. E. Petersen, and J. Pedersen. 2006. Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. Protein Expr. Purif. 48:1-13. [DOI] [PubMed] [Google Scholar]
- 3.Bagby, S., T. S. Harvey, S. G. Eagle, S. Inouye, and M. Ikura. 1994. NMR-derived three-dimensional solution structure of protein S complexed with calcium. Structure 2:107-122. [DOI] [PubMed] [Google Scholar]
- 4.di Guan, C., P. Li, P. D. Riggus, and H. Inouye. 1988. Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene 67:21-30. [DOI] [PubMed] [Google Scholar]
- 5.Harlocker, S. L., L. Bergstrom, and M. Inouye. 1995. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J. Biol. Chem. 270:26849-26856. [DOI] [PubMed] [Google Scholar]
- 6.Hochuli, E., H. Döbeli, and A. Schacher. 1987. New metal chelate adsorbent selective for proteins and peptides containing neighbouring histidine residues. J. Chromatogr. 411:177-184. [DOI] [PubMed] [Google Scholar]
- 7.Hwang, J., and M. Inouye. 2001. An essential GTPase, Der, containing double GTP-binding domains from Escherichia coli and Thermotoga maritima. J. Biol. Chem. 276:31415-31421. [DOI] [PubMed] [Google Scholar]
- 8.Inouye, M., S. Inouye, and D. R. Zusman. 1979. Biosynthesis and self-assembly of protein S, a development-specific protein of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:209-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kishii, R., L. Falzon, T. Yoshida, H. Kobayashi, and M. Inouye. 2007. Structural and functional studies of the HAMP domain of EnvZ, an osmosensing transmembrane histidine kinase in Escherichia coli. J. Biol. Chem. 282:26401-26408. [DOI] [PubMed] [Google Scholar]
- 10.Malakhov, M. P., M. R. Mattern, O. A. Malakhova, M. Drinker, S. D. Weeks, and T. R. Butt. 2004. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genomics 5:75-86. [DOI] [PubMed] [Google Scholar]
- 11.McCleary, W. R., B. Esmon, and D. R. Zusman. 1991. Myxococcus xanthus protein C is a major spore surface protein. J. Bacteriol. 173:2141-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishihara, K., M. Kanemori, M. Kitagawa, H. Yanagi, and T. Yura. 1998. Chaperone coexpression plasmids: differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl. Environ. Microbiol. 64:1694-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otani, M., M. Inouye, and S. Inouye. 1995. Germination of myxospores from the fruiting bodies of Myxococcus xanthus. J. Bacteriol. 177:4261-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park, H., S. K. Saha, and M. Inouye. 1998. Two-domain reconstitution of a functional protein histidine kinase. Proc. Natl. Acad. Sci. USA 95:6728-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qing, G., L. C. Ma, A. Khorchid, G. V. Swapna, T. K. Mal, M. M. Takayama, B. Xia, S. Phadtare, H. Ke, T. Acton, G. T. Montelione, M. Ikura, and M. Inouye. 2004. Cold-shock induced high-yield protein production in Escherichia coli. Nat. Biotechnol. 22:877-882. [DOI] [PubMed] [Google Scholar]
- 16.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 17.Stevens, R. C. 2000. Design of high-throughput methods of protein production for structural biology. Structure 8:R177-R185. [DOI] [PubMed] [Google Scholar]
- 18.Wenk, M., R. Baumgartner, T. A. Holak, R. Huber, R. Jaenicke, and E. M. Mayr. 1999. The domains of protein S from Myxococcus xanthus: structure, stability and interactions. J. Mol. Biol. 286:1533-1545. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida, T., L. Qin, L. A. Egger, and M. Inouye. 2006. Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J. Biol. Chem. 281:17114-17123. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida, T., L. Qin, and M. Inouye. 2002. Formation of the stoichiometric complex of EnvZ, a histidine kinase, with its response regulator, OmpR. Mol. Microbiol. 46:1273-1282. [DOI] [PubMed] [Google Scholar]