Abstract
DNA extraction bias is a frequently cited but poorly understood limitation of molecular characterizations of environmental microbial communities. To assess the bias of a commonly used soil DNA extraction kit, we varied the cell lysis protocol and conducted multiple extractions on subsamples of clay, sand, and organic soils. DNA, as well as bacterial and fungal ribosomal gene copies as measured by quantitative PCR, continued to be isolated in successive extractions. When terminal restriction fragment length polymorphism was used, a significant shift in community composition due to extraction bias was detected for bacteria but not for fungi. Pyrosequencing indicated that the relative abundances of sequences from rarely cultivated groups such as Acidobacteria, Gemmatimonades, and Verrucomicrobia were higher in the first extraction than in the sixth but that the reverse was true for Proteobacteria and Actinobacteria. This suggests that the well-known phylum-level bacterial cultivation bias may be partially exaggerated by DNA extraction bias. We conclude that bias can be adequately reduced in many situations by pooling three successive extractions, and additional measures should be considered when divergent soil types are compared or when comprehensive community analysis is necessary.
The vast majority of soil bacteria (1, 7, 27) and fungi (13, 29) cannot be cultured via traditional laboratory techniques and must be identified using molecular methods. Successful characterization of microbial communities is therefore often dependent on DNA that is extracted from the environment. However, extraction of high-quality DNA from soil can be problematic (8, 11, 22, 26). Commercial DNA extraction kits are now commonly used in the assessment of taxonomic and functional diversity, community composition, and population abundance (e.g., references 19, 21, 23, 25, and 31). Studies comparing various kits (18, 32) or comparing commercial kits to other methods (2, 10, 24) have shown that DNA yield and purity vary depending on methodology and soil type. While these comparative studies are valuable, it is still unclear to what extent these protocols yield genomic DNA representative of the microbial community found within soil.
Our objective in this study was to optimize and assess the bias of a widely used commercial soil DNA extraction kit. We hypothesized that cell lysis would be enhanced and DNA would be removed from adsorption sites by conducting multiple extractions on a single sample, thereby increasing genomic DNA yield and obtaining a more complete survey of microbial taxa. This hypothesis was tested by (i) varying the extraction protocol and measuring DNA yield for three soils with differing characteristics and (ii) examining extraction bias in the genomic DNA obtained from successive extractions by using an improved method. Analytical replicates rather than biological replicates were used in order to focus strictly on variation and bias introduced through methodology, although multiple soil types were analyzed to determine whether biases detected were consistent.
Soil collection.
Soil was obtained from three forested locations with sterile sampling instruments and stored at −20°C. Soil properties are described in Table 1.
TABLE 1.
Soil properties
| Soil type | % Clay | % Sand | % Organic C | pH | Depth (cm) | Location | Coordinates |
|---|---|---|---|---|---|---|---|
| Clay | 40.6 | 0.0 | 6.0 | 6.7 | 10 | Toledo Metroparks, OH | 41°38′N, 83°26′W |
| Sanda | NDb | 72 | 4.4 | 3.9 | 10 | Manistee National Forest, MI | 44°9′N, 85°54′W |
| Organic | 5 | 70 | 10.9 | 5.9 | 2 | Kent State University, OH | 41°09′N, 81°20′W |
Data from Zak and Pregitzer (33).
ND, not determined.
DNA extraction and quantification.
Soil DNA was extracted using a PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA) in accordance with the manufacturer's instructions except as noted below. For each soil type, we extracted DNA from two analytical replicate samples (250 mg [fresh weight]). An initial extraction, followed by five successive extractions, was conducted on each replicate sample. A successive extraction involved adding new aliquots of bead solution (not including beads) and solution C1 to the soil pellet after initial lysis, centrifugation, and removal of supernatant containing crude DNA extract (i.e., after step 7 in the manufacturer's instructions). Lysis and centrifugation steps were then repeated, resulting in a new supernatant that was subsequently processed separately from previous supernatants. Four cell lysis protocols (treatments) were tested as alternatives to step 5 in the manufacturer's instructions (Table 2). All treatments involved incubating soil at 70°C and then bead beating; we also experimented with extended incubation and bead-beating times, freeze treatment at −80°C before incubation at 70°C, and the use of a GenoGrinder (SPEX CertiPrep, Metuchen, NJ) in place of a vortexer for bead beating. The volumes of unused supernatant at different steps were measured so that final soil DNA concentrations could be corrected for this loss.
TABLE 2.
Alternative cell lysis procedures (DNA extraction treatments)
| Treatment | Time (min)
|
|||
|---|---|---|---|---|
| Incubation before bead beating
|
Bead beating procedure
|
|||
| 70°C | −80°C | Vortexera | GenoGrinderb | |
| 1 | 10 | 5 | ||
| 2 | 20 | 10 | ||
| 3 | 10 | 5 | 5 | |
| 4 | 10 | 5 | 1 | |
Vortexer set at maximum speed.
GenoGrinder set at 1,500 strokes per min.
All subsequent assays were performed on extracts from both analytical replicate soil samples for each soil type. Extracted DNA concentration was quantified with Quant-iT PicoGreen fluorescent stain (Invitrogen, Carlsbad, CA) by using a Synergy 2 microplate reader (BioTek, Winooski, VT) in accordance with the manufacturer's instructions. All DNA extracts were diluted to 1.25 ng/μl for subsequent analyses.
Pulsed-field gel electrophoresis.
DNA shearing was assessed using a CHEF-DR III pulsed-field electrophoresis system (Bio-Rad, Hercules, CA). Samples were run in a 1.0% pulsed-field certified agarose gel (Bio-Rad) in 0.5× Tris-borate-EDTA buffer for 15 h at 6 V/cm and an angle of 120°, using a 1-s initial switch time and a 6-s final switch time. DNA fragment size was estimated by comparison to the Lambda ladder pulsed-field gel marker (1,018.5 to 48.5 kbp) and the Lambda DNA/PstI (11.5 to 2.4 kbp) markers (New England Biolabs, Ipswich, MA).
qPCR.
Copy numbers of bacterial and fungal small subunit ribosomal genes were quantified by quantitative PCR (qPCR) with an Mx3005P thermocycler (Stratagene, La Jolla, CA). The qPCR conditions included 0.2 ng genomic DNA/μl, 0.025 U/μl Taq DNA polymerase (GeneChoice, Frederick, MD), 3 mM MgCl2, 1× ammonium polymerase buffer, 0.16 mM each deoxynucleoside triphosphate (dNTP; New England BioLabs), 10 μM each primer (Integrated DNA Technologies, Coralville, IA), and 0.1 μg/μl bovine serum albumin. SYBR green I was added to give a final concentration of 0.167×. The primers used to amplify small subunit ribosomal fragments in the bacterial assay were Eub338F and Eub518R (12). In the fungal assay, the primers were FF390 and FR1 (28). After an initial denaturation (3 min at 95°C), the PCR program consisted of 40 cycles, including an 88°C step for fluorescence quantification (30 s at 94°C, 30 s at 57°C, 90 s at 72°C, and 33 s at 88°C), followed by a 7-min extension at 72°C and a melting curve analysis. Copy number was quantified by comparing the cycle at which fluorescence crossed a threshold to a standard curve constructed using a serial dilution of a plasmid containing an appropriate template. Assays were performed in duplicate for each sample, with standards and negative controls included in each run. Efficiency of amplification in each reaction was estimated using the method of Kontanis and Reed (15).
Community analysis by T-RFLP.
PCR for terminal restriction fragment length polymorphism (T-RFLP) was run in a DNA Engine Dyad cycler (Bio-Rad). Amplification of the bacterial 16S gene was conducted with the primers 1392R and Eub338F-0-III, the latter of which was labeled with 6-carboxyfluorescein (6). PCR was performed using 24 to 27 cycles under conditions identical to those described above for qPCR except that SYBR green I was not included and there was no 88°C step. Amplification of the fungal internal transcribed spacer region was conducted using primers NLB4R and NSI1F, the latter of which was labeled with 6-carboxyfluorescein (20). Fungal PCR was conducted using 0.02 to 0.12 ng of genomic DNA/μl, 0.03 U/μl Taq DNA polymerase, 2 mM MgCl2, 1× ammonium polymerase buffer, 0.2 mM each dNTP, and 0.5 μg/μl bovine serum albumin. The PCR program consisted of a 3-min initial denaturation at 95°C; 30 to 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 90 s; and a 7-min final extension at 72°C. Cycle number was varied for samples to obtain a strong band without nonspecific amplification. PCR products were run on a 1.5% agarose gel to confirm successful amplification. Triplicate PCRs were pooled prior to restriction digestion.
PCR product was digested overnight at 37°C with 10 units of HaeIII (New England Biolabs). Digests were purified with QIAquick nucleotide removal kits (Qiagen, Valencia, CA). Digested PCR products were sent to the Ohio State Plant Microbe Genomics Facility for fragment analysis by use of an Applied Biosystems 3730 DNA analyzer with a LIZ1200 size standard and minimum peak height of 50 fluorescence units. Peaks between 50 and 600 bp were included in the analysis if they represented >1% of the cumulative peak height for the sample.
Pyrosequencing.
Partial bacterial 16S rRNA gene sequences were obtained from each replicate of the first and the sixth extracts of sand and clay soils by using the coded-primer approach to multiplex pyrosequencing (3). PCR amplification of the hypervariable V4 region of the 16S rRNA gene was performed using 8-bp key-tagged eubacterial primers 563F and 802R (http://wildpigeon.cme.msu.edu/pyro/help.jsp). PCR mixtures contained 1 μM each primer (Integrated DNA Technologies), 1.8 mM MgCl2, 0.2 M dNTPs, 1.5× bovine serum albumin (New England Biolabs), 1 unit of FastStart high-fidelity PCR system enzyme blend (Roche Applied Science, Indianapolis, IN), and 10 ng of a DNA template. The PCR program consisted of a 3-min initial denaturation at 95°C; 30 cycles of 95°C for 45 s, 57°C for 45 s, and 72°C for 1 min; and a 4-min final extension at 72°C. For each sample, amplicons of three replicated PCRs were recovered using a QIAquick gel extraction kit followed by a QIAquick PCR purification kit (Qiagen). Equimolar amplicons were combined and submitted to pyrosequencing using a Genome Sequencer FLX system (454 Life Sciences, Branford, CT) at the Michigan State University Genomics Technology Support Facility. Sequences were excluded from analysis if the read length was less than 150 bp or if primer sequences contained errors. Raw sequences were processed through the Ribosomal Database Project (RDP) pyrosequencing pipeline (http://wildpigeon.cme.msu.edu/pyro/index.jsp). Qualified sequences were clustered into operational taxonomic units (OTUs) defined by 95% similarity using complete-linkage clustering and were assigned to phyla by the RDP-II classifier using a 50% confidence threshold (30). Sequences that could not be classified into a phylum at this level of confidence were excluded from subsequent phylum composition analyses.
Statistical analysis.
Number of ribosomal copies per ng DNA and weighted average fungal-to-bacterial-gene-copy ratio were analyzed by mixed-model analysis of variance using SAS Proc Mixed (SAS Institute, Cary, NC). Soil subsamples were designated as subjects, with individual extractions treated as repeated measurements. qPCR data were log transformed prior to analysis in order to stabilize variance. The effects of extraction step, soil type, and the interaction between the extraction step and the soil on the community composition (T-RFLP profile, phylum, and OTU composition) were analyzed by redundancy analysis with Canoco software (Microcomputer Power, Ithaca, NY). Relative abundances were square root transformed, resulting in analysis of Hellinger distances between samples (5, 17). Statistical significance was determined using 999 random permutations of sample identity.
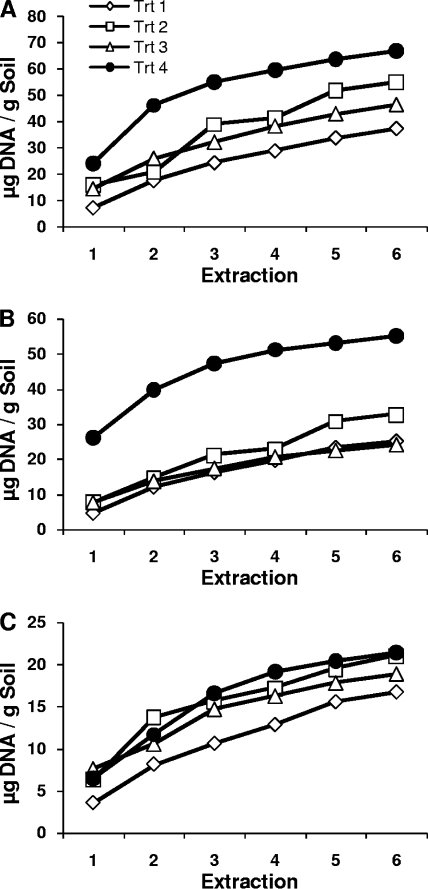
DNA yield and shearing from four DNA extraction methods.
Regardless of soil type or treatment, substantial quantities of DNA were isolated with successive extraction steps (Fig. 1). For organic and clay soils, treatment 4 (using a GenoGrinder and freeze-thawing) cumulative yields were higher than those obtained with all other treatments for all extraction steps. Treatment 4 cumulative yields also approached an asymptote more quickly than those obtained with other extraction treatments, with ∼80% of maximum DNA yield being reached in three extraction steps (Fig. 1). Liquid was entrained in the soil pellet after each extraction (22%, 26%, and 35% for clay, sand, and organic soils, respectively) and could therefore contribute DNA to the next extraction. However, DNA concentrations were consistently twice as high as would be expected due to carryover of DNA for clay and sand soils and 1.5 times as high for organic soils, indicating substantial amounts of DNA released from newly lysed cells at each extraction step. In addition, reduced yields were obtained when, after a single, extended lysis step, repeated “washing” of the soil pellet with extraction buffer was performed in place of heating or bead beating (data not shown).
FIG. 1.
Cumulative DNA yields in successive extractions conducted using four cell lysis treatments. (A) Organic soil; (B) clay soil; (C) sand soil.
These results are similar to those reported by Bürgmann et al. (9), who also performed multiple extractions on individual soil samples. In our case, however, the more vigorous procedure resulted in only slightly more shearing of genomic DNA visible on pulsed-field gel electrophoresis gels in first extracts, and less shearing in sixth extracts, than the vortexing procedure recommended by the manufacturer (all fragments between 5 and 50 kb; data not shown). Total extracted DNA has been used as an index of soil microbial biomass, but Leckie et al. (16) found no correlation between DNA extracted and biomass as measured by phospholipid fatty acid or chloroform fumigation extraction. Our results suggest that this lack of correlation may be due to different degrees of incomplete cell lysis in single DNA extractions in different soils.
More-detailed characterizations, described below, were performed on DNA extracts from treatment 4 because this treatment performed the best at quantitatively extracting DNA from soil.
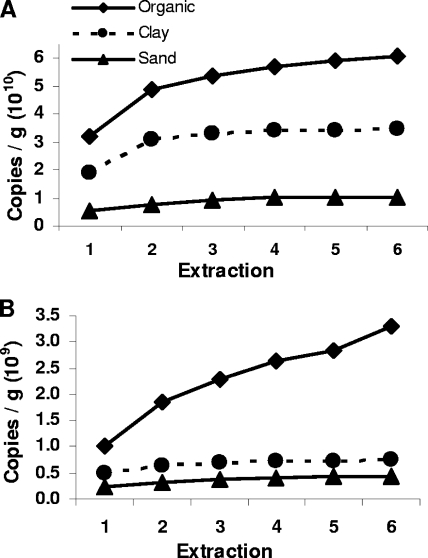
Bacterial DNA in successive extractions.
Similar to the case for DNA yield, numbers of cumulative bacterial ribosomal gene copies/g soil leveled off after one to three extraction steps (Fig. 2A). Bacterial qPCR efficiency was not significantly affected by soil type or extraction step (mean = 0.88). The only indication of potential damage to DNA in later extractions was a significant (P = 0.003) decline in number of bacterial ribosomal copies/ng genomic DNA (data not shown). However, these declines in number of ribosomal copies/ng genomic DNA could also result from a shift in the microbial community from which DNA was extracted, for example, toward organisms with fewer ribosomal gene copies.
FIG. 2.
Cumulative numbers of microbial gene copies/g soil in successive extractions. (A) Bacterial 16S gene; (B) fungal 18S gene.
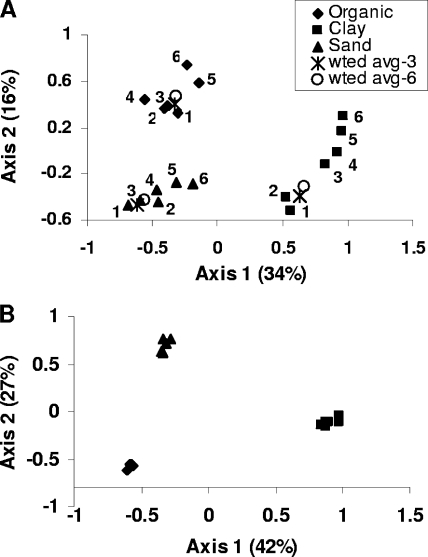
Analysis of T-RFLP profiles indicated that extraction of bacterial genomic DNA was biased, particularly in clay and sand soils. Extraction step explained a significant portion of bacterial community profile variability (P = 0.012; variance explained = 13%), with soil type also explaining a large portion (P = 0.01; variance explained = 47%). This is evident in the canonical ordination plot displaying soil type and extraction step effects (Fig. 3A), where gradients in clay and sand ordination scores are directly related to extraction step. The interaction between the soil and the extraction step was not significant for bacterial community profiles. When soils were analyzed individually, extraction step was significant in clay and sand soils (P < 0.05) but not significant in organic soil. The theoretical T-RFLP profile that would have resulted if DNA extractions were pooled prior to PCR is represented by the average T-RFLP profile weighted by the proportion of DNA obtained in each extract. Figure 3A shows that the weighted average profile of all six extractions lies between the profiles of extractions 1, 2, and 3 and is very similar to the weighted average profile of the first three extractions.
FIG. 3.
Canonical principal component plot of T-RFLP profiles derived from redundancy analysis. Each point represents the centroid of profiles from duplicate soil samples. Soil type and extraction are significant, as described in the text. (A) Bacterial profiles; (B) fungal profiles. “wted avg-3” is the weighted average of the first three T-RFLP profiles; “wted avg-6” is the weighted average of all six T-RFLP profiles.
To obtain a detailed taxonomic understanding of the bacterial community composition bias associated with DNA extraction, we performed pyrosequencing of 16S rRNA gene PCR amplicons from the first and sixth DNA extracts of duplicate subsamples of clay and sand soil. Soil type and extraction step affected the taxonomic composition of DNA extracts at the phylum and OTU levels. The interaction between the soil and the extraction step was significant only at the level of OTUs (Table 3). There was a greater level of unexplained variation in OTU composition than there was in phylum composition, primarily due to the greater amount of variation explained by the extraction step for phylum composition (Table 3). For both clay and sand soil, the phyla Acidobacteria, Gemmatimonades, Nitrospira, Verrucomicrobia, OD1, TM7, and WS3 were strongly affiliated with the first extraction, whereas Actinobacteria and Planctomycetes were affiliated with the sixth extraction (Table 4). Proteobacteria abundance was also higher in the sixth extraction, although this was not significant at the P level of 0.05. It is interesting to note that phyla representing two of the historically important cultivatable groups from soil, Actinobacteria and Proteobacteria (14), had much higher abundance in the sixth extraction than in the first (Table 4). In contrast, groups often detected using molecular methods but rarely cultivated from soil were approximately half as abundant in the sixth extraction as they were in the first (except Planctomycetes), suggesting that some measurements of cultivation bias in soil bacteria may be enhanced by a reciprocal DNA extraction bias. Between 1,531 and 3,106 sequences were obtained per sample, resulting in 547 to 996 OTUs per sample. The average sequence length was 207 bp.
TABLE 3.
Variance partitioning of bacterial sequence community compositiona
| Phylogenetic resolution | Soil % | Extraction % | Soil-extract interaction % |
|---|---|---|---|
| OTU | 35.9* | 21.1* | 13.1* |
| Phylum | 36.4* | 39.3* | NS |
*, P < 0.05; NS, not significant.
TABLE 4.
Response of bacterial phyla composition to extraction step revealed through redundancy analysisa
| Phylumb | Avg % abundancec
|
Cumulative fit (%)d | t | |
|---|---|---|---|---|
| Extract 1 | Extract 6 | |||
| Acidobacteria | 32.8 | 17.3 | 61.8 | 0.20 |
| Actinobacteria | 5.5 | 11.2 | 32.8 | −0.26 |
| Bacteroidetes | 1.0 | 0.8 | 12.1 | 1.20 |
| BRC1 | <0.1 | <0.1 | 16.6 | 1.34 |
| Chlamydia | 0.7 | 0.5 | 3.5 | 1.26 |
| Chloroflexi | 0.6 | 1.1 | 1.8 | −1.55 |
| Firmicutes | 1.1 | 1.0 | 14.5 | 1.13 |
| Fusobacteria | <0.1 | 0 | 14.3 | 1.25 |
| Gemmatimonades | 1.0 | 0.6 | 34.6 | 0.58 |
| Nitrospira | 0.1 | <0.1 | 44.5 | 0.51 |
| OD1 | 0.5 | 0.1 | 47.1 | 0.35 |
| OP10 | <0.1 | <0.1 | 9.8 | 1.77 |
| Planctomycetes | 1.8 | 10.0 | 80.9 | −0.07 |
| Proteobacteria | 20.8 | 31.9 | 23.9 | −0.88 |
| Spirochaetes | <0.1 | 0 | 14.3 | 1.25 |
| Thermomicrobia | 0 | <0.1 | 14.3 | −1.25 |
| TM7 | 0.1 | <0.1 | 70.7 | 0.32 |
| Verrucomicrobia | 17.2 | 9.8 | 21.9 | 0.55 |
| WS3 | 0.6 | 0.2 | 51.3 | 0.46 |
Bold type indicates phyla that have critical t values closer to 0 than the t value for the extraction step in this redundancy analysis (0.63). This indicates that the extraction step is a significant predictor for the abundance of bolded phyla.
Sequence classification into phyla was based on the RDP-II classifier at 50% confidence (30).
Relative abundances are averaged across sand and clay soil samples.
Percentage of variability in phylum abundance as explained by extraction step 1 versus extraction step 6.
Fungal DNA in successive extractions.
Results for fungi were quite different from those for bacteria. Cumulative numbers of fungal ribosomal gene copies/g soil reached an asymptote by three extractions in clay and sand soil but continued to increase in organic soil (Fig. 2B). Numbers of fungal gene copies/ng genomic DNA decreased with successive extractions in clay and sand soil but increased in organic soil (data not shown), indicating that there was a bias against extraction from fungi in the early extraction steps in this soil. This effect of organic soil is unlikely to be due to binding of released DNA to organic matter, because total DNA and bacterial ribosomal copy number/g soil both reached asymptotes for organic soil. We hypothesize that there is a greater degree of protection of fungi in organic soil due to growth within small organic particles that must be disrupted before the fungal cells are lysed. Alternatively, the fungal community in organic soil may be such that their cell walls are more difficult to disrupt than those of fungi in other soils, although there is no a priori reason to expect that this would be the case. Organic soil had significantly greater numbers of fungal gene copies/ng DNA than other soils (P < 0.0001); however, the extraction step and the interactions between the extraction step and the soil were not significant, primarily due to variability between organic soil replicates. Fungal qPCR efficiency was not significantly affected by soil type or extraction step (mean = 0.76).
No extraction bias in fungal community composition was detected using T-RFLP (Fig. 3B). Soil type had a significant effect on fungal T-RFLP profiles (P = 0.001; variance explained = 65%), whereas extraction step had no effect (P = 0.645) (Fig. 3B). The interactions between the soil and the extraction step were not significant for fungal community profiles, and the extraction step was also not significant for any soil analyzed individually.
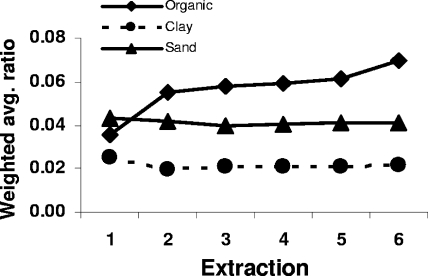
Fungus-to-bacterium ratio.
Despite the continued increase of fungal ribosomal copy numbers in organic soil, the weighted average ratio of fungal to bacterial ribosomal copies was quite stable in all soils after the second extraction, although it increased slightly for organic soil at extraction step 6 (Fig. 4). Due to differences between bacteria and fungi in PCR efficiency and ribosomal gene copy numbers per genome, we interpret the ratio of fungal to bacterial ribosomal genes as an index of fungus-to-bacterium ratio for comparative purposes rather than an indication of actual biomass ratio. Weighted average fungus-to-bacterium ratio was significantly affected by soil (P < 0.0001). The effects of the extraction step and the interaction between the soil and the extraction step were not significant.
FIG. 4.
Weighted average ratios of fungal to bacterial ribosomal gene copies. Significant differences are described in the text.
Conclusions.
We have found that substantial quantities of DNA are not extracted with a commonly used DNA extraction procedure. This results in biased estimates of DNA quantity, ribosomal copy number, and bacterial community composition. Although the composition of the sixth extraction represents a low overall proportion of the total DNA that can be extracted from a soil, this is the first suggestion that the reported severity of phylum-level cultivation bias (14) may be inflated by DNA extraction bias.
Because the majority of DNA is obtained within the first few extractions, this bias can be greatly reduced for some analyses by pooling three successive extractions. The weighted average T-RFLP profile of all six extractions was closely approximated by the weighted average of the first three extractions. Also, except in one case, cumulative ribosomal gene copy number did not increase substantially after three extractions. Comparisons of soils based on single DNA extractions may still be valid but should be recognized as representing an easily lysed portion of the community and possibly confounded by differences in extraction bias between mineral and organic soils. In addition, a complete characterization of the bacterial diversity or taxonomic makeup of a soil sample through sequencing (e.g., reference 23) would not be possible without analysis of additional DNA extractions. Diversity (e.g., OTU richness) present in a late, low-concentration extraction is not masked (or diluted out) by pooling with other DNA extracts in the same way as in community profiles (4), because counts and presence/absence are not weighted by abundance.
Nucleotide sequence accession numbers.
Sequences have been submitted to GenBank under accession numbers FJ240440 to FJ247115, FJ250290 to FJ261916, and GQ217543 to GQ219563.
Footnotes
Published ahead of print on 26 June 2009.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbeli, Z., and C. L. Fuentes. 2007. Improved purification and PCR amplification of DNA from environmental samples. FEMS Microbiol. Lett. 272:269-275. [DOI] [PubMed] [Google Scholar]
- 3.Binladen, J., M. T. Gilbert, J. P. Bollback, F. Panitz, C. Bendixen, R. Nielsen, and E. Willerslev. 2007. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS ONE 2:e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwood, C. B., D. Hudleston, D. R. Zak, and J. S. Buyer. 2007. Interpreting ecological diversity indices applied to T-RFLP data: insights from simulated microbial communities. Appl. Environ. Microbiol. 73:5276-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwood, C. B., T. Marsh, S.-H. Kim, and E. A. Paul. 2003. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 69:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackwood, C. B., A. Oaks, and J. S. Buyer. 2005. Phylum- and class-specific PCR primers for general microbial community analysis. Appl. Environ. Microbiol. 71:6193-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borneman, J., P. W. Skroch, K. M. O'Sullivan, J. A. Palus, N. G. Rumjanek, J. L. Jansen, J. Nienhuis, and E. W. Triplett. 1996. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl. Environ. Microbiol. 62:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braid, M. D., L. M. Daniels, and C. L. Kitts. 2003. Removal of PCR inhibitors from soil DNA by chemical flocculation. J. Microbiol. Methods 52:389-393. [DOI] [PubMed] [Google Scholar]
- 9.Bürgmann, H., M. Pesaro, F. Widmer, and J. Zeyer. 2001. A strategy for optimizing quality and quantity of DNA extracted from soil. J. Microbiol. Methods 45:7-20. [DOI] [PubMed] [Google Scholar]
- 10.Carrigg, C., O. Rice, S. Kavanagh, G. Collins, and V. O'Flaherty. 2007. DNA extraction method affects microbial community profiles from soils and sediment. Appl. Microbiol. Biotechnol. 77:955-964. [DOI] [PubMed] [Google Scholar]
- 11.de Lipthaya, J. R., C. Enzingerb, K. Johnsena, J. Aamanda, and S. J. Sørensen. 2004. Impact of DNA extraction method on bacterial community composition measured by denaturing gradient gel electrophoresis. Soil Biol. Biochem. 36:1607-1614. [Google Scholar]
- 12.Fierer, N., J. A. Jackson, R. Vilgalys, and R. B. Jackson. 2005. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71:4117-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawksworth, D. L., and A. Y. Rossman. 1997. Where are all the undescribed fungi? Phytopathology 87:888-891. [DOI] [PubMed] [Google Scholar]
- 14.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kontanis, E. J., and F. A. Reed. 2006. Evaluation of real-time PCR amplification efficiencies to detect PCR inhibitors. J. Forensic Sci. 51:795-804. [DOI] [PubMed] [Google Scholar]
- 16.Leckie, S. E., C. E. Prescott, S. J. Grayston, J. D. Neufeld, and W. W. Mohn. 2004. Comparison of chloroform fumigation-extraction, phospholipid fatty acid, and DNA methods to determine microbial biomass in forest humus. Soil Biol. Biochem. 36:529-532. [Google Scholar]
- 17.Legendre, P., and E. D. Gallagher. 2001. Ecologically meaningful transformations for ordination of species data. Oecologia 129:271-280. [DOI] [PubMed] [Google Scholar]
- 18.Lloyd-Jones, G., and D. W. F. Hunter. 2001. Comparison of rapid DNA extraction methods applied to contrasting New Zealand soils. Soil Biol. Biochem. 33:2053-2059. [Google Scholar]
- 19.Lord, N. S., C. W. Kaplan, P. Shank, C. L. Kitts, and S. Elrod. 2002. Assessment of fungal diversity using terminal restriction fragment (TRF) pattern analysis: comparison of 18S and ITS ribosomal regions. FEMS Microbiol. Ecol. 42:327-337. [DOI] [PubMed] [Google Scholar]
- 20.Martin, K. J., and P. T. Rygiewicz. 2005. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Brien, H. E., J. L. Parrent, J. A. Jackson, J. M. Moncalvo, and R. Vilgalys. 2005. Fungal community analysis by large-scale sequencing of environmental samples. Appl. Environ. Microbiol. 71:5544-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robe, P., R. Nalin, C. Capellano, T. M. Vogel, and P. Simonet. 2003. Extraction of DNA from soil. Eur. J. Soil Biol. 39:183-190. [Google Scholar]
- 23.Roesch, L. F. W., R. R. Fulthorpe, A. Riva, G. Casella, A. K. M. Hadwin, A. D. Kent, S. H. Daroub, F. A. O. Camargo, W. G. Farmerie, and E. W. Triplett. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roh, C., F. Villatte, B.-G. Kim, and R. D. Schmid. 2006. Comparative study of methods for extraction and purification of environmental DNA from soil and sludge samples. Appl. Biochem. Biotechnol. 134:97-112. [DOI] [PubMed] [Google Scholar]
- 25.Shanks, O. C., J. W. S. Domingo, R. Lamendella, C. A. Kelty, and J. E. Graham. 2006. Competitive metagenomic DNA hybridization identifies host-specific microbial genetic markers in cow fecal samples. Appl. Environ. Microbiol. 72:4054-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tebbe, C. C., and W. Vahjen. 1993. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl. Environ. Microbiol. 59:2657-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torsvik, V., and L. Øvreås. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5:240-245. [DOI] [PubMed] [Google Scholar]
- 28.Vainio, J., and J. Hantula. 2000. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol. Res. 104:927-936. [Google Scholar]
- 29.van Elsas, J. D., G. Frois-Duarte, A. Keijzer-Wolters, and E. Smit. 2000. Analysis of the dynamics of fungal communities in soil via fungal-specific PCR of soil DNA followed by denaturing gradient gel electrophoresis. J. Microbiol. Methods 43:133-151. [DOI] [PubMed] [Google Scholar]
- 30.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wawrik, B., L. Kerkhof, J. Kukor, and G. Zylstra. 2005. Effect of different carbon sources on community composition of bacterial enrichments from soil. Appl. Environ. Microbiol. 71:6776-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitehouse, C. A., and H. E. Hottel. 2007. Comparison of five commercial DNA extraction kits for the recovery of Francisella tularensis DNA from spiked soil samples. Mol. Cell. Probes 21:92-96. [DOI] [PubMed] [Google Scholar]
- 33.Zak, D. R., and K. S. Pregitzer. 1990. Spatial and temporal variability of nitrogen cycling in northern lower Michigan. Forest Sci. 36:367-380. [Google Scholar]