Abstract
We determined the prevalence of Escherichia coli O157:H7 in organically and naturally raised beef cattle at slaughter and compared antibiotic susceptibility profiles of the isolates to those of isolates from conventionally raised beef cattle. The prevalences of E. coli O157:H7 were 14.8 and 14.2% for organically and naturally raised cattle, respectively. No major difference in antibiotic susceptibility patterns among the isolates was observed.
Many cattle producers have adopted production methods termed niche marketing to meet consumer demand for safe and healthy beef. The two main niches for beef cattle producers are organic and natural production (3). Organic beef cattle production, regulated by the U.S. Department of Agriculture, requires feeding with certified organic feed (16) and raising cattle without the use of antibiotics, hormones, and other veterinary products (3). Guidelines for producers to label the product as “natural” differ among natural beef programs, and such programs are administered and regulated by the company or organization that owns the brand name rather than the U.S. Department of Agriculture (11). Natural production guidelines often include a complete restriction on the use of antibiotics and growth-promoting hormones, but unlike guidelines for organic production, they allow feed from nonorganic sources (11). Escherichia coli O157:H7 is a major food-borne pathogen that causes outbreaks of hemorrhagic enteritis, which often leads to hemolytic uremic syndrome in children and the elderly (10). Cattle are major reservoirs of E. coli O157:H7, which colonizes the hindgut, specifically the rectoanal mucosal region. Cattle feces are the major source of food and water contamination (10). The impact of organic production methods on the prevalence of food-borne pathogens, including E. coli O157:H7 and Campylobacter spp. in dairy cattle (7, 14) and Campylobacter and Salmonella spp. in chickens (6, 19), has been studied previously. However, there is no published study on the prevalence of E. coli O157:H7 in organically and naturally raised beef cattle. Additionally, nothing is known regarding the effects of organic and natural production methods on the antibiotic susceptibilities of E. coli O157:H7 in beef cattle. Our objectives were to determine the prevalence of E. coli O157:H7 in the feces of organically and naturally raised beef cattle at slaughter and compare the antibiotic susceptibilities of isolates from organically, naturally, and conventionally raised beef cattle.
Cattle included in this study were from three types of production systems, organic, natural, and conventional. Organically raised beef cattle were from farms that were certified by the National Organic Program (17). The naturally raised beef cattle were from farms that were certified by the All Natural Source Verified Beef Program (17). The collection of samples from these cattle occurred in an abattoir. Samples from conventionally raised cattle from two feedlots were collected in a different abattoir so that the antibiotic susceptibilities of their isolates could be compared with those of isolates from organically and naturally raised cattle. Fecal samples were obtained by cutting open the rectum and spooning out the contents. The mucosa of the rectum was then rinsed with water until free of visible fecal material and swabbed with a sterile foam-tipped applicator (4). The isolation and identification of E. coli O157 and PCR detection of major virulence genes (eae, stx1, stx2, hlyA, and fliC) were carried out as described by Reinstein et al. (13). A subset of 60 isolates, 20 (10 from fecal samples and 10 from rectoanal mucosal swabs [RAMS]) from each production system, was randomly chosen to determine the antibiotic susceptibility patterns by the broth microdilution method (9). The antibiotics (all from Sigma-Aldrich) tested were amikacin, amoxicillin (amoxicilline), ampicillin, apramycin, bacitracin, cefoxitin, ceftazidime, ceftriaxone, cephalothin (cefalotin), chloramphenicol, chlortetracycline, ciprofloxacin, enrofloxacin, erythromycin, florfenicol, gentamicin, kanamycin, lincomycin, monensin, nalidixic acid, neomycin, norfloxacin, novobiocin, oxytetracycline, penicillin, rifampin (rifampicin), spectinomycin, streptomycin, tetracycline, tilmicosin, trimethoprim, tylosin, and vancomycin. The MIC was defined as the lowest concentration of an antibiotic that prevented visible growth of the organism. Each concentration of the antibiotic compound was duplicated in the microtiter plate, and the MIC determination was repeated with a different inoculum preparation. Logistic regression was performed using the PROC GENMOD procedure in the SAS system (SAS Institute, Cary, NC) to compare the prevalences of E. coli O157:H7 (with binomial distribution of outcomes) in fecal samples, RAMS samples, and fecal or RAMS samples (overall animal level prevalence). The MICs of antibiotics for E. coli O157:H7 isolates were analyzed using a nonparametric survival test in the PROC LIFETEST program of SAS to determine the effects of the production system (natural, organic, or conventional). Data were right censored when necessary (when the organism was resistant to the highest concentration evaluated). The Wilcoxon test was utilized to determine the effect of the production system on MICs.
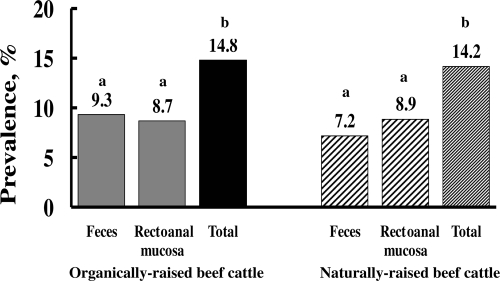
Samples from a total of 553, 506, and 322 organically, naturally, and conventionally raised cattle, respectively, were collected. In organically raised cattle, the prevalence of E. coli O157:H7 in fecal samples ranged from 0 to 24.4% across sampling days, with an average of 9.3%, and the prevalence in RAMS ranged from 0 to 30.9%, with an average of 8.7% (Fig. 1). In naturally raised cattle, the prevalence of E. coli O157:H7 in fecal samples ranged from 0 to 20.3%, with an average of 7.2%, and the prevalence in RAMS ranged from 0 to 23.8%, with an average of 8.9% (Fig. 1). In both organically and naturally raised cattle, the prevalence (total) detected by both sampling methods together was greater (P < 0.05) than the prevalence detected by either method alone (Fig. 1). Samples (either feces or RAMS) from 36 (11.2%) of 322 conventionally raised feedlot cattle were culture positive for E. coli O157:H7. The fecal prevalence of E. coli O157:H7 was 6.5%, and the prevalence determined by the RAMS sampling method was 7.1%. Most isolates (66.7% from organically raised beef cattle and 77.8% from naturally raised beef cattle) were positive for eae, stx2, hlyA, and fliC but negative for stx1. The stx2 gene was present in 100 and 95% of isolates from organically and naturally raised cattle, respectively. The prevalences of E. coli O157:H7 that we observed in organically and naturally raised beef cattle were similar to the previously reported prevalence in conventionally raised cattle (1). Our study did not include a statistical comparison of the prevalence data because of a number of differences, particularly in diet, among the organic, natural, and conventional production systems. Organically and naturally raised cattle are either required to graze a pasture or fed a forage-based diet. Although conflicting data exist (1), studies have shown that cattle fed a forage diet have both higher levels and longer durations of fecal shedding of E. coli O157:H7 than cattle fed a grain diet (18).
FIG. 1.
Prevalences of E. coli O157:H7 in organically and naturally raised beef cattle at slaughter. For each production system, bars not labeled with the same letter represent significantly different levels at P of <0.05.
None of the tested isolates from the three production systems were susceptible to bacitracin, lincomycin, monensin, novobiocin, tilmicosin, tylosin, and vancomycin (MICs > 50 μg/ml). The MICs of 12 antibiotics (amikacin, apramycin, cefoxitin, ceftriaxone, gentamicin, kanamycin, nalidixic acid, neomycin, penicillin, rifampin, streptomycin, and tetracycline) for isolates collected from different production systems were significantly different (P < 0.05). MICs of gentamicin and neomycin for E. coli O157:H7 isolates from conventionally raised cattle were higher (P < 0.05) than those for isolates from naturally and/or organically raised cattle (Table 1). However, MICs of amikacin, apramycin, cefoxitin, ceftriaxone, kanamycin, nalidixic acid, penicillin, rifampin, and tetracycline for isolates from conventionally fed cattle were lower (P < 0.05) than those for isolates from naturally and/or organically raised cattle (Table 1). Among the 60 isolates tested for antibiotic susceptibilities, 6 isolates (10%) were susceptible to all antibiotics included in the study, excluding the seven antibiotics to which all isolates were resistant. Forty-two isolates (70%) were resistant to one antibiotic (MIC, >50 μg or >50 IU/ml), nine isolates (15%) were resistant to two antibiotics, and two isolates (3%) were resistant to five antibiotics. One isolate from the organically raised cattle group was resistant to 10 (amoxicillin, ampicillin, cefoxitin, cephalothin, chloramphenicol, florfenicol, oxytetracycline, penicillin, streptomycin, and tetracycline) of the 26 antibiotics that were inhibitory to other isolates. We have presented the data as the median MICs for each production system. In some instances, the median values were the same but the actual MIC data differed between production systems. This effect occurred because the data were right censored if isolates were not susceptible at 50 μg or 50 IU/ml. If more isolates from a particular production system than from another are censored, it may lead to statistical differences. This pattern justifies the use of survival analysis for this type of data. There were differences between MICs of many antibiotics (cefoxitin, ceftriaxone, gentamicin, nalidixic acid, neomycin, penicillin, rifampin, and tetracycline) for isolates from organically raised cattle and conventionally raised cattle. Similarly, there were differences between MICs of many antibiotics (amikacin, apramycin, ceftriaxone, kanamycin, nalidixic acid, and rifampin) for isolates from naturally raised cattle and conventionally raised cattle. For many of these antibiotics, MICs for isolates from organically or naturally raised cattle were greater than those for isolates from conventionally raised cattle. Resistance genes can be transferred among the enteric pathogen populations in food animals and humans (8), and it is possible that resistance genes from other bacteria in the gastrointestinal system of cattle may be acquired by E. coli O157:H7. For cattle, heavy metals like copper and zinc, which are also antimicrobial, are included in diets at concentrations in excess of the nutritional requirements, often replacing conventional antibiotics, to achieve growth promotion (5). Feeding with metals also results in the emergence of bacterial populations resistant to metals (5), which in some instances may lead to resistance to antibiotics. Mechanisms of resistance to copper at concentrations above those usually tolerated by normal cellular processes have been found on plasmids linked to resistance to antibiotics in some bacteria (5). Therefore, it is possible that isolates from organically or naturally raised cattle that are not exposed to antibiotics still may become resistant to antibiotics.
TABLE 1.
MICs of antimicrobials for E. coli O157:H7 isolates from conventionally, naturally, and organically raised beef cattle
| Antibiotic agent | Median MICa (95% confidence interval) for isolates from:
|
P value (Wilcoxon test) | ||
|---|---|---|---|---|
| Conventionally raised cattle (n = 20) | Naturally raised cattle (n = 20) | Organically raised cattle (n = 20) | ||
| Amikacin | 2.5 (2.3-3.1)* | 3.9 (3.1-4.7)† | 2.7 (2.3-3.1)* | <0.01 |
| Apramycin | 9.4 (8.6-9.4)* | 12.5 (9.4-15.6)† | 6.3 (6.3-9.4)* | <0.01 |
| Cefoxitin | 7.8 (6.3-7.8)* | 7.8 (6.3-9.4)*† | 8.2 (7.8-10.9)† | 0.08 |
| Ceftriaxone | 0.04 (0.04-0.05)* | 0.05 (NE)† | 0.05 (NE)† | 0.02 |
| Gentamicin | 0.6 (0.4-0.6)† | 0.6 (0.5-0.8)† | 0.4 (0.3-0.5)* | <0.01 |
| Kanamycin | 3.0 (2.3-3.1)* | 3.9 (2.7-4.7)† | 2.3 (2.0-3.1)* | <0.01 |
| Nalidixic acid | 3.1 (3.1-3.9)* | 4.7 (3.9-6.3)† | 4.7 (3.1-6.3)† | <0.01 |
| Neomycin | 1.6 (1.2-1.6)† | 1.6 (1.2-2.3)† | 1.0 (0.8-1.2)* | <0.01 |
| Penicillin | 50.0 (NE)* | 50.0 (NE)*† | 50.0 (NE)† | 0.02 |
| Rifampin | 6.3 (5.5-6.3)* | 6.3 (NE)† | 6.3 (6.3-12.5)† | <0.01 |
| Streptomycin | 9.4 (9.4-12.5)*† | 9.4 (9.4-12.5)† | 7.8 (6.3-9.4)* | 0.04 |
| Tetracycline | 3.1 (NE)* | 3.1 (3.1-4.7)*† | 4.7 (3.1-4.7)† | 0.02 |
MICs of all antibiotics are expressed as micrograms per milliliter, except those of penicillin, which are in international units per milliliter. For each row, values not labeled with the same symbol (* or †) are significantly different (P < 0.05) as determined by survival analysis (Wilcoxon test). NE, not estimable.
Information on the prevalence and antibiotic susceptibilities of food-borne pathogens in organic or natural livestock production systems is limited and variable. In a study of organic and conventional dairy cattle farms, conventional farms were found to be more likely than organic farms to have at least one Salmonella isolate resistant to antibiotics (12). Kuhnert et al. (7) observed no difference between the prevalences of E. coli O157:H7 in samples from organic and conventional dairy farms. Sato et al. reported that E. coli isolates from conventional dairies had significantly higher rates of resistance to certain antibiotics than isolates from organic dairies (15). Cho et al. (2) compared the antibiotic susceptibilities of Shiga toxin-producing O157 and non-O157 isolates from organic and conventional dairy farms and concluded that there was no overall significant difference in resistance between isolates from the two production systems.
Although organic and natural beef production systems are becoming popular, little is known about the effects of these production systems on food-borne pathogens. Because the safety of the food supply is crucial, further investigation into these production systems and their potential for altering the risk of human illness is warranted. Our study found similar prevalences of E. coli O157:H7 in the feces of organically and naturally raised beef cattle, and our prevalence estimates for cattle in these types of production systems are similar to those reported previously for conventionally raised feedlot cattle.
Acknowledgments
We thank Neil Wallace, Kip Butler, Nathan Hoffman, and Meredith Lee for help in the laboratory and in sample collection.
Funding for this study was provided in part by a U.S. Department of Agriculture grant (2006-34359-06177).
Footnotes
Published ahead of print on 19 June 2009.
Contribution no. 08-294-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Callaway, T. R., R. O. Elder, J. E. Keen, R. C. Anderson, and D. J. Nisbet. 2003. Forage feeding to reduce preharvest Escherichia coli populations in cattle: a review. J. Dairy Sci. 86:852-860. [DOI] [PubMed] [Google Scholar]
- 2.Cho, S., C. P. Fossler, F. Diez-Gonzales, S. J. Wells, C. W. Hedberg, J. B. Kaneene, P. L. Ruegg, L. D. Warnick, and J. B. Bender. 2008. Antimicrobial susceptibility of Shiga toxin-producing Escherichia coli isolated from organic dairy farms, conventional dairy farms, and county fairs in Minnesota. Foodborne Pathog. Dis. 4:178-186. [DOI] [PubMed] [Google Scholar]
- 3.Fox, J. T., S. Reinstein, M. E. Jacob, and T. G. Nagaraja. 2008. Niche marketing production practices for beef cattle in the United States and prevalence of foodborne pathogens. Foodborne Pathog. Dis. 5:1-11. [DOI] [PubMed] [Google Scholar]
- 4.Fox, J. T., X. Shi, and T. G. Nagaraja. 2008. Escherichia coli O157 in the rectoanal mucosal region of cattle. Foodborne Pathog. Dis. 5:69-77. [DOI] [PubMed] [Google Scholar]
- 5.Hasman, H., S. Franke, and C. Rensing. 2006. Resistance to metals used in agriculture production, p. 99-114. In F. Aarestrup (ed.), Antimicrobial resistance in bacteria of animal origin. ASM Press, Washington, DC.
- 6.Heuer, O. E., K. Pedersen, J. S. Andersen, and M. Madsen. 2001. Prevalence and antimicrobial susceptibility of thermophilic Campylobacter in organic and conventional broiler flocks. Lett. Appl. Microbiol. 33:269-274. [DOI] [PubMed] [Google Scholar]
- 7.Kuhnert, P., C. R. Dubosson, M. Roesch, E. Homfeld, M. G. Doherr, and J. W. Blum. 2005. Prevalence and risk-factor analysis of Shiga toxigenic Escherichia coli in faecal samples of organically and conventionally farmed dairy cattle. Vet. Microbiol. 109:37-45. [DOI] [PubMed] [Google Scholar]
- 8.Mathew, A., R. Cissell, and S. Liamthong. 2007. Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Foodborne Pathog. Dis. 4:115-133. [DOI] [PubMed] [Google Scholar]
- 9.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial susceptibility testing. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 10.Rangel, J. M., P. H. Sparling, C. Crowe, P. M. Griffin, and D. L. Swerdlow. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 11:603-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rawls, E., L. Meyer, and K. Burdine. 10 January 2008, accession date. Managing for today's cattle market and beyond. Niche marketing of cattle/beef. Department of Agricultural Economics, Kansas State University, Manhattan. http://www.agecon.ksu.edu/livestock/Master%20Web%20Page%20Folder/Extension%20Bullentins.Research/ManageForTodaysCattleMkt/Niche_Marketing.pdf.
- 12.Ray, K. A., L. D. Warnick, R. M. Mitchell, J. B. Kaneene, P. L. Ruegg, S. J. Wells, C. P. Fossler, L. W. Halbert, and K. May. 2006. Antimicrobial susceptibility of Salmonella from organic and conventional dairy farms. J. Dairy Sci. 89:2038-2050. [DOI] [PubMed] [Google Scholar]
- 13.Reinstein, S., J. T. Fox, X. Shi, and T. G. Nagaraja. 2007. Prevalence of Escherichia coli O157 in gall bladders of beef cattle. Appl. Environ. Microbiol. 73:1002-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato, K., P. C. Bartlett, J. B. Kaneene, and F. P. Downes. 2004. Comparison of prevalence and antimicrobial susceptibilities of Campylobacter spp. isolates from organic and conventional dairy herds in Wisconsin. Appl. Environ. Microbiol. 70:1442-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato, K., P. C. Bartlett, and M. A. Saeed. 2005. Antimicrobial susceptibility of Escherichia coli isolates from dairy farms using organic versus conventional production methods. J. Am. Vet. Med. Assoc. 226:589-594. [DOI] [PubMed] [Google Scholar]
- 16.Troxel, T. R. 10 January 2007, accession date. Natural and organic beef. FSA3103.University of Arkansas Cooperative Extension Service, Little Rock. http://www.uaex.edu/Other_Areas/publications/PDF/FSA-3103.pdf.
- 17.USDA Agricultural Marketing Service. 10 January 2008, accession date. National organic program. USDA Agricultural Marketing Service, Washington, DC. http://www.ams.usda.gov/AMSv1.0/getfile?dDocName=STELDEV3004443&acct=nopgeninfo.
- 18.Van Baale, M. J., J. M. Sargeant, D. P. Gnad, B. M. DeBey, K. F. Lechtenberg, and T. G. Nagaraja. 2004. Effect of forage or grain diets with or without monensin on ruminal persistence and fecal Escherichia coli O157:H7 in cattle. Appl. Environ. Microbiol. 70:5336-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Overbreke, I., L. Duchateau, L. De Zutter, G. Albers, and R. Ducatelle. 2006. A comparison survey of organic and conventional broiler chickens for infectious agents affecting health and food safety. Avian Dis. 50:196-200. [DOI] [PubMed] [Google Scholar]