Abstract
Bacillus thuringiensis has been used as a bioinsecticide to control agricultural insects. Bacillus cereus group genomes were found to have a Bacillus enhancin-like (bel) gene, encoding a peptide with 20 to 30% identity to viral enhancin protein, which can enhance viral infection by degradation of the peritrophic matrix (PM) of the insect midgut. In this study, the bel gene was found to have an activity similar to that of the viral enhancin gene. A bel knockout mutant was constructed by using a plasmid-free B. thuringiensis derivative, BMB171. The 50% lethal concentrations of this mutant plus the cry1Ac insecticidal protein gene were about 5.8-fold higher than those of the BMB171 strain. When purified Bel was mixed with the Cry1Ac protein and fed to Helicoverpa armigera larvae, 3 μg/ml Cry1Ac alone induced 34.2% mortality. Meanwhile, the mortality rate rose to 74.4% when the same amount of Cry1Ac was mixed with 0.8 μg/ml of Bel. Microscopic observation showed a significant disruption detected on the midgut PM of H. armigera larvae after they were fed Bel. In vitro degradation assays showed that Bel digested the intestinal mucin (IIM) of Trichoplusia ni and H. armigera larvae to various degrading products, similar to findings for viral enhancin. These results imply Bel toxicity enhancement depends on the destruction of midgut PM and IIM, similar to the case with viral enhancin. This discovery showed that Bel has the potential to enhance insecticidal activity of B. thuringiensis-based biopesticides and transgenic crops.
Bacillus thuringiensis is a ubiquitous gram-positive, spore-forming soil bacterium and produces insecticidal crystal proteins during the sporulation phase of its growth cycle. Because these insecticidal crystal proteins have activity against certain insect species, B. thuringiensis has been extensively used as a biopesticide to control crop pests in commercial agriculture and forest management. It is also a key source of genes for transgenic expression and provides pest resistance in plants (2, 20, 30).
The viral enhancin protein was originally described for granuloviruses (GVs) as a 126-kDa protein that showed an ability to enhance the infectivity of nucleopolyhedroviruses (NPVs) (36, 37, 39). It has also been found in several other GVs (13) and NPVs (19, 27). Considered a pathogenicity factor, it is not essential for growth of viruses in cell culture or infected insects but has the function of facilitating GV and NPV infection and decreasing larval survival time (14, 17, 19, 27).
The widely accepted action mode of the viral enhancin protein, which has been identified as a metalloprotease (17), is that it can disrupt the protective peritrophic matrix (PM), allowing virion access to the underlying epithelial cells of the insect gut (17). The PM has a lattice structure formed by chitin and insect intestinal mucin (IIM), and the viral enhancin protein targets the IIM for degradation (33).
Enhancin-like genes with 24 to 25% nucleotide identity to viral enhancin genes have been found in Yersinia pestis, Bacillus anthracis, Bacillus thuringiensis, and Bacillus cereus genome sequences (16, 25, 28). When B. cereus enhancin-like protein was expressed in recombinant Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) budded viruses and polyhedral inclusion bodies, it was found to be cytotoxic compared to viral enhancin protein. However, larval bioassays indicated that this enhancin-like protein did not enhance infection (8). Hajaij-Ellouze et al. (12) isolated a B. thuringiensis enhancin-like gene from a 407 crystal-minus strain and found that this enhancin-like protein has a typical metalloprotease zinc-binding domain (HEIAH) and belongs to the PlcR regulon. When the enhancin-like mutant was fed to Galleria mellonella larvae, no significant reduction in virulence was observed.
In the present study, we report a B. thuringiensis enhancin-like gene (bel) encoding a protein (Bel) that has 20 to 30% identity to the viral enhancin protein and 95% identity to bacterial enhancin-like proteins. Therefore, Bel function may have a synergistic action similar to that of the virus enhancin protein. To understand the biochemical activity of this novel bacterial gene, bel was knocked out in the plasmid-free strain BMB171. We expected that this bel mutant would have no significant reduction in toxicity according to the reports of Galloway et al. (8) and Hajaij-Ellouze et al. (12). However, the bel mutant surprisingly resulted in dramatically reduced Cry1Ac toxicity to Helicoverpa armigera larvae. To further confirm this result, purified Bel was fed together with the Cry1Ac protein to H. armigera larvae. We found that Bel can function as a synergist of Cry1Ac toxicity against H. armigera. In vivo and in vitro observations showed that Bel can disrupt the insect midgut PM and degrade IIM of insect midgut PM. The target of Bel is the IIM of PM, similar to the results found with viral enhancin.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in this study are listed in Table 1. All Escherichia coli strains and B. thuringiensis strains were maintained on Luria-Bertani (LB) agar plates (1% tryptone, 0.5% yeast extract, 0.5% NaCl, and 1.5% agar) and supplemented with appropriate antibiotics at 37°C and 28°C, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Origin or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169 (ϕ80 lacZΔM15) hsd R17 recA1 endA1 gyrA96thi−1relA1 | Amersham Biosciences |
| BL21(DE3) | T7 promoter | Amersham Biosciences |
| Bacillus thuringiensis | ||
| BMB171 | Nonplasmid and acrystalliferous mutant, subsp. kurstaki, no Cry protein | 18 |
| YBT1520 | Wild type, subsp. kurstaki cry1Aa cry1Ab cry1Ac cry2Aa cry2Ab | 38 |
| Plasmids | ||
| pHT304 | E. coli and B. thuringiensis shuttle vector; Ampr Ermr ori1030 ori ColEI, 6.7 kb | 1 |
| pDG646 | ori E. coli, Ampr Ermr, 4.3 kb | 10 |
| pDG780 | ori E. coli, Ampr Kanr, 4.4 kb | 10 |
| pBMB0631 | Derivative of pDG780, containing temp-sensitive replicon, 6.9 kb | 21 |
| pGEX-6P-1 | ori E. coli, Ampr, 4.9 kb | Amersham Biosciences |
Ampr, ampicillin resistance; Ermr, erythromycin resistance; Kanr, kanamycin resistance.
Cloning of the bel gene from B. thuringiensis strains YBT-1520 and BMB171.
The B. thuringiensis bel genes were amplified from strains YBT-1520 and BMB171, respectively, by PCR using genomic DNA as the template with a pair of primers, which were designed based on the bel gene sequence of YBT-1520, belP1 (5′-GCCGGATCCATGTATACAATGTTTTTCCTC-3′) and belP2 (5′-GGCGAATTCTTATTCATTATATAAGCTATC-3′) (Table 2).
TABLE 2.
Primers used in this study
| Primer | Sequencea | Product size |
|---|---|---|
| EUP1 | 5′-GCAGGATCCACCGCCAGCACCAGATAT-3′ | 840 bp |
| EUP2 | 5′-GCATCTAGAGTAACTATAATAATTACACT-3′ | |
| EDP1 | 5′-GCAGAATTCCCTTCCATTTCCTTCGTAT-3′ | 1.1 kb |
| EDP2 | 5′-GCAGGATCCATATCTGGTGCTGGCGGT-3′ | |
| belP1 | 5′-GCCGGATCCATGTATACAATGTTTTTCCTC-3′ | 2.2 kb |
| belP2 | 5′-GGCGAATTCTTATTCATTATATAAGCTATC-3′ |
The restriction sites included in the oligonucleotide sequence, used during the cloning experiments, are underlined.
Construction of the bel knockout mutant of strain BMB171.
An 840-bp BamHI-XbaI fragment and an 1,100-bp BamHI-EcoRI DNA fragment, corresponding to the chromosomal DNA regions upstream and downstream of the open reading frame of the bel gene in BMB171, respectively, were generated by PCR using the oligonucleotide pairs EUP1-EUP2 and EDP1-EDP2 (Table 2). The two amplified DNA fragments were digested with BamHI-XbaI and BamHI-EcoRI, respectively, and cloned into the temperature-sensitive plasmid pBMB0631. Then, the erythromycin resistance cassette (1.6 kb) from plasmid pDG646 was digested with BamHI and inserted between the two amplified DNA fragments. The resulting plasmid, pBMB1083, was transformed into strain BMB171 by electroporation. These transformants were cultivated in LB medium (containing 10 μg/ml erythromycin) for 8 h. Then, the transformants were cultivated at 42°C for 4 days to eliminate the unintegrated temperature-sensitive plasmid pBMB1083. Erythromycin-resistant (MIC of 25 μg/ml) but kanamycin-sensitive (MIC of 50 μg/ml) colonies were harvested. The correct mutant strain was named BMB1084.
Expression and purification of the Bel protein.
The gene bel, amplified from YBT-1520, was inserted into the BamHI and EcoRI sites of the expression vector pGEX-6P-1 to create the recombinant expression vector pEMB0717. Then, the recombinant plasmid was transferred into the E. coli strain BL21(DE3), resulting in the recombinant EMB0717. The Bel protein was overexpressed in E. coli as a fusion protein with a glutathione S-transferase (GST) tag. Soluble-state GST-Bel was purified on a GSTrap FF column as described by the GST.bind kit protocol (Amersham Biosciences, Uppsala, Sweden) and then dialyzed (against 10 mM Tris-HCl, pH 8.0, at 4°C), quantified (4), and stored at −80°C.
Cry1Ac crystal protein preparation and bioassay.
The Cry1Ac crystal protein was purified from recombinant strain BMB1087 (a BMB171 derivative containing the full cry1Ac gene in the vector pHT304 [Table 1]) as described by Luo et al. (22).
Bioassays were carried out by the diet incorporation method as described by Gunning et al. (11). Bel was mixed with the Cry1Ac toxin at different ratios (one group received Bel:Cry1Ac at the following ratios: 0:30, 1:30, 2:30, 3:30, 4:30, 6:30, and 8:30; the other group received Bel:Cry1Ac at the following ratios: 1:0, 1:15, 1:30, and 1:60) in phosphate-buffered saline buffer. The number of neonate larva used per dilution was 24, and three replicates were conducted with each dilution. After 3 days of incubation at 25°C, mortalities for each treatment were recorded. Data were analyzed by the general linear model method and Tukey's test at an α value of 0.05 for the nominal criterion level, using the SAS 9.1 software program (SAS Institute, Cary, NC). All the bioassays were conducted three times for each treatment.
Enhancin isolation.
The enhancin (En-Tn) from Trichoplusia ni granulovirus (TnGV) was isolated from capsules as described previously (7, 9).
PM and IIM preparation and degradation assays.
Midguts were isolated from mid-fifth-instar H. armigera and T. ni larvae and stored at −80°C. PMs were prepared from midguts of H. armigera and T. ni according to the methods described by Tanada et al. (32) and stored at −80°C in a solution of 300 mM mannitol, 5 mM EGTA, and 17 mM Tris at pH 7.5 until needed. IIMs were isolated from PMs of H. armigera and purified as described by Wang et al. (33). Degradation assays were conducted using the methods described by Wang et al. (33) by using prepared PMs and IIMs.
Effects of Bel on PM of H. armigera.
Purified Bel was mixed with an artificial diet at a concentration of 1 μg/ml. Then, 48 fifth-instar H. armigera larvae were individually fed this artificial diet in a tube (30 ml) for 1.5 days at 28°C for each group. Next, intact midguts of H. armigera larvae were removed as described above and stored in primary buffer at 4°C (1.5% formaldehyde and 2.5% gluteraldehyde in 0.1 M phosphate buffer, pH 7.4) (6). Selected midguts were rinsed three times for 5 min (each) at room temperature in 0.1 M cacodylate buffer (pH 7.4). Samples were secondarily fixed in 1% osmium tetroxide for 1 h, rinsed three times in cacodylate buffer for 5 min (each), and dehydrated through an ethanol series (15). Following ethanol dehydration, midgut samples from three to four larvae were freeze-fractured in liquid nitrogen for scanning electron microscopy (SEM) as described by Plymale et al. (26).
SDS-PAGE and Western blot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were performed as described by Bourgouin et al. (3). Proteins were separated by SDS-PAGE on a 10% gel and stained with Coomassie brilliant blue R250 after electrophoresis. Both an antiserum to T. ni IIM and an antibody to H. armigera IIM were used for detection in Western blotting. The former was kindly provided by Ping Wang, and the latter was isolated as described previously (34).
Nucleotide sequence accession numbers.
The bel nucleotide sequences of YBT-1520 and BMB171 have been deposited in the GenBank database under the accession numbers FJ644934 and FJ644935, respectively.
RESULTS
Isolation and characterization of the bel gene from B. thuringiensis.
The bel genes were amplified from B. thuringiensis strain YBT1520 and a plasmid-free strain, BMB171. Both bel genes revealed an open reading frame of 2,202 nucleotides that encodes a protein containing 733 amino acids with a molecular mass of 84 kDa. Sequence comparison showed the two Bel proteins share 92% identity and 95% similarity. A comparison among bel, viral enhancin genes, and other bacterial enhancin-like genes was conducted at the amino acid level (Table 3). The bel protein showed 22% to 25% identity to viral enhancin proteins, 30% identity with the enhancin-like protein from Yersinia pestis, and 95% to 96% identity to the B. cereus group enhancin-like proteins.
TABLE 3.
Identities and similarities of various kinds of enhancin proteinsa
| Bacterium or virus | % Identity/similarity of enhancin protein to protein from:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| B. cereus | B. anthracis | B. thuringiensis | LdNPV | PsunGV | HearGV | Y. pestis | CfMNPV | |
| B. cereus | 100/100 | 95/98 | 95/97 | 22/41 | 24/42 | 25/42 | 30/50 | 23/41 |
| B. anthracis | 100/100 | 96/98 | 21/24 | 24/42 | 25/42 | 30/51 | 24/42 | |
| B. thuringiensis | 100/100 | 22/41 | 24/42 | 25/42 | 30/51 | 24/41 | ||
| LdNPV | 100/100 | 31/47 | 31/47 | 24/41 | 25/42 | |||
| PsunGV | 100/100 | 80/89 | 24/42 | 23/39 | ||||
| HearGV | 100/100 | 23/41 | 22/39 | |||||
| Y. pestis | 100/100 | 24/43 | ||||||
| CfMNPV | 100/100 | |||||||
LdNPV, Lymantria dispar NPV; CfMNPV, Choristoneura fumiferana multicapsid NPV; PsunGV, Pseudaletia unipuncta GV; HearGV, Helicoverpa armigera GV.
Amino acid sequence alignment revealed that the homology among Bel, bacterial enhancin-like proteins, and viral enhancin proteins was high in the N-terminal regions of the proteins. In the 255- to 259-amino-acid sequence of Bel, a conserved metal binding motif (HEIAH) similar to that in the reported bacterial enhancin-like proteins was found (HEXXH in viral enhancin protein) (8) (Fig. 1A). There is also a conserved glutamic acid in Bel, 20 residues downstream from the first histidine within the zinc binding sequence (21 residues in Y. pestis enhancin-like protein; 20 residues in B. cereus enhancin-like protein; 19 residues in TnGV enhancin protein) (8). The presence of these features indicates that the Bel protein has the structural requirements defining a metalloprotease, similar to the viral enhancin proteins identified by Lepore et al. (17) and Li et al. (19).
FIG. 1.
(A) Partial alignment of Bel protein from YBT-1520 (from position 227 to 281) with enhancin family proteins. The highly conserved metalloprotease zinc-binding domain (HEIAH) in the region of positions 255 to 259 is shown boxed, whereas the conserved glutamic acid (E) is underlined. This was analyzed by using the ClustalX software program. (B) Phylogenetic tree of enhancin family. The enhancin amino acid sequences were aligned using ClustalX, and a neighbor-joining tree was generated. The final phylogenetic tree was generated using MEGA software, and bootstrap analysis values are displayed for each tree branch. Numbers on each branch indicate the frequency of a given tree branch after bootstrap analysis (out of 100 replicates). The bar at the bottom of the figure represents the number of amino acid substitutions per site.
To assess evolutionary relatedness, a phylogenetic tree was generated using the MEGA software program. In this analysis, enhancin proteins from Pseudaletia unipuncta GV, apple stem GV, H. armigera GV, Lymantria dispar multicapsid nucleopolyhedrovirus, TnGV, and Xestia c-nigrum GV were grouped together while enhancin proteins from NPVs (Mamestra configurata NPV and Choristoneura fumiferana multicapsid nucleopolyhedrovirus) were clustered together with the bacterial enhancin-like proteins (Fig. 1B).
B. thuringiensis bel mutant decreases toxicity of the Cry1Ac protein to H. armigera larvae.
To understand the biochemical activity of this novel bacterial bel gene, bel was knocked out in the plasmid-free B. thuringiensis strain BMB171, resulting in the bel mutant strain BMB1084. Then, pBMB1085, which contains the cry1Ac gene, was transferred to BMB171 and BMB1084 to generate the recombinant strains BMB1087 and BMB1088, respectively. They were the same in their growth curves, spore formation, and crystal production (data not shown). Their toxicity to H. armigera larvae was detected with their sporulation cultures at different concentrations. When the bel gene was knocked out, a significant reduction in the toxicity of Cry1Ac was found in the mutant strain BMB1088. Bioassay results showed that the 50% lethal concentrations (LC50s) of BMB1087 and BMB1088 for H. armigera larvae were 0.46 ± 0.05 μl/ml and 2.73 ± 0.36 μl/ml, respectively (regression equations of toxicity, y = 1.3324x + 5.4396 and y = 1.6982x + 4.2599, respectively). (The LC50 unit is the amount of fermented cultures per milliliter of contaminated diet). The relative ratio was 5.8-fold. These results indicated that deletion of the bel gene tends to decrease Cry1Ac protein virulence toward H. armigera larvae.
The Bel protein enhances toxicity of the Cry1Ac protein to H. armigera larvae.
To gain further insight into the biological function of the bel gene, purified Bel was used to test its enhancement of Cry1Ac toxicity to H. armigera larvae. Figure 2 shows the SDS-PAGE analysis of the soluble fractions from the expression of pEMB0717 (Fig. 2, lane 4). Major bands of approximately 112 kDa, corresponding to the expected molecular masses of GST-Bel fusion proteins, were observed. Purified GST-Bel fusion proteins were then obtained by affinity chromatography with glutathione Sepharose 4B (Fig. 2, lane 5).
FIG. 2.
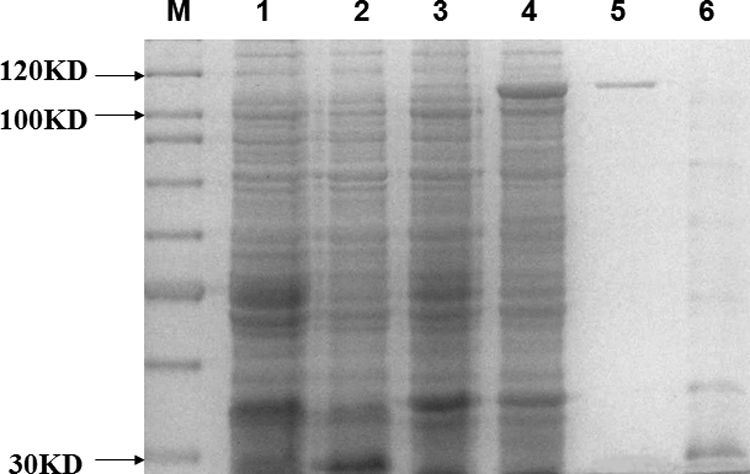
SDS-PAGE map of GST-Bel protein expression. M, protein marker SM0661 (Fermentas); lane 1, E.coli BL21(DE3) containing pGEX-6p-1 vector without isopropyl-β-d-thiogalactopyranoside (IPTG) induction, cell pellet; lane 2, BL21(DE3) containing pGEX-6p-1 vector with 0.8 mM IPTG induction, cell pellet; lane 3, E. coli BL21(DE3) containing pEMB0717 vector without IPTG induction, cell pellet; lane 4, E. coli BL21(DE3) containing pEMB0717 vector with 0.8 mM IPTG induction, cell pellet; lane 5, purified GST-Bel fusion protein eluted from the glutathione Sepharose bulk; lane 6, purified GST tag peptide eluted from the glutathione Sepharose bulk. The acrylamide percentage of the SDS-PAGE gels is 10%.
During insect bioassays, the enhancing effect depended on Cry1Ac, the Bel dosage, and the ratio of the Cry1Ac protein to the Bel protein. The Bel protein was mixed with the Cry1Ac protein at different ratios. Cry1Ac alone (3 μg/ml) was found to induce 34.2% insect mortality, while when the same amount of toxin was mixed with 0.1 μg/ml to 0.8 μg/ml of Bel, larval mortality reached 52.5% to 74.4%, respectively. This indicates that the mortality caused by the Cry1Ac protein plus the Bel protein was significantly different from that caused by the Cry1Ac protein alone; the mortality caused by the Cry1Ac protein was increased with an increase in the Bel protein concentration and finally reached saturation (Fig. 3A). On the other hand, at different Cry1Ac protein doses, the addition of 0.2 μg/ml Bel can increase Cry1Ac toxicity significantly. At the largest Cry1Ac protein doses (12.5 μg/ml), larval mortality reached 91.7%, which was increased about 15% over that with the control. At small Cry1Ac protein doses (3 μg/ml), larval mortality reached 65.3%, which was an increase of about 91% (Fig. 3B). Therefore, the Cry1Ac protein was more significantly virulent at a low concentration than at a high concentration. In controls, insecticidal activity was not observed when insects were fed GST or GST-Bel at the highest concentration used (from 10 to 50 μg/ml) alone. This means that Cry1Ac toxicity enhancement was due to a synergistic rather than direct effect of the Bel protein, so a significant enhancement of Cry1Ac toxicity to H. armigera larvae would be expected if the Bel protein is added.
FIG. 3.
(A) The Bel protein at different concentrations enhances Cry1Ac protein (3 μg/ml) toxicity toward H. armigera in diet incorporation method bioassays. The Cry1Ac protein was mixed with purified Bel at Bel:Cry1Ac ratios of 0:30, 1:30, 2:30, 3:30, 4:30, 6:30, and 8:30. (B) The Bel protein enhances Cry1Ac protein toxicity toward H. armigera in diet incorporation method bioassays. The Cry1Ac protein was mixed with purified Bel at Bel:Cry1Ac ratios of 1:0, 1:15, 1:30, and 1:60. Control treatments included the Cry1Ac protein at different concentrations. The figure indicates that the mortality with the Cry1Ac protein plus Bel protein treatment showed significant differences from that with the Cry1Ac treatment alone at the same Cry1Ac dosage.
Bel destroyed PM of H. armigera in vivo.
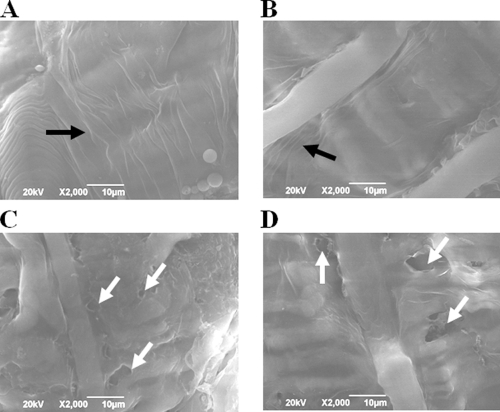
Viral enhancin could disrupt the protective PM, allowing virion access to the underlying epithelial cells of the insect gut (17), and target the IIM for degradation (33). Here, to understand the mode of action of the Bel protein during the Cry1Ac protein infection process, in vivo pathogenesis assays with fifth-instar H. armigera larvae treated with the Bel protein were conducted. The SEM observation showed that the PM of larvae fed an artificial diet plus Bel was extremely thin, to the point of complete degradation in some cases, and had a highly porous surface (Fig. 4C and D). These results demonstrated that the Bel protein can destroy the PM of H. armigera in vivo, similarly to the metalloprotease viral enhancin. In contrast, the PMs from larvae after 1.5 days of feeding on an artificial diet mixed with GST did not reveal any obvious differences in surface characteristics compared to the nontreatment groups (Fig. 4A and B).
FIG. 4.
Scanning electron micrographs of PM from Helicoverpa armigera larvae, revealing the influence of Bel on PM structure. (A) artificial diet fed; (B) artificial diet-plus-GST fed; (C and D) artificial diet-plus-Bel fed; black arrow points to the PM, white arrow points to the porous surface. The PM surface was visibly degraded by ingested Bel in artificial diet-fed larvae. SEM images were taken from the midguts of fifth-instar H. armigera per treatment.
IIM of H. armigera is target for Bel action.
The above data showed that Bel has an ability to attack the PM of H. armigera in vivo similar to that of viral enhancins, but the destruction pattern and target are not clear. Here, in vitro IIM degradation assays were conducted with purified Bel on H. armigera and T. ni. The phosphate-buffered saline buffer and T. ni enhancin protein (En-Tn) were used as negative and positive controls, respectively (Fig. 5). Bel protein exhibited degradation activity on PM in the midgut of T. ni (Fig. 5A) and the IIM of H. armigera (Fig. 5B), which is similar to the functions of the viral enhancin protein (En-Tn) (35). These results indicated that the Bel protein has the ability to degrade H. armigera IIM.
FIG. 5.
(A) In vitro fifth-instar Trichoplusia ni larval PM assay for degradation by Bel. The degradation of PM in the larval was analyzed by Western blot analysis using anti-IIM antiserum. Lane 1, T. ni PM treated with Bel; lane 2, T. ni PM treated with En-Tn; lane 3, T. ni PM. (B) In vitro HaIIM86 assay for degradation by Bel. The degradation of HaIIM86 was analyzed by Western blot analysis using anti-HaIIM86 antiserum. Lane 1, HaIIM86 treated by Bel; lane 2, HaIIM86 treated by En-Tn; lane 3, HaIIM86.
DISCUSSION
Previous studies indicated that B. cereus and B. thuringiensis enhancin-like proteins have no effects on the enhancement of virus infection and B. thuringiensis virulence (8, 12). However, in the present study, we found the Bel protein from B. thuringiensis significantly enhanced the effect of Cry1Ac toxin against H. armigera lepidopteran larvae. Discrepancies existing between the conclusion from our study and previous reports may be due to whether spore germination occurs in the intestine of insects or there is an amino acid sequence difference in Bel genes. When Hajaij-Ellouze et al. (12) performed bioassays with Galleria mellonella, spores of Bt-407 Cry− or mutant strains with the Cry1C crystal protein were used as toxins. However, some Cry− spores of B. thuringiensis were not able to germinate in the normal intestinal tract of insects (5). Perhaps bel may not have been expressed and therefore the Bel protein was not assessed in the experiment of Hajaij-Ellouze et al. Galloway et al. (8) found that purified bacterial enhancin-like protein did not show IIM degradation activity in vitro. There were differences between our protein and their protein, because the Bel protein used in this study was truncated N-terminally by 9 amino acids compared with the bacterial enhancin-like protein.
The biological activity of viral enhancin protein in terms of its ability to increase the efficacy of NPV and GV infection of insect larvae has been well documented (17, 33). Two mechanisms have been suggested for viral enhancins activity, namely, enhancement of virus-host midgut cell fusion and degradation of peritrophic matrix proteins by metalloprotease activity. In the present study, we found bel gene deletion reduced Cry1Ac toxicity to H. armigera larvae (Table. 4) while the purified Bel protein can function as a synergist of Cry1Ac toxicity against H. armigera (Fig. 3A and B). An explanation of the observed synergism is that the Bel protein can attack the PM of H. armigera to increase toxin permeativeness. In fact, significant perforation (Fig. 4C and D) was detected by SEM on the midgut PM of H. armigera larvae that were fed the Bel protein, increasing the ability of the crystal protein to gain access to the epithelial membrane, where its receptors are located (24), similar to the report of Regev et al. (29). Further, in vitro degradation assays also showed that the Bel protein can degrade IIM of T. ni and H. armigera larvae as did the viral enhancin protein (Fig. 5). These results indicated that the toxicity enhancement of the Bel protein is dependent on the destruction of midgut PM of insect larvae. This situation is similar to that of viral enhancin protein in terms of increasing the efficacy of NPV infection of insect larvae. The Bel protein can also degrade IIM of different lepidopteran species (Fig. 5A and B). It is interesting to note that Bel has a greater synergistic effect on Cry1Ac toxicity when the toxin concentrations are relatively low (Fig. 5B), suggesting that PM degradation by Bel is less important at higher Cry1Ac concentrations (greater than 10 μg/ml).
Furthermore, based on the phylogenetic analysis of all the enhancin proteins (Fig. 1B), the B. cereus group contains other closely related organisms: B. cereus, an opportunistic pathogen of humans; B. anthracis, a mammalian pathogen; and B. thuringiensis, an insect pathogen (8, 16, 28, 31). The presence of bel genes in the B. cereus group led to the suggestion that these organisms evolved from a common ancestor and the insect intestine may be the natural habitat for the ancestor. The relationship of the B. cereus gorup to insects was discussed in some previous reports. For example, Ivanova et al. (16), based on the genome sequence and the deduced metaboic analysis, showed that B. thuringiensis and other B. cereus group members have fewer genes for carbohydrate catabolism and more genes for amino acid metabolism than B. subtilis. These observations suggest that proteins, peptides, and amino acids may be a preferred nutrient source for the B. cereus group (16) and support the observations of Margulis et al. (23) that the insect intestine could have been the natural habitat for the common ancestor of the B. cereus group.
In the present article, we report that the Bel protein in B. thuringiensis can degrade the PM and IIM of different lepidopteran species (Fig. 5A and B), which is similar to the case with of viral enhancins. If the B. cereus group ancestor resided in the guts of insects and was this insect virus's host, bel genes in the B. cereus group and the viral enhancin gene originated with a common ancestor. The Y. pestis enhancin-like gene is flanked by a tRNA gene and transposase fragments, which may suggest that this bacterium obtained its enhancin-like gene via horizontal transfer (8). However, tRNA gene and transposase fragments like that of the Y. pestis enhancin-like gene do not flank bacterial bel genes in the B. cereus group. In any event, the origin of bacterial bel genes in the B. cereus group and whether bacterial bel genes appeared earlier than the viral enhancin gene require further study.
In summary, the present study reported that the Bel protein enhances Cry1Ac toxicity to H. armigera larvae by degrading the IIM. Our discovery has important implications related to the use of this protein to enhance insecticidal activity of B. thuringiensis toxin-based biopesticides and transgenic Bt crops.
Acknowledgments
We thank the Hubei Academy of Agricultural Sciences for providing the H. armigera larvae. We thank Ping Wang for discussions and suggestions concerning the research and for kindly providing the antibody.
This study was supported by grants from the National High Technology Research and Development project (863) of China (no. 2006AA02Z174 and 2006AA10A212), the National Basic Research Program (973) of China (no. 2009CB118902), and the Research Fund for the Doctoral Program of Higher Education of China (no. 20050504025).
Footnotes
Published ahead of print on 19 June 2009.
REFERENCES
- 1.Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115-119. [DOI] [PubMed] [Google Scholar]
- 2.Baum, J. A., T. B. Johnson, and B. C. Carlton. 1999. Bacillus thuringiensis natural and recombinant bioinsecticide products, p. 189-209. In F. R. Hall and J. J. Menn (ed.), Methods in biotechnology, vol. 5. Biopesticides: use and delivery. Humana Press, Totowa, NJ. [Google Scholar]
- 3.Bourgouin, C., A. Delecluse, and F. de la Terre. 1990. Transfer of the toxin protein genes of Bacillus sphaericus into Bacillus thuringiensis subsp. israelensis and their expression. Appl. Environ. Microbiol. 56:340-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Du, C., and K. W. Nickerson. 1996. Bacillus thuringiensis HD-73 spores have surface-localized Cry1Ac toxin: physiological and pathogenic consequences. Appl. Environ. Microbiol. 62:3722-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dykstra, M. J. 1992. Biological electron microscopy: theory, techniques and troubleshooting. Plenum Press, New York, NY.
- 7.Gallo, L. G., B. G. Corsaro, P. R. Hughes, and R. R. Granados. 1991. In vivo enhancement of baculovirus infection by the viral enhancing factor of a granulosis virus of the cabbage looper Trichoplusia ni (Lepidoptera Noctuidae). J. Invertebr. Pathol. 58:203-210. [Google Scholar]
- 8.Galloway, C. S., P. Wang, D. Winstanley, and I. M. Jones. 2005. Comparison of the bacterial enhancin-like proteins from Yersinia and Bacillus spp. with a baculovirus enhancin. J. Invertebr. Pathol. 90:134-137. [DOI] [PubMed] [Google Scholar]
- 9.Granados, R. R., Y. Fu, B. Corsaro, and P. R. Hughes. 2001. Enhancement of Bacillus thuringiensis toxicity to lepidopterous species with the enhancin from Trichoplusia ni granulovirus. Biol. Control 20:153-159. [Google Scholar]
- 10.Guérout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 11.Gunning, R. V., H. T. Dang, F. C. Kemp, I. C. Nicholson, and G. D. Moores. 2005. New resistance mechanism in Helicoverpa armigera threatens transgenic crops expressing Bacillus thuringiensis Cry1Ac toxin. Appl. Environ. Microbiol. 71:2558-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajaij-Ellouze, M., S. Fedhila, D. Lereclus, and C. Nielsen-LeRoux. 2006. The enhancin-like metalloprotease from the Bacillus cereus group is regulated by the pleiotropic transcriptional activator PlcR but is not essential for larvicidal activity. FEMS Microbiol. Lett. 260:9-16. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto, Y., B. G. Corsaro, and R. R. Granados. 1991. Location and nucleotide sequence of the gene encoding the viral enhancing factor of the Trichoplusia ni granulosis virus. J. Gen. Virol. 72:2645-2651. [DOI] [PubMed] [Google Scholar]
- 14.Hukuhara, T., and A. Wijonarko. 2001. Enhanced fusion of a nucleopolyhedrovirus with cultured cells by a virus enhancing factor from an entomopoxvirus. J. Invertebr. Pathol. 77:62-67. [DOI] [PubMed] [Google Scholar]
- 15.Hunter, E. E. 1993. Practical electron microscopy, 2nd ed. Cambridge University Press, New York, NY.
- 16.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 17.Lepore, L. S., P. R. Roelvink, and R. R. Granados. 1996. Enhancin, granulosis virus protein that facilitates nucleopolyhedrovirus (NPV) infections, is a metalloprotease. J. Invertebr. Pathol. 68:131-140. [DOI] [PubMed] [Google Scholar]
- 18.Li, L., C. Yang, Z. Liu, F. Li, and Z. N. Yu. 2000. Screening of acrystalliferous mutants from Bacillus thuringiensis and their transformation properties. Acta Microbiol. Sin. 5:395-399. [PubMed] [Google Scholar]
- 19.Li, Q., L. Li, K. Moore, C. Donly, D. A. Theilmann, and M. Erlandson. 2003. Characterization of Mamestra configurata nucleopolyhedrovirus enhancin and its functional analysis via expression in an Autographa californica M nucleopolyhedrovirus recombinant. J. Gen. Virol. 84:123-132. [DOI] [PubMed] [Google Scholar]
- 20.Li, X. Q., A. Tan, M. Voegtline, S. Bekele, C. S. Chen, and R. V. Aroian. 2008. Expression of Cry5B protein from Bacillus thuringiensis in plant roots confers resistance to root-knot nematode. Biol. Control 1:97-102. [Google Scholar]
- 21.Liu, X. J., S. F. Zhu, W. X. Ye, L. F. Ruan, Z. N. Yu, C. M. Zhao, and M. Sun. 2008. Genetic characterization of two putative toxin antitoxin systems on cryptic plasmids from Bacillus thuringiensis strain YBT-1520. J. Microbiol. Biotechnol. 18:1630-1633. [PubMed] [Google Scholar]
- 22.Luo, K., D. Banks, and M. J. Adang. 1999. Toxicity, binding, and permeability analyses of four Bacillus thuringiensis Cry1 delta-endotoxins using brush border membrane vesicles of Spodoptera exigua and Spodoptera frugiperda. Appl. Environ. Microbiol. 65:457-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margulis, L., J. Z. Jorgensen, S. Dolan, R. Kolchinsky, F. A. Rainey, and S. C. Lo. 1998. The Arthromitus stage of Bacillus cereus: intestinal symbionts of animals. Proc. Natl. Acad. Sci. USA 95:1236-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardo-López, L., C. Muñoz-Garay, H. Porta, C. Rodríguez-Almazán, M. Soberón, and A. Bravo. 2009. Strategies to improve the insecticidal activity of Cry toxins from Bacillus thuringiensis. Peptides 30:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeño-Tárraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 26.Plymale, R., M. J. Grove, D. Cox-Foster, N. Ostiguy, and K. Hoover. 2008. Plant-mediated alteration of the peritrophic matrix and baculovirus infection in lepidopteran larvae. J. Insect Physiol. 54:737-749. [DOI] [PubMed] [Google Scholar]
- 27.Popham, H. J., D. S. Bischoff, and J. M. Slavicek. 2001. Both Lymantria dispar nucleopolyhedrovirus enhancin genes contribute to viral potency. J. Virol. 75:8639-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlande, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 29.Regev, A., M. Keller, N. Strizhov, B. Sneh, E. Prudovsky, I. Chet, I. Ginzberg, Z. Koncz-Kalman, C. Koncz, J. Schell, and A. Zilberstein. 1996. Synergistic activity of a Bacillus thuringiensis delta-endotoxin and a bacterial endochitinase against Spodoptera littoralis larvae. Appl. Environ. Microbiol. 62:3581-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slavicek, J. M., and H. J. Popham. 2005. The Lymantria dispar nucleopolyhedrovirus enhancins are components of occlusion-derived virus. J. Virol. 79:10578-10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanada, Y., H. Inoue, R. T. Hess, and E. M. Omi. 1980. Site of action of a synergistic factor of a granulosis virus of the armyworm, Pseudaletia unipuncta. J. Invertebr. Pathol. 35:249-255. [Google Scholar]
- 33.Wang, P., and R. R. Granados. 1997. An intestinal mucin is the target for a baculovirus enhancin. Proc. Natl. Acad. Sci. USA 94:6977-6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, P., and R. R. Granados. 2003. Rapid and efficient isolation of highly specific antibodies from an antiserum against a pool of proteins. Biotech. Histochem. 78:201-205. [DOI] [PubMed] [Google Scholar]
- 35.Wang, P., D. A. Hammer, and R. R. Granados. 1994. Interaction of enhancin, a viral encoded protein, from the granulosis virus of Trichoplusia ni with the midgut epithelium and peritrophic membrane of four lepidopteran insects. J. Gen. Virol. 75:1961-1967. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto, T., and Y. Tanada. 1978. Phospholipid, an enhancing component in the synergistic factor of a granulosis virus of the armyworm, Pseudaletia unipuncta. J. Invertebr. Pathol. 31:48-56. [Google Scholar]
- 37.Yamamoto, T., and Y. Tanada. 1980. Physiochemical properties and location of capsule components, in particular the synergistic factor, in the occlusion body of a granulosis virus of the armyworm, Pseudaletia unipuncta. Virology 107:434-440. [DOI] [PubMed] [Google Scholar]
- 38.Zhao, C. M., C. X. Song, Y. Luo, Z. N. Yu, and M. Sun. 2008. L-2, 3-diaminopropionate: one of the building blocks for the biosynthesis of Zwittermicin A in Bacillus thuringiensis subsp. kurstaki strain YBT-1520. FEBS Lett. 582:3125-3131. [DOI] [PubMed] [Google Scholar]
- 39.Zhu, Y., T. Hukuhara, and K. Tamura. 1989. Location of a synergistic factor in the capsule of a granulosis virus of the armyworm, Pseudaletia unipuncta. J. Invertebr. Pathol. 54:49-56. [DOI] [PubMed] [Google Scholar]