Abstract
A collection of 54 clinical and agricultural isolates of Burkholderia cenocepacia was analyzed for genetic relatedness by using multilocus sequence typing (MLST), pathogenicity by using onion and nematode infection models, antifungal activity, and the distribution of three marker genes associated with virulence. The majority of clinical isolates were obtained from cystic fibrosis (CF) patients in Michigan, and the agricultural isolates were predominantly from Michigan onion fields. MLST analysis resolved 23 distinct sequence types (STs), 11 of which were novel. Twenty-six of 27 clinical isolates from Michigan were genotyped as ST-40, previously identified as the Midwest B. cenocepacia lineage. In contrast, the 12 agricultural isolates represented eight STs, including ST-122, that were identical to clinical isolates of the PHDC lineage. In general, pathogenicity to onions and the presence of the pehA endopolygalacturonase gene were detected only in one cluster of related strains consisting of agricultural isolates and the PHDC lineage. Surprisingly, these strains were highly pathogenic in the nematode Caenorhabditis elegans infection model, killing nematodes faster than the CF pathogen Pseudomonas aeruginosa PA14 on slow-kill medium. The other strains displayed a wide range of pathogenicity to C. elegans, notably the Midwest clonal lineage which displayed high, moderate, and low virulence. Most strains displayed moderate antifungal activity, although strains with high and low activities were also detected. We conclude that pathogenicity to multiple hosts may be a key factor contributing to the potential of B. cenocepacia to opportunistically infect humans both by increasing the prevalence of the organism in the environment, thereby increasing exposure to vulnerable hosts, and by the selection of virulence factors that function in multiple hosts.
The betaproteobacterium Burkholderia cenocepacia, 1 of now 17 classified species belonging to the Burkholderia cepacia complex (BCC), is ubiquitous and extremely versatile in its metabolic capabilities and interactions with other organisms (38, 40, 57, 58). Strains of B. cenocepacia are pathogens of onion and banana plants, opportunistic pathogens of humans, symbionts of numerous plant rhizospheres, contaminants of pharmaceutical and industrial products, and inhabitants of soil and surface waters (14, 29, 33, 34, 37, 45). Originally described as a pathogen of onions (8), organisms of the BCC emerged in the past 3 decades as serious human pathogens, capable of causing devastating chronic lung infections in persons with cystic fibrosis (CF) or chronic granulomatous disease (21, 24, 28). Infections due to BCC are a serious concern to CF patients due to their inherent antibiotic resistance and high potential for patient-to-patient transmission (23). Although 16 of the BCC species have been recovered from respiratory secretions of CF patients in many countries (46, 58), B. cenocepacia has been the most common species isolated in North America, detected in 50% of 606, 83% of 447, and 45.6% of 1,218 patients in recent studies (35, 46, 52).
The epidemiology of infectious disease caused by B. cenocepacia appears to involve patient-to-patient spread of genetically distinct lineages. B. cenocepacia lineages, such as ET12, Midwest, and PHDC, have been identified from large numbers of individuals in disease outbreaks in North America and Europe (11, 32, 54). A recently developed multilocus sequence typing (MLST) scheme has been shown to be a reliable epidemiologic tool for differentiating between the five subgroups (IIIA to IIIE) of B. cenocepacia, and strains representing three of these subgroups (IIIA, IIIB, and IIID) have been recovered from CF patients (2). Outside of the patient-to-patient transmission of clonal lineages, the mode of acquisition of strains causing sporadic cases of B. cenocepacia in CF patients remains unclear, although environmental sources are a logical reservoir for infection. Previously, an isolate of B. cenocepacia indistinguishable from the PHDC epidemic clonal lineage by using standard typing methods (e.g., repetitive-sequence-based PCR, randomly amplified polymorphic DNA, pulsed-field gel electrophoresis) was detected in an agricultural soil sample (34). Similarly, three distinct MLST sequence types containing both clinical and environmental (plant and soil) B. cenocepacia isolates were identified (1). These findings suggest that natural populations of B. cenocepacia in soil or associated with plants are a potential reservoir for the emergence of new human pathogenic lineages.
Experimental models for the study of virulence potential and traits of B. cenocepacia include mouse and rat models with genetic defects allowing chronic lung infections to be established (e.g., see reference 48). Nematode (Caenorhabditis elegans), alfalfa (Medicago sativa), and onion (Allium cepa) models have also been routinely utilized for the identification of virulence factors (5, 29, 31). C. elegans has been extensively used to study the pathogenesis and virulence factors of a wide variety of bacterial and fungal pathogens (9, 15, 42, 51, 56). In several pathogens, including Pseudomonas (56) and Burkholderia (20), putative virulence factors important for the pathogenesis in mammalian systems (15, 51) have been identified using the C. elegans model. The C. elegans model might be limited in the detection of host-specific virulence factors; however, several attributes, such as small size and rapid development, make it an excellent whole animal model for pathogenesis research (16, 51).
The evidence that individual strains of B. cenocepacia can be pathogenic to both plants and humans and are prevalent in various environmental niches has provoked particular interest in elucidating the clinical pathogenic potential of environmental isolates. The basis of this study was to examine whether genetically related B. cenocepacia strains exhibit shared characteristics that contribute to their pathogenicity in multiple hosts and to examine the potential for circulating environmental isolates to emerge as new clinical pathogens. Here, we tested the degree of virulence in animal (nematode) and plant (onion) infection models, the production of antifungal activity, and the genetic relatedness of clinical and environmental B. cenocepacia subgroup IIIB strains predominantly isolated from Michigan.
MATERIALS AND METHODS
Bacterial strains, nematode strains, and growth conditions.
The 56 BCC strains utilized in this study are listed in Table 1. Among the 54 B. cenocepacia strains were 29 clinical isolates that were isolated from 29 persons with CF in Michigan and Ohio between 1987 and 2000 (11 of which were previously identified as being members of the Midwest clonal lineage [13]), 13 recent clinical isolates from North America and Europe that were genotyped as distinct from members of the Midwest clonal lineage, and 12 environmental isolates collected from the onion or maize rhizosphere or onion field soil in Michigan and New York. All B. cenocepacia strains were routinely cultured on either King's medium B (KB) (30), lysogeny broth (LB) (41), Todd-Hewitt broth, or Mueller-Hinton II medium (Becton Dickinson, Franklin Lakes, NJ) at 37°C for clinical isolates and at 28°C for environmental isolates. The remaining two strains included the environmental isolates Burkholderia cepacia ATCC 25416 and Burkholderia ambifaria AMMD, which were used as controls.
TABLE 1.
The 42 clinical and 14 environmental BCC strains used in this study
| No. | Strain | Yr collected | Source(s) | Geographic origin | STa | esmRb | Source or reference |
|---|---|---|---|---|---|---|---|
| 1 | 6RP195 | 2005 | Rhizosphere, onion | Allegan County, MI | 125 | − | 29 |
| 2 | AU10310 | 2005 | CF clinical isolate | Massachusetts | 125 | − | LiPumac |
| 3 | 3RP82 | 2005 | Rhizosphere, onion | Eaton County, MI | 428 | + | 29 |
| 4 | 5ST6 | 2005 | Soil, onion field | Allegan County, MI | 430 | + | 29 |
| 5 | 1SP104 | 2005 | Soil, onion field | Eaton County, MI | 467 | + | 29 |
| 6 | 2RT145 | 2005 | Rhizosphere, onion | Eaton County, MI | 429 | + | 29 |
| 7 | 3RP107 | 2005 | Rhizosphere, onion | Eaton County, MI | 427 | + | 29 |
| 8 | 3RP193 | 2005 | Rhizosphere, onion | Eaton County, MI | 427 | + | 29 |
| 9 | 6RP111 | 2005 | Rhizosphere, onion | Allegan County, MI | 427 | + | 29 |
| 10 | 6RT130 | 2005 | Rhizosphere, onion | Allegan County, MI | 122 | − | 29 |
| 11 | 6RT131 | 2005 | Rhizosphere, onion | Allegan County, MI | 122 | − | 29 |
| 12 | AU1054 | CF clinical isolate | United States | 122 | − | LiPuma | |
| 13 | HI2424 | Soil, onion field | New York | 122 | − | 34 | |
| 14 | AU9629 | 2005 | CF clinical isolate | Virginia | 332 | + | LiPuma |
| 15 | MCO-3 | 2004 | Rhizosphere, maize | Ingham County, MI | 281 | + | Rametted |
| 16 | AU2765 | 2001 | CF clinical isolate | Arizona | 433 | + | LiPuma |
| 17 | AU6690 | 2004 | CF clinical isolate | Texas | 435 | + | LiPuma |
| 18 | AU0279 | 1997 | CF clinical isolate | Michigan | 40 | − | 13 |
| 19 | AU0280 | 1997 | CF clinical isolate | Michigan | 40 | − | 13 |
| 20 | AU0281 | 1997 | CF clinical isolate | Michigan | 40 | − | 13 |
| 21 | AU0283 | 1997 | CF clinical isolate | Michigan | 40 | + | 13 |
| 22 | AU0284 | 1997 | CF clinical isolate | Michigan | 40 | − | 13 |
| 23 | AU0407 | 1998 | CF clinical isolate | Michigan | 40 | − | 13 |
| 24 | AU0408 | 1998 | CF clinical isolate | Michigan | 40 | − | 13 |
| 25 | AU0418 | 1998 | CF clinical isolate | Michigan | 40 | + | 13 |
| 26 | AU0419 | 1998 | CF clinical isolate | Michigan | 40 | + | 13 |
| 27 | AU0738 | 1998 | CF clinical isolate | Michigan | 40 | + | 13 |
| 28 | AU1601 | 1999 | CF clinical isolate | Michigan | 40 | + | 13 |
| 29 | CF1 | 1988 | CF clinical isolate | Ingham County, MI | 40 | + | Hardene |
| 30 | CF2 | 1988 | CF clinical isolate | Ingham County, MI | 40 | + | Harden |
| 31 | CF17 | 1988 | CF clinical isolate | Ingham County, MI | 40 | + | Harden |
| 32 | CF20 | 1988 | CF clinical isolate | Ingham County, MI | 40 | + | Harden |
| 33 | CF23 | 1988 | CF clinical isolate | Ingham County, MI | 40 | − | Harden |
| 34 | CF38 | 1988 | CF clinical isolate | Ingham County, MI | 40 | − | Harden |
| 35 | CF39 | 1988 | CF clinical isolate | Ingham County, MI | 40 | + | Harden |
| 36 | CF44 | 1988 | CF clinical isolate | Ingham County, MI | 40 | + | Harden |
| 37 | CF51 | 1988 | CF clinical isolate | Ingham County, MI | 40 | − | Harden |
| 38 | CF72 | 1989 | CF clinical isolate | Ingham County, MI | 40 | + | Harden |
| 39 | CF157 | 1989 | CF clinical isolate | Grand Rapids, MI | 40 | + | Harden |
| 40 | CF160 | 1989 | CF clinical isolate | Cleveland, OH | 40 | + | Harden |
| 41 | MDCH2 | 1991-1992 | CF clinical isolate | Michigan | 40 | − | MDCHf |
| 42 | MDCH989 | 1991-1992 | CF clinical isolate | Michigan | 40 | + | MDCH |
| 43 | MDCH991-1 | 1991-1992 | CF clinical isolate | Michigan | 40 | + | MDCH |
| 44 | MDCH992 | 1991-1992 | CF clinical isolate | Michigan | 40 | + | MDCH |
| 45 | PC184 | <1986 | CF clinical isolate | Cleveland, OH | 40 | + | LiPuma |
| 46 | AU2289 | 2000 | CF clinical isolate | Michigan | 249 | − | LiPuma |
| 47 | AU9710 | 2005 | Non-CF clinical isolate | Nebraska | 469 | + | LiPuma |
| 48 | AU3343 | 2001 | Non-CF clinical isolate | Texas | 263 | + | LiPuma |
| 49 | AU6100 | 2003 | CF clinical isolate | New York | 434 | − | LiPuma |
| 50 | AU6880 | 2004 | CF clinical isolate | Colorado | 330 | + | LiPuma |
| 51 | AU7610 | 2004 | CF clinical isolate | Alabama | 468 | − | LiPuma |
| 52 | J2315 | 1988 | CF, clinical, sputum | Edinburgh, United Kingdom | 28 | + | 23 |
| 53 | CF164 | 1989 | CF clinical isolate | Cleveland, OH | 32 | + | Harden |
| 54 | AU10575 | 2006 | Non-CF clinical isolate | Texas | 470 | − | LiPuma |
| 55 | ATCC25416 | 1948 | Onion | United States | 10 | − | ATCCg |
| 56 | AMMD | 1985 | Rhizosphere, pea | Wisconsin | 77 | − | LiPuma |
Multilocus sequence type (ST).
Presence (+) or absence (−) of the esmR epidemic strain marker.
J. J. LiPuma, U.S. Burkholderia cepacia Research Laboratory and Repository, University of Michigan, Ann Arbor.
A. Ramette, Center for Microbial Ecology, Michigan State University.
D. Harden, formerly of the Department of Pediatrics, Michigan State University.
Michigan Department of Community Health, Lansing, MI.
American Type Culture Collection, Manassas, VA.
Strain selection and genotypic characterization.
Previously published oligonucleotide primers and PCR conditions were used in the genotypic analyses of all 56 Burkholderia strains (Table 2). To confirm isolates as members of the BCC, we performed PCR on bacterial cell lysates by using primers that amplify a 320-bp region of the 16S rRNA gene (3, 45). Species assignments were made based on the restriction fragment length polymorphism (RFLP) analysis of an amplified 1,040-bp fragment of the recA gene, followed by digestion with the restriction enzymes HaeIII and MnlI (38, 45). The final B. cenocepacia strain set utilized in this study included mostly isolates belonging to the IIIB subgroup (n = 52), with a high representation of isolates of the Midwest epidemic lineage (n = 29), and isolates belonging to the IIIA subgroup (n = 2).
TABLE 2.
B. cenocepacia gene targets, PCR primers, and corresponding references
| Gene | Primer | Sequencea | Product size (bp) | Reference |
|---|---|---|---|---|
| 16S rRNA | Burkf | 5′-GGCGAAAGCCGGATTAATACC-3′ | 320 | 45 |
| 16S rRNA | CeMuVi-16-2 | 5′-CCGRCTGTATTAGAGCCA-3′ | 3 | |
| recA | BCR1 | 5′-TGACCGCCGAGAAGAGCAA-3′ | 1,040 | 45 |
| recA | BCR2 | 5′-CTCTTCTTCGTCCATCGCCTC-3′ | ||
| esmR | BCESM 1 | 5′-CCACGGACGTGACTAACA-3′ | 1,400 | 39 |
| esmR | BCESM 2 | 5′-CGTCCATCCGAACACGAT-3′ | ||
| pehA | peh F | 5′-CGAATTGCGACTCATAGGTTAC-3′ | 1,600 | 22 |
| pehA | peh R | 5′-TGTCAGCTGTCATCGTGGTTGC-3′ | ||
| prnD | PRND1 | 5′-GGGGCGGGCCGTGGTGATGGA-3′ | 800 | 18 |
| prnD | PRND2 | 5′-YCCCGCSGCCTGYCTGGTCTG-3′ | ||
| trpB | trpB-F2 | 5′- CTGGGTCACGAACATCGA-3′ | 301 | This study |
Underlined base represents the nucleotide altered from the original sequencing primer.
In addition to the recA genotypic analysis involved with the initial selection of isolates, PCR was used to evaluate the presence of certain virulence factors associated with infection in various hosts. The gene targets for PCR included the esmR epidemic strain marker which encodes a putative regulatory gene within a 31.7-kb pathogenicity island (39); the plasmid-borne pehA gene which encodes endopolygalacturonase, a degradative enzyme required for maceration of onion tissue (22); and the prnD gene encoding an enzyme involved in the biosynthesis of pyrrolnitrin, a compound with broad-spectrum antifungal activity (18).
MLST.
An established MLST system (2) was applied to all 56 BCC strains by amplifying and sequencing PCR fragments from the following housekeeping genes: atpD (ATP synthase beta chain), gltB (glutamate synthase large subunit), gyrB (DNA gyrase subunit B), lepA (GTP binding protein), phaC (acetoacetyl coenzyme A reductase), recA (recombinase A), and trpB (tryptophan synthase subunit B). In brief, genomic DNA used for PCR amplification of the seven MLST genes was isolated from overnight cultures grown in Mueller-Hinton II broth, using the Purgene DNA isolation kit (Gentra Systems Inc., Minneapolis, MN). PCR products were purified using the QIAquick PCR purification kit (Qiagen Inc., Valencia, CA), and sequencing reactions were prepared according to the GenomeLab Dye terminator cycle sequencing quick start kit protocol (Beckman Coulter Inc., Fullerton, CA). For the environmental isolates, a modified trpB forward sequencing primer was used, due to observed ambiguities in the primer site of these strains (Table 2). Sequencing reactions were separated on a CEQ2000XL DNA sequencer (Beckman Coulter Inc.), and sequences were analyzed using the CEQ2000XL software and then exported for editing in the SeqMan module of Lasergene (DNASTAR Inc., Madison, WI). The BCC MLST webpage (http://pubmlst.org/bcc/) was used for allele and sequence type (ST) assignments (2); any isolates containing new alleles or new allelic profiles were submitted to the PubMLST database and subsequently given ST designations by the database curator.
Phylogenetic and sequence analyses.
The MEGA4 software package was used to create bootstrap (1,000 replicates) neighbor-joining trees of the MLST sequence data, based on the p-distance model (55). The neighbor-net algorithm in the SplitsTree 4 software (27) was used to create phylogenetic networks from the MLST sequences. To test for the role of past recombination in generating allelic variation, the pairwise homoplasy index (Phi) was calculated using SplitsTree 4 (7, 27).
The contribution of natural selection on allelic variation was inferred from the number of synonymous substitutions per synonymous site (dS) and the number of nonsynonymous substitutions per nonsynonymous site (dN) estimated by the modified Nei-Gojobori method using MEGA4 (55). In addition, allelic sequences were fit to a nucleotide substitution model, and the single likelihood ancestor counting (SLAC) method (http://www.datamonkey.org/) was used to estimate the ratio of dN/dS with a 95% confidence interval and to determine selective pressures acting on the individual codons (44).
Onion pathogenesis assays.
Assays to determine pathogenicity of B. cenocepacia isolates in an onion plant model were performed as previously described (29). Briefly, individual quartered onion scales were wounded on the interior surface with a sterile pipette tip (1- to 200-μl volume), and 5 μl of a bacterial suspension (107 CFU/ml) was inoculated into the wound. Onion scales were incubated at 30°C for 48 h, after which the degree of maceration was estimated by probing the onion tissue with a toothpick. A visual rating scale of 0 to 3 was used to indicate the area of affected plant tissue exhibiting water-soaking symptoms, or water-soaking symptoms together with maceration symptoms. The rating scale was applied as follows: 0 indicated water-soaking without maceration, 1 indicated 1% to 33% macerated tissue, 2 indicated 34% to 66% macerated tissue, and 3 indicated 67% to 100% macerated tissue. Onion pathogenicity assays were conducted with each isolate, in two or more independent experiments with a minimum of six replications. Experimental negative controls consisted of no wounding and inoculation with the onion-pathogenic control strain B. cepacia ATCC 25416 (no. 55), wounding and no inoculation, and wounding and inoculation with sterile KB broth. The positive control consisted of wounding followed by inoculation with B. cepacia strain ATCC 25416 (no. 55).
Fungal antibiosis assays.
The ability of B. cenocepacia strains to inhibit the growth of the fungus Rhizoctonia solani AG4 was determined in a plate assay. Bacteria were grown overnight in KB broth cultures and then spotted (2 μl) in quadruplicate onto a potato dextrose agar (Sigma Chemical Co., St. Louis, MO) plate. A grid map was used to spot the bacteria in a square pattern, and the bacteria were incubated at 37°C (clinical isolates) or 28°C (environmental isolates) for 48 h. A 7-mm plug of R. solani AG4 (obtained from W. Kirk) was inoculated onto the potato dextrose agar plate in a central location equidistant from each bacterial inoculum spot. The plates were incubated for 96 h at 25°C, after which growth of the fungus was assessed visually. A rating scale was developed to qualitatively determine the extent of fungal inhibition. Three independent experiments with four replicates per experiment were performed with each B. cenocepacia isolate. Positive and negative control strains in this experiment were B. ambifaria AMMD (no. 56), a known biological control strain with antifungal properties, and an R. solani AG4 strain inoculated alone without bacteria, respectively.
Nematode pathogenesis assays.
C. elegans strain SS104 [glp-4(bn2)I], which is sterile at 25°C but reproduces normally at 15°C (4), was obtained from the Caenorhabditis Genetics Center, University of Minnesota (Minneapolis, MN) and used in the B. cenocepacia pathogenic bioassays. Green fluorescent protein (GFP)-labeled strains of Escherichia coli OP50 and Pseudomonas aeruginosa PA14 were used as controls. GFP labeling was performed by inserting a mini Tn7 transposon containing Ptac::GFP into the bacterial strains by a triparental mating method as described elsewhere (12).
The nematode pathogenicity bioassays were conducted on nematode growth medium (NGM) and peptone glucose sorbitol (PGS) medium, which promotes slow and fast killing, respectively, in P. aeruginosa (56). The known CF pathogen P. aeruginosa PA14 (56) was used as a positive control, and E. coli OP50, a food source for C. elegans (6), served as the negative control. Routinely, the nematodes were maintained on lawns of E. coli OP50 on NGM plates (53) at 15°C. Prior to the bioassays, the nematodes were synchronized by collecting eggs from the gravid nematodes, using bleach-alkali solution. The eggs were then seeded onto the lawns of E. coli OP50 on NGM plates, incubated at 25°C for 36 to 38 h, after which the early fourth larval stage (L4 stage) of nematode was used for the pathogenicity assays.
The bacterial isolates were cultured from the freezer stocks on LB and incubated overnight at 37°C. Liquid cultures were grown in LB for 14 to 16 h at 37°C, shaking at 250 rpm. Seventy-five microliters of these liquid cultures were spread onto 60- by 15-mm petri plates containing NGM or PGS medium and incubated at 37°C for 16 to 18 h. Forty to sixty early-L4-stage C. elegans nematodes were added to the plates containing bacterial lawns, and the number of survivors after 3 days and 7 days was determined as those moving or responding to touch. Three independent experiments consisting of three replicates were performed.
The nematode killing data for each isolate obtained from the described assays were analyzed for variance by analysis of variance, and Tukey's honestly significant difference test was used to compare the means at a P value of 0.01. To estimate the pathogenicity of the isolates from the nematode survival data on NGM and PGS medium at 3 days and 7 days more broadly, a composite survival score (CSS) was calculated by a mixed model, using SAS statistical software (SAS Institute Inc., Cary, NC), which assumed the following: nematode survival = intercept + a × batch + b × medium + c × bacteria + d × day + error, where a, b, c, and d were respective constants; bacteria, day, and media were considered the fixed effects; and the replicates (batch) were considered the random effects. Each isolate was compared to E. coli OP50, for calculation of a CSS, and to each of the other isolates by using Tukey-Kramer adjustment to determine significant differences between the isolates (P < 0.0001).
RESULTS
Genetic analyses of isolates.
All 56 Burkholderia isolates used in this study were confirmed as members of the BCC and identified to the species level based on 16S rRNA gene and recA RFLP analyses, respectively. Of the 54 B. cenocepacia isolates, 40 clinical and 12 environmental isolates were confirmed as belonging to the B. cenocepacia IIIB subgroup. Two of the clinical isolates, AU3343 (no. 48 [Table 1]) and AU10575 (no. 54), yielded variant RFLP patterns but appeared to be most closely related to B. cenocepacia IIIB.
In screening for the presence of the esmR epidemic strain marker, 26 of 42 clinical isolates, including 64% of the Midwest clonal isolates, were positive (Table 1). The frequency of clinical isolates carrying esmR dropped from 75% of isolates collected in 1988 to 1989 to 45% of Midwest clone isolates collected in 1997 to 2000. Among the environmental isolates, 67% were positive for esmR.
MLST and phylogenetic analyses.
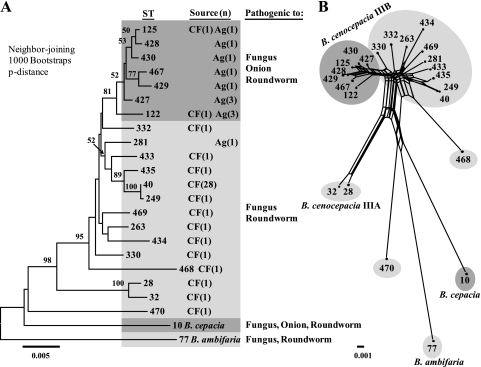
MLST analysis of the 56 BCC strains represented in this study resolved 23 distinct combinations of alleles or STs. The neighbor-joining phylogenetic tree revealed a distinct cluster (>80% bootstrap support) containing 11 environmental isolates recovered from onion fields (no. 1, 3 to 11, and 13, which are STs 122, 125, 427 to 430, and 467) and two clinical isolates, including the PHDC epidemic clone strain AU1054 (no. 12; ST-122) (Fig. 1A). One environmental isolate, MCO-3 (no. 15), isolated from the maize rhizosphere in Michigan, resided outside of this main cluster and matched the known clinical and environmental genotype ST-281. The ST-40 clones (no. 18 to 45), known as the Midwest epidemic lineage, revealed a minor allelic variant, ST-249 (no. 46), which was isolated from a CF patient in Michigan, and another closely related ST, ST-435 (no. 47), was isolated from a CF patient in Texas (Fig. 1A). Overall, the neighbor-joining phylogenetic analysis confirmed the close relatedness among B. cenocepacia IIIB strains; however, relatively few significant groupings were observed between strains, as indicated by the low bootstrap values (values of >50% are shown in Fig. 1A).
FIG. 1.
(A) Phylogenetic relationships among 23 distinct multilocus STs from the BCC (numbers at the branch nodes represent relationships with >50% bootstrap support). Groups are shaded according to which hosts the genotypes that are generally pathogenic. Ag, agricultural source; CF, CF or other clinical source. (B) Neighbor-net analysis revealed extensive recombination among the 21 STs representing the B. cenocepacia isolates (illustrated as parallelograms).
We investigated the possibility that recombination was the underlying reason for the weak phylogenetic relationships among strains by using neighbor-net phylogenetic analysis and the Phi test for genetic recombination. In contrast to the neighbor-joining trees we generated, the neighbor-net analysis does not restrict the sequence data into a typical bifurcating tree, but allows for multiple paths of recurrent mutation and recombination in the generation of new STs. A high degree of reticulation indicative of recombination was observed among the B. cenocepacia IIIB strains, confirming the conflicting phylogenies observed in the neighbor-joining tree (Fig. 1B). The Phi test result for genetic recombination among the 23 STs was highly significant (P < 0.001), suggesting that the genetic diversity observed between B. cenocepacia IIIB isolates likely resulted from recombination, parallel mutations, or lateral gene transfer events (13).
Distribution of sequence types.
Among the 23 STs identified through MLST, 11 STs were novel variants identified in this study, 5 of which were Michigan environmental isolates (STs 427 to 430 and 467) (Fig. 1A and B; Table 1). Two of the B. cenocepacia IIIB environmental isolates, 6RT130 (no. 10) and 6RT131 (no. 11), obtained from the onion rhizosphere matched the ST-122 genotype, known as the PHDC epidemic clone, which is a human pathogen that has been recovered from CF patients in North America and Europe and also previously isolated from soil in New York (34). Additionally, ST-125 was represented in our collection by both a clinical isolate, AU10310 (no. 2), recovered from a CF patient in Massachusetts, and an environmental isolate, 6RP195 (no. 1), obtained from the onion rhizosphere in Michigan. The 29 Michigan clinical isolates (no. 18 to 45) recovered from CF patients over the period of 1988 to 1999 were all genotyped as ST-40, the Midwest clone. A single ST-40 clonal variant, AU2289 (no. 46; ST-249), isolated in 2000 from a CF patient, contained a single nucleotide difference in the phaC gene. Two clinical isolates of more recent origin, AU7610 (no. 51) from a CF patient in Alabama and AU10575 (no. 54) from a non-CF patient in Texas, were highly divergent by MLST from the other B. cenocepacia IIIB isolates and also represented novel STs 468 and 470, respectively.
Genetic variability in seven MLST loci.
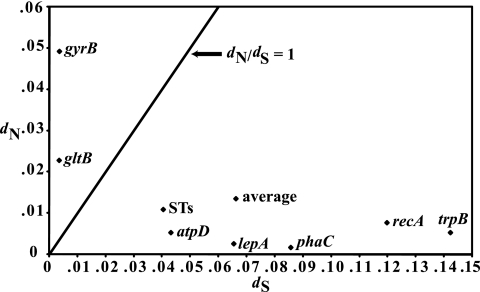
The sequence data for the 23 STs identified by MLST was analyzed further to uncover the extent of genetic variation present among the different alleles. Sequence analysis revealed 282 variable nucleotide sites among all seven concatenated housekeeping gene sequences (2,773 bp), resulting in a range of 9 to 17 distinct alleles per MLST gene (Table 3). To determine the action of natural selection on generating allelic variability, the number of synonymous (dS) and nonsynonymous (dN) amino acid substitutions and the dN/dS ratio were determined. The number of synonymous substitutions (dS) between allelic variants ranged from 0.04% for gltB to 1.42% for trpB, with an average of 0.664% synonymous substitution sites for all seven MLST loci (Table 3). The number of nonsynonymous substitutions (dN) was lower overall, with an average of 0.14% for the seven MLST loci, and ranged from 0.02% for phaC to 0.50% for gyrB (Table 3). The ratio of dN/dS found using SLAC analysis was high for both gltB (dN/dS = 10.20) and gyrB (dN/dS = 7.14), while the remaining MLST genes contained dN/dS ratios below 0.1, which is typically observed for conserved MLST genes (Table 3; Fig. 2). These findings suggest that gltB and gyrB are not evolving under the same evolutionary pressures as the other five MLST loci and may impact the phylogenetic inferences in B. cenocepacia. Although omitting these genes from the phylogenetic analysis did not change the major grouping of strains, due to the behavior of these genes and the amount of recombination observed, precaution was used in making strong evolutionary conclusions.
TABLE 3.
Sequence variation in seven MLST genes from 23 BCC STs
| MLST locus | Size (bp) | No. of alleles | No. of variable sites | dS × 100 (mean ± SE) | dN × 100 (mean ± SE) | dN/dS (95% CI) |
|---|---|---|---|---|---|---|
| atpD | 443 | 12 | 25 | 0.431 ± 0.0101 | 0.053 ± 0.030 | 0.093 (0.043, 0.174) |
| gltB | 400 | 12 | 27 | 0.035 ± 0.0024 | 0.228 ± 0.0049 | 10.202 (6.769, 14.747) |
| gyrB | 454 | 17 | 66 | 0.036 ± 0.020 | 0.492 ± 0.0071 | 7.139 (5.859, 8.597) |
| lepA | 397 | 15 | 41 | 0.654 ± 0.0113 | 0.026 ± 0.0015 | 0.029 (0.010, 0.063) |
| phaC | 385 | 9 | 29 | 0.856 ± 0.0165 | 0.0017 ± 0.0011 | 0.016 (0.002, 0.050) |
| recA | 393 | 12 | 53 | 1.197 ± 0.0181 | 0.077 ± 0.0011 | 0.021 (0.007, 0.044) |
| trpB | 301 | 12 | 41 | 1.422 ± 0.0255 | 0.053 ± 0.0035 | 0.018 (0.006, 0.039) |
| Average | 396.1 | 12.7 | 40.3 | 0.662 ± 0.0123 | 0.135 ± 0.0035 | 2.494 |
FIG. 2.
Plot of nonsynonymous substitution rate versus synonymous substitution rate for each MLST gene, the concatenated gene sequences (ST), and the average for all seven loci. The line plotted represents a dN-to-dS ratio of 1; conserved genes are expected to fall below this line. The relatively high values for gltB and gyrB suggest these genes are not evolving under the same evolutionary pressures as are the other five MLST loci.
In addition, the SLAC analysis was used to test the selective pressures (e.g., positive, negative, or neutral) acting on individual codon sites within the seven MLST genes. Sixteen negatively selected codon sites (two in atpD, two in lepA, four in recA, and eight in trpB) and one positively selected codon site in gyrB were identified. Surprisingly, the sequenced gltB gene fragment was interrupted by an in-frame stop codon within 6 of the 12 allelic variants which are likely to render this gene nonfunctional in these strains. These findings also indicate that the current seven MLST genes may not be the most appropriate housekeeping genes for phylogenetic analysis; however, the established MLST system has been proven reliable in differentiating Burkholderia isolates.
Onion pathogenesis assays.
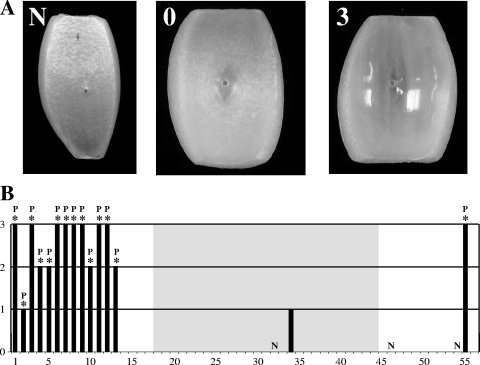
The 54 B. cenocepacia strains examined in this study were easily distinguishable in pathogenicity to onions. Strains originally isolated from the onion rhizosphere or onion field soil (e.g., no. 1 and 3 to 11) were all pathogenic to onions and capable of causing water-soaking and maceration of onion tissue in the assays (Fig. 3). Ratings of 2 or 3, differentiated by the degree of onion tissue maceration, were given to all onion-associated strains and to the control onion pathogen strain B. cepacia ATCC 25416 (no. 55). The environmental strain MCO-3 (no. 15), which was isolated from the maize rhizosphere, was not pathogenic to onions and did not carry the pehA gene. The clinical isolate AU1054 (no. 12), which shared the same genotype (ST-122) as the onion rhizosphere or onion field soil isolates 6RT130 (no. 10) and 6RT131 (no. 11), also was pathogenic to onions, having a rating of 3. All other clinical isolates (e.g., no. 14 and 16 to 54), except for AU10310 (no. 2) and CF38 (no. 34), had values of N for a null reaction or 0 for a small area of water-soaked tissue. All isolates capable of causing onion tissue maceration, except for CF38, harbored the pehA gene, as assessed by PCR (Fig. 3B).
FIG. 3.
Pathogenicity of B. cenocepacia and control strains to onions. (A) Rating scale. N indicates a null reaction and no affected tissue, 0 indicates water-soaking without maceration, 1 indicates 1% to 33% water-soaked/macerated tissue, 2 indicates 34% to 66% water-soaked/macerated tissue, and 3 indicates 67% to 100% water-soaked/macerated tissue. Examples of the rating scale are as follows: N, inoculated with AU10575 (no. 54); 0, inoculated with B. cenocepacia CF20 (no. 32); 3, inoculated with B. cenocepacia AU1054 (no. 12). (B) Onion pathogenesis ratings for 56 BCC strains. Strain order (no. 1 to 56) is as listed in Table 1, and the shaded box indicates strains belonging to the ST-40 Midwest epidemic clone lineage. All affected onion tissue exhibited water-soaking symptoms; the presence of an asterisk above the bar for a particular strain indicates positive tissue maceration, and a P indicates a PCR-positive result for pehA.
Fungal antibiosis assays.
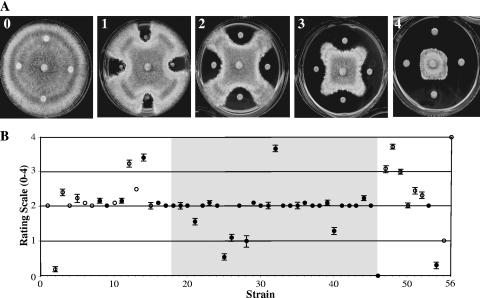
Five distinct patterns of inhibition of the fungus R. solani AG4 were observed when challenged by B. cenocepacia isolates (Fig. 4A). If we observed no inhibition of the fungus, a rating of 0 was assigned. Four other distinct qualitative zones of inhibition were observed and designated ratings of 1 through 4. The control strain, B. ambifaria AMMD (no. 56), a known antifungal biological control agent, exhibited significant inhibition of growth of R. solani AG4 and had a rating of 4 (Fig. 4B). All isolates originally recovered from the onion rhizosphere or onion field soil (no. 1 and 3 to 11) inhibited growth of R. solani AG4 to a moderate degree and exhibited average antifungal ratings of ≥2. Most clinical isolates (29 out of 42 strains; e.g., no. 16 to 23) also showed a moderate degree of fungal antibiosis although antibiosis varied in different strains of the same ST (e.g., ST-40; strains AU0418 [no. 25] and CF20 [no. 32] had antifungal activities of 0.5 and 3.75, respectively). Eight of 42 clinical isolates were less capable of impeding the growth of R. solani AG4 and had average antifungal ratings between 0 and 1.58 (e.g., strains AU10310 [no. 2] and AU0418 [no. 25]). The presence of the prnD gene required for synthesis of the antifungal compound pyrrolnitrin was not correlated with antifungal activity against R. solani AG4, as seven out of nine strains with the high antifungal ratings did not harbor prnD, and prnD was detected in six out of eight strains with low antifungal activity (Fig. 4B).
FIG. 4.
Antibiosis activity of B. cenocepacia and control strains against Rhizoctonia solani AG4. (A) Qualitative rating scale (0 to 4) for fungal inhibition. Examples of the rating scale are as follows: 0, B. cenocepacia AU10310 (no. 2); 1, B. cenocepacia AU0283 (no. 21); 2, B. cenocepacia 6RT131 (no. 11); 3, B. cenocepacia AU1054 (no. 12); 4, B. cenocepacia CF20 (no. 32). (B) Antibiosis activity ratings for 54 B. cenocepacia strains, B. cepacia 25416 (no. 55), and B. ambifaria AMMD (no. 56). The strain order (no. 1 to 56) is as listed in Table 1, the shaded box indicates strains belonging to the ST-40 Midwest epidemic clone lineage, and filled circles indicate the presence of the prnD gene required for pyrrolnitrin biosynthesis. Bars shown depict the standard error of the mean values. If no bars are present, the standard error was either 0 or smaller than the size of the symbol.
Nematode pathogenesis assays.
Nematode pathogenesis assays were conducted on bacterial lawns grown on two different media, NGM and PGS, for which P. aeruginosa PA14 exhibits slow or fast killing, respectively. Thus, the number of surviving nematodes was counted at 3 and 7 days after nematodes were added.
Most STs (17 out of 23), consisting of 23 environmental and clinical strains (e.g., no. 1 to 16), exhibited rapid nematode killing on NGM. A significant reduction (P < 0.01) in nematode survival was observed for these strains (<20%) at 3 days compared to the 33.8% survival for PA14 (Fig. 5A; also see Table S1 in the supplemental material). Later, at day 7, few or no nematodes survived (<1.3%) on the lawns of these 23 strains or on PA14 (0.15%). In contrast, nematode virulence exhibited by the 28 clinical strains (no. 18 to 45) of the Midwest lineage (ST-40) on NGM at day 3 was variable and on average moderate, with 80.9% nematode survival. For example, five strains, AU0279 (no. 18), AU0418 (no. 25), CF51 (no. 37), CF157 (no. 39), and MDCH2 (no. 41), exhibited nematode survival similar to that of the negative control strain, E. coli OP50 (95.5%), at day 3, whereas 23 other ST-40 strains exhibited significantly less nematode survival on day 3 (54.1 to 87%) than did E. coli OP50 (Fig. 5A; see also Table S1 in the supplemental material). Later, at day 7, two ST-40 strains, AU0281 (no. 20) and AU0284 (no. 22), exhibited a slow-kill reaction where nematode survival on day 7 (<11%) was much lower than on day 3 (81.3 to 85%). Twenty-two ST-40 strains exhibited a modest slow-kill reaction and decreased nematode survival by >20% from day 3 to day 7. However, three ST-40 strains, CF2 (no. 30), CF157 (no. 39), and CF160 (no. 40), exhibited low or no nematode virulence (>80% survival) on day 7, similar to the negative control OP50. In summary, most STs exhibited fast nematode killing on NGM (on which PA14 kills slowly), but a more modest and variable virulence was observed for ST-40 clinical strains.
FIG. 5.
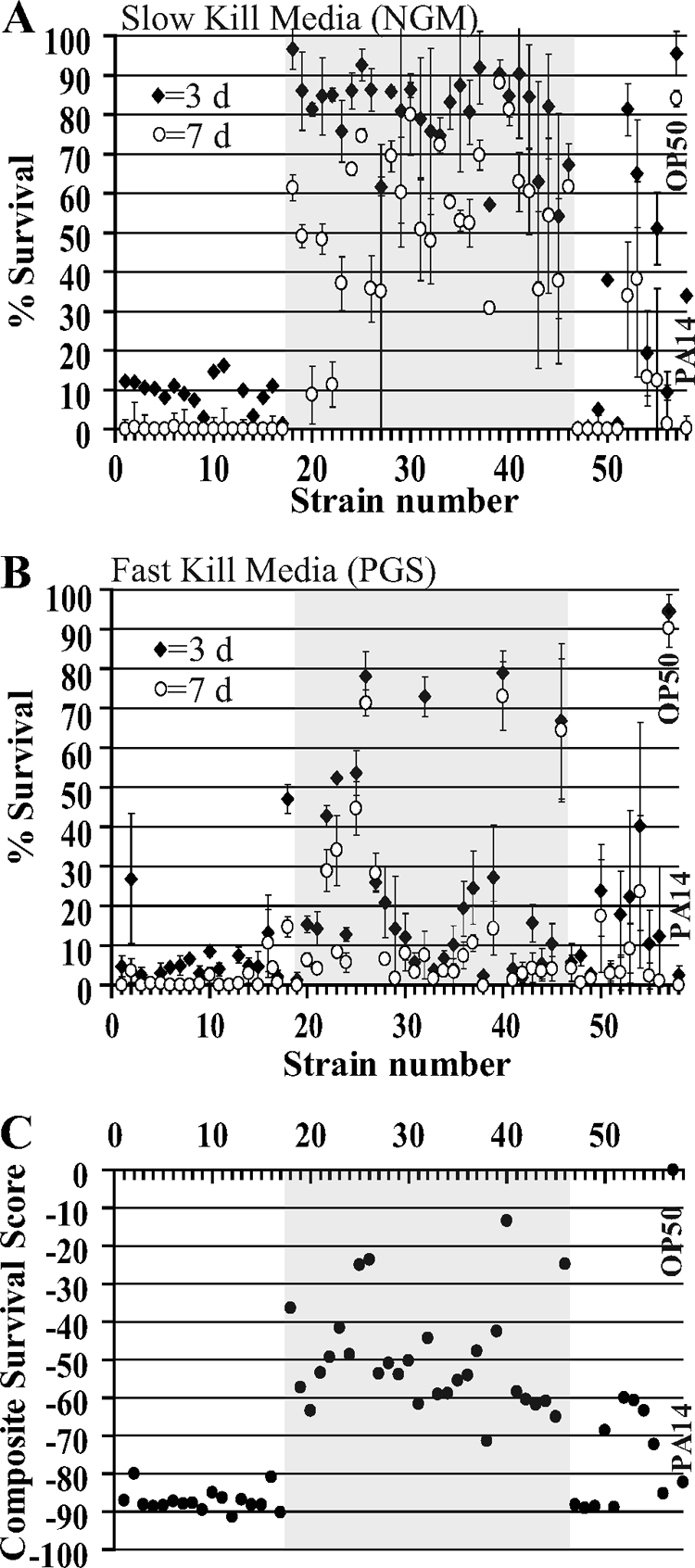
Pathogenicity of B. cenocepacia and control strains to C. elegans. Survival of early-L4-stage C. elegans at days 3 and 7 after addition to Burkholderia lawns growing on slow-kill (NGM) (A) or fast-kill (PGS) (B) media. (C) CSS calculated by the mixed model analysis. The x-axes in panels A to C indicates the strain numbers listed in Table 1, and the shaded boxes indicate strains belonging to the ST-40 Midwest epidemic clone lineage. Closed diamonds and open circles represent worm survival data at day 3 and day 7, respectively. Bars shown depict the standard error of the mean values. If no bars are present, the standard error was either 0 or smaller than the size of the symbol.
On the high-osmolarity PGS medium, most BCC strains (46 out of 56 strains) and PA14 exhibited fast killing (Fig. 5A and B; also see Table S1 in the supplemental material). Twenty-six BCC strains (e.g., no. 3 to 16 and 19 to 21) exhibited 0 to 26.9% nematode survival at day 3 on PGS medium, of which 16 strains representing 13/23 STs resulted in survival similar (P < 0.01) to that of the positive control, PA14 (2.4%). At day 7 on PGS medium, when few or no nematodes survived on most strains, >64% of worms survived on three strains, AU0419 (no. 26; ST-40), CF160 (no. 40; ST-40), and AU2289 (no. 46; ST-249). Eight ST-40 Midwest lineage strains showed high virulence to nematodes, similar to PA14, at day 3, whereas the remaining ST-40 strains showed 10% to 78% nematode survival. Strains AU0279 (no. 18) and CF20 (no. 32) exhibited slow killing on PGS medium, where >20% nematodes were killed between days 3 and 7. In summary, most BCC strains killed nematodes fast on PGS medium, including some of the ST-40 strains that failed to kill on NGM. An exception is the ST-40 strain AU0419 (no. 40) that failed to kill nematodes on both NGM and PGS medium.
To quantify the overall virulence of bacterial strains to nematodes on two different media and time points, we calculated a CSS. The CSS values were negative, indicating decreased worm survival compared to that of the negative control strain, OP50 (P < 0.0001). A large negative CSS value indicates low nematode survival and high virulence (Fig. 5C; see also Table S1 in the supplemental material). Overall virulence of the BCC strains to nematodes clustered into three groups: high virulence (CSS < −66), moderate virulence (CSS from −66 to −33), and low virulence (CSS > −33). Most of the STs exhibited high virulence to nematodes, as evidenced by the observation that 25 clinical and environmental strains (e.g., no. 1 to 17) belonging to 19 STs (ST-10, ST-40, ST-77, ST-122, ST-125, ST-263, ST-281, ST-330, ST-332, ST-427, ST-428, ST-429, ST-430, ST-433, ST-434, ST-435, ST-467, ST-468, and ST-469) grouped into the high-virulence group (CSS < −66), which included the positive control strain, PA14 (CSS = −82.2). Only one ST-40 strain, CF72 (no. 38), exhibited high nematode virulence (CSS = −71.3). The PHDC epidemic clone strain AU1054 (no. 12, ST-122) had the highest virulence (CSS = −91.3). The moderate-virulence group (CSS from −66 to −33) consisted of 27 clinical strains (e.g., no. 27 to 39) distributed among four ST genotypes (ST-28, ST-32, ST-40, ST-470), which consisted of 24 out of 27 strains of the ST-40 Midwest genotype. The low-virulence group (CSS > −33) consisted of four clinical strains, AU0418 (no. 25), AU0419 (no. 26), CF160 (no. 40), and AU2289 (no. 46), and the negative control strain, OP50 (Fig. 5C; see also Table S1 in the supplemental material). In summary, most STs represented in this study exhibited high virulence to nematodes, while different ST-40 clinical strains exhibited low, moderate, or high virulence.
Overall, most of the tested BCC strains were more virulent to nematodes than was PA14 on NGM and showed high pathogenicity by exhibiting <20% survival on both media at day 3. This is supported by the CSS analysis results, which showed that strains representing 19 out of 23 STs clustered into the high-virulence group with CSS values of <−66. Midwest lineage ST-40 strains showed moderate or low pathogenicity to the nematodes and displayed strain-specific variability in the virulence characteristics.
DISCUSSION
The objectives of this study were to determine phylogenetic relationships and survey virulence potential of clinical and environmental isolates of B. cenocepacia. The species B. cenocepacia is an ideal organism to study opportunistic pathogenicity, because it is ubiquitous in the environment, including Michigan onion agricultural fields (29); includes the Midwest epidemic clone still present in Michigan CF patients (13); and carries large multireplicon genomes, several which have been sequenced (40). From this study, we determined that most B. cenocepacia STs and strains exhibited multihost pathogenicity against C. elegans nematodes and the fungus R. solani AG4. The characteristic of onion pathogenicity differentiated two groups of genetically related strains. Surprisingly, because of the prevalence of onion agriculture in Michigan and elsewhere in the Midwest, all but one of the Midwest epidemic clone strains and 12 other related STs isolated from sporadic infections from persons with CF lacked pathogenicity to onion. It is possible that onion agriculture is not a source for these Midwest clinical STs or that onion pathogenicity is lost after human infection. Thus, it remains to be determined what the environmental source of the Midwest epidemic clone is and why the PHDC epidemic clone found in environmental isolates from Michigan is not appearing in persons with CF in the Midwest.
MLST analysis revealed a high degree of nucleotide variability among the B. cenocepacia IIIB strains evaluated in this study, particularly among the Michigan agricultural strains. All of the onion isolates clustered together, and several new STs were identified. Two distinct STs, ST-122 and ST-125, included both clinical and onion isolates. Interestingly, B. cenocepacia ST-125 was also recovered from the maize rhizosphere in southern Italy (14), and ST-122 has been isolated from soil in New York and from CF and non-CF patients in the United States and Canada, respectively (1), indicating the wide geographical distribution of this ST. One additional environmental strain obtained from the maize rhizosphere, MCO-3 (no. 15; ST-281), grouped with the majority of clinical isolates examined. Of these clinical isolates, two variants, AU7610 (no. 51; ST-468) and AU10575 (no. 54; ST-470), grouped separately.
The current MLST system has been shown to be a reliable method for the identification of Burkholderia isolates (2). However, due to the complexity of the Burkholderia genome and the high degree of recombination observed between strains, the current MLST system using only seven genes does not provide a solid framework for uncovering evolutionary relationships. As indicated by SLAC analysis performed in this study, gltB and gyrB MLST loci appear to be evolving at different rates on the chromosomal backbone; therefore, these genes should be used with caution when making phylogenetic inferences for Burkholderia. Increasing the number of MLST loci used for phylogenetic inferences or developing a more sensitive method (i.e., single nucleotide polymorphism typing) may improve the resolution of the phylogenetic relationships of strains belonging to the BCC. In addition, the differences in pathogenicity to nematodes and antifungal activity exhibited by some ST-40 strains (discussed below) suggest that caution should be used when inferring phenotypic traits and virulence potential from MLST analysis results.
In general, pathogenicity to onion was correlated with the source from which the strains were isolated and the presence of the pehA gene encoding endopolygalacturonase. Onion pathogenicity and pehA were both absent in 41 isolates representing 14 STs mostly isolated from CF patients, while a related group consisting of 13 isolates representing seven STs from onion field soil or rhizosphere (except AU1054 [no. 12]) were pathogenic to onion and contained pehA. The characteristic of onion pathogenicity and the presence of the pehA gene could be explained by acquisition of pehA by onion isolates or the loss of ancestral pehA from most clinical isolates. Onion pathogenicity of 475 of 623 B. ambifaria isolates (a BCC species member that is usually avirulent in onions) obtained from onion fields suggests that acquisition of pehA can occur in the onion rhizosphere (29).
High to moderate virulence to nematodes and antifungal activity were pathogenic characteristics demonstrated by most strains in our BCC strain collection, with notable exceptions. Many of the strains exhibited high nematode virulence with lower CSSs (e.g., AU1054 [no. 12], CSS = −91) than those of P. aeruginosa PA14, primarily due to lower nematode survival after day 3 (e.g., 0% for AU1054) on NGM compared to that on PA14 (34%). The fast killing of nematodes on NGM could result from a more systemic or intense infection of the nematode intestine, increased production of toxic compounds on NGM compared to that on PA14, or production of virulence factors or toxic metabolites more lethal to the nematode than those produced by PA14.
Fast killing on PGS medium by certain strains of P. aeruginosa (e.g., PA01) requires the production of cyanide (19). We assayed for cyanide production by using a colorimetric assay (10), in which a blue color is formed in the presence of cyanide. Cyanide production was not detected in 48-h lawns of AU1054 (no. 12) on LB and was detected in cyanogenic P. aeruginosa PA01 under the same conditions (data not shown). Recently, cyanide production was detected in B. cenocepacia grown on LB at levels comparable to those detected in PA01 (when normalized for viable cell number) by using a different detection method (47). However, we cannot rule out the possibility that cyanide is produced by these strains on NGM or in infected nematodes. It remains to be determined what the mechanism(s) is for the rapid killing of nematodes by AU1054 on NGM.
Notably, several ST-40 or Midwest epidemic lineage strains differed from other ST-40 strains by exhibiting moderate nematode pathogenicity and antifungal activity. Three of the ST-40 strains and one closely related strain, AU2289 (no. 46; ST-249), exhibited very low nematode pathogenicity and did not differ significantly from the negative control, E. coli OP50, and also demonstrated poor antifungal activity. It is possible that these strains lost genes required for nematode pathogenicity and antifungal activity during infection of CF patients. Loss of pathogenicity in these strains might be analogous to social cheaters that arise during P. aeruginosa infection of CF patients and are attenuated in virulence (49). In contrast, strain CF72 (no. 38; ST-40) had high virulence (CSS = −71) that was not significantly different from that of the control strain, P. aeruginosa PA14 (CSS = −82). The variability in nematode virulence exhibited by some ST-40 strains is similar to that observed previously in Burkholderia (9). The moderate nematode virulence exhibited by most ST-40 strains on NGM might be analogous to the slow killing of C. elegans by PA14 that requires infection of the nematode intestine (56).
Pathogenicity to multiple hosts might be a key factor contributing to the potential of B. cenocepacia strains to become opportunistic pathogens, as this characteristic is shared by other opportunistic pathogens, such as P. aeruginosa, which is commonly isolated from CF patients and exhibits pathogenicity to fungi, plants, nematodes, flies, and mice (16, 17, 25, 36, 43, 56). Burkholderia spp. are also known to infect multiple hosts such as nematodes and insects (26, 50). Common virulence factors are often employed and important for virulence in multiple hosts (56, 59). The ability to interact with multiple hosts may contribute to the potential of an environmental microbe to function as an opportunistic pathogen by increasing the prevalence of the pathogen in the environment and, as a consequence, exposure to potential hosts and selection for virulence factors that function in multiple hosts.
In summary, our collection of B. cenocepacia IIIB strains appears to include two clusters of related strains—one primarily composed of onion agriculture isolates and pathogenic to onions, nematodes, and fungi, and another cluster composed primarily of CF isolates that lacked onion pathogenicity, but exhibited pathogenicity to nematodes and fungi. Some strains, including the CF isolate AU1054, were exceptionally pathogenic to nematodes on NGM. Future elucidation of genes required for pathogenicity and their distribution in B. cenocepacia strains should increase our understanding of the evolution of opportunistic and environmental pathogens.
Supplementary Material
Acknowledgments
We thank Joshua Buysse, Anthony Fasi, and Nikola Vuljaj for technical assistance and Chandni Bhan from the Center for Statistical Training and Consulting (CSTAT), Michigan State University, for help with the statistics and nematode data analysis.
This work was supported by the Michigan State University Center for Microbial Pathogenesis, Michigan State University Project GREEEN, and the Michigan Agricultural Experiment Station.
We dedicate this paper to the late Tom Whittam (1954 to 2008), a dear colleague and friend.
Footnotes
Published ahead of print on 19 June 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baldwin, A., E. Mahenthiralingam, P. Drevinek, P. Vandamme, J. R. Govan, D. J. Waine, J. J. LiPuma, L. Chiarini, C. Dalmastri, D. A. Henry, D. P. Speert, D. Honeybourne, M. C. J. Maiden, and C. G. Dowson. 2007. Environmental Burkholderia cepacia complex isolates in human infections. Emerg. Infect. Dis. 13:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin, A., E. Mahenthiralingam, K. M. Thickett, D. Honeybourne, M. C. J. Maiden, J. R. Govan, D. P. Speert, J. J. LiPuma, P. Vandamme, and C. G. Dowson. 2005. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J. Clin. Microbiol. 43:4665-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., I. Schneider, R. Jungwirth, and C. Roller. 1999. Discrimination of Burkholderia multivorans and Burkholderia vietnamensis from Burkholderia cepacia genomovars I, III, and IV by PCR. J. Clin. Microbiol. 37:1335-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beanan, M. J., and S. Strome. 1992. Characterization of a germ-line proliferation mutation in Caenorhabditis elegans. Development 116:755-766. [DOI] [PubMed] [Google Scholar]
- 5.Bernier, S. P., L. Silo-Suh, D. E. Woods, D. E. Ohman, and P. A. Sokol. 2003. Comparative analysis of plant and animal models for characterization of Burkholderia cepacia virulence. Infect. Immun. 71:5306-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruen, T. C., H. Philippe, and D. Bryant. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkholder, W. H. 1950. Sour skin, a bacterial rot of onion bulbs. Phytopathology 40:115-118. [Google Scholar]
- 9.Cardona, S. T., J. Wopperer, L. Eberl, and M. A. Valvano. 2005. Diverse pathogenicity of Burkholderia cepacia complex strains in the Caenorhabditis elegans host model. FEMS Microbiol. Lett. 250:97-104. [DOI] [PubMed] [Google Scholar]
- 10.Castric, K. F., and P. Castric. 1983. Method for rapid detection of cyanogenic bacteria. Appl. Environ. Microbiol. 45:701-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J. S., K. A. Witzmann, T. Spilker, R. J. Fink, and J. J. LiPuma. 2001. Epidemicity and inter-city spread of Burkholderia cepacia genomovar III in cystic fibrosis. J. Pediatr. 139:643-649. [DOI] [PubMed] [Google Scholar]
- 12.Ciche, T., and S. K. Goffredi. 2007. General methods to investigate microbial symbioses, p. 394-419. In C. R. Reddy, T. J. Beveridge, J. A. Breznak, G. A. Marzluf, T. Schmidt, and L. R. Snyder (ed.), Methods in general and molecular microbiology. ASM Press, Washington, DC.
- 13.Coenye, T., and J. J. LiPuma. 2003. Population structure analysis of Burkholderia cepacia genomovar III: varying degrees of genetic recombination characterize major clonal complexes. Microbiology 149:77-88. [DOI] [PubMed] [Google Scholar]
- 14.Dalmastri, C., A. Baldwin, S. Tabacchioni, A. Bevivino, E. Mahenthiralingam, L. Chiarini, and C. Dowson. 2007. Investigating Burkholderia cepacia complex populations recovered from Italian maize rhizosphere by multilocus sequence typing. Environ. Microbiol. 9:1632-1639. [DOI] [PubMed] [Google Scholar]
- 15.Darby, C. 2005. Interactions with microbial pathogens. In The C. elegans Research Community (ed.), WormBook. doi: 10.1895/wormbook.1.21.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 16.Darby, C., C. L. Cosma, J. H. Thomas, and C. Manoil. 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:15202-15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Argenio, D. A., L. A. Gallagher, C. A. Berg, and C. Manoil. 2001. Drosophila as a model host for Pseudomonas aeruginosa infection. J. Bacteriol. 183:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Souza, J. T., and J. M. Raaijmakers. 2003. Polymorphisms within the prnD and pltC genes from pyrrolnitrin and pyoluteorin-producing Pseudomonas and Burkholderia spp. FEMS Microb. Ecol. 43:21-34. [DOI] [PubMed] [Google Scholar]
- 19.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gan, Y. H., K. L. Chua, H. H. Chua, B. Liu, C. S. Hii, H. L. Chong, and P. Tan. 2002. Characterization of Burkholderia pseudomallei infection and identification of novel virulence factors using a Caenorhabditis elegans host system. Mol. Microbiol. 44:1185-1197. [DOI] [PubMed] [Google Scholar]
- 21.Goldmann, D. A., and J. D. Klinger. 1986. Pseudomonas cepacia: biology, mechanisms of virulence, epidemiology. J. Pediatr. 108:806-812. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez, C. F., E. A. Pettit, V. A. Valadez, and E. Provin. 1997. Mobilization, cloning, and sequence determination of a plasmid-encoded polygalacturonase from a phytopathogenic Burkholderia (Pseudomonas) cepacia. Mol. Plant-Microbe Interact. 10:840-851. [DOI] [PubMed] [Google Scholar]
- 23.Govan, J. R. W., C. J. Doherty, J. W. Nelson, P. H. Brown, A. P. Greening, J. Maddison, M. Dodd, and A. K. Webb. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15-19. [DOI] [PubMed] [Google Scholar]
- 24.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogan, D. A., and R. Kolter. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296:2229-2232. [DOI] [PubMed] [Google Scholar]
- 26.Huber, B., F. Feldmann, M. Kothe, P. Vandamme, J. Wopperer, K. Riedel, and L. Eberl. 2004. Identification of a novel virulence factor in Burkholderia cenocepacia H111 required for efficient slow killing of Caenorhabditis elegans. Infect. Immun. 72:7220-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 28.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levision. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 29.Jacobs, J. L., A. C. Fasi, A. Ramette, J. J. Smith, R. Hammerschmidt, and G. W. Sundin. 2008. Identification and onion pathogenicity of Burkholderia cepacia complex isolates from the onion rhizosphere and onion field soil. Appl. Environ. Microbiol. 74:3121-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the detection of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 31.Köthe, M., M. Antl, B. Huber, K. Stoecker, D. Ebrecht, I. Steinmetz, and L. Eberl. 2003. Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell. Microbiol. 5:343-351. [DOI] [PubMed] [Google Scholar]
- 32.Kumar, A., S. Dietrich, W. Schneider, R. Jacobson, F. Pouch Downes, B. E. Robinson-Dunn, R. Honicky, J. Smith, and R. Martin. 1997. Genetic relatedness of Burkholderia (Pseudomonas) cepacia isolates from five cystic fibrosis centers in Michigan. Respir. Med. 91:485-492. [DOI] [PubMed] [Google Scholar]
- 33.Lee, Y. A., and C. W. Chan. 2007. Molecular typing and presence of genetic markers among strains of banana finger-tip rot pathogen, Burkholderia cenocepacia, in Taiwan. Phytopathology 97:195-201. [DOI] [PubMed] [Google Scholar]
- 34.LiPuma, J. J., T. Spilker, T. Coeyne, and C. F. Gonzalez. 2002. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 359:2002-2003. [DOI] [PubMed] [Google Scholar]
- 35.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell III, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 164:92-96. [DOI] [PubMed] [Google Scholar]
- 36.Mahajan-Miklos, S., L. G. Rahme, and F. M. Ausubel. 2000. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol. Microbiol. 37:981-988. [DOI] [PubMed] [Google Scholar]
- 37.Mahenthiralingam, E., A. Baldwin, and C. G. Dowson. 2008. Burkholderia cepacia complex bacteria: opportunistic bacteria with important natural biology. J. Appl. Microbiol. 104:1539-1551. [DOI] [PubMed] [Google Scholar]
- 38.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. VanDamme. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahenthiralingam, E., D. Simpson, and D. P. Speert. 1997. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J. Clin. Microbiol. 35:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144-156. [DOI] [PubMed] [Google Scholar]
- 41.Miller, J. H. 1972. Experiments in molecular genetics, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Peleg, A. Y., E. Tampakakis, B. B. Fuchs, G. M. Eliopoulos, R. C. Moellering, Jr., and E. Mylonakis. 2008. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 105:14585-14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plotnikova, J. M., L. G. Rahme, and F. M. Ausubel. 2000. Pathogenesis of the human opportunistic pathogen Pseudomonas aeruginosa PA14 in Arabidopsis. Plant Physiol. 124:1766-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pond, S. L., and S. D. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531-2533. [DOI] [PubMed] [Google Scholar]
- 45.Ramette, A., J. J. LiPuma, and J. M. Tiedje. 2005. Species abundance and diversity of Burkholderia cepacia complex in the environment. Appl. Environ. Microbiol. 71:1193-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reik, R., T. Spilker, and J. J. LiPuma. 2005. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. J. Clin. Microbiol. 43:2926-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryall, B., X. Y. Lee, J. E. A. Zlosnik, S. Hoshino, and H. D. Williams. 2008. Bacteria of the Burkholderia cepacia complex are cyanogenic under biofilm and colonial growth conditions. BMC Microbiol. 8:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sajjan, U., G. Thanassoulis, V. Cherapanov, A. Lu, C. Sjolin, B. Steer, Y. J. Wu, O. D. Rotstein, G. Kent, C. McKerlie, J. Forstner, and G. P. Downey. 2001. Enhanced susceptibility to pulmonary infection with Burkholderia cepacia in Cftr−/− mice. Infect. Immun. 69:5138-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandoz, K. M., S. M. Mitzimberg, and M. Schuster. 2007. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. USA 40:15876-15881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seed, K. D., and J. J. Dennis. 2008. Development of Galleria mellonella as an alternative infection model for the Burkholderia cepacia complex. Infect. Immun. 76:1267-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sifri, C. D., J. Begun, and F. M. Ausubel. 2005. The worm has turned—microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 3:119-127. [DOI] [PubMed] [Google Scholar]
- 52.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sulston, J., and J. Hodgkin. 1988. Methods, p. 587-606. In W. B. Wood (ed.), The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 54.Sun, L., R. Jiang, S. Steinbach, A. Holmes, C. Campanelli, J. Forstner, Y. Tan, M. Riley, and R. Goldstein. 1995. The emergence of a highly transmissible lineage of cbl+ Pseudomonas (Burkholderia) cepacia causing CF centre epidemics in North America and Britain. Nat. Med. 1:661-666. [DOI] [PubMed] [Google Scholar]
- 55.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 56.Tan, M., S. M. Milkos, and F. M. Ausubel. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vanlaere, E., J. J. LiPuma, A. Baldwin, D. Henry, E. DeBrandt, E. Mahenthiralingam, D. Speert, C. Dowson, and P. Vandamme. 2008. Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int. J. Syst. Evol. Microbiol. 58:1580-1590. [DOI] [PubMed] [Google Scholar]
- 58.Vanlaere, E., A. Baldwin, D. Gevers, D. Henry, E. De Brandt, J. J. LiPuma, E. Mahenthiralingam, D. P. Speert, C. Dowson, and P. Vandamme. 2009. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59:102-111. [DOI] [PubMed] [Google Scholar]
- 59.Yorgey, P., L. G. Rahme, M. W. Tan, and F. M. Ausubel. 2001. The roles of mucD and alginate in the virulence of Pseudomonas aeruginosa in plants, nematodes and mice. Mol. Microbiol. 41:1063-1076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.