Abstract
Dead-end ultrafiltration (DEUF) is an alternative approach to tangential-flow hollow-fiber ultrafiltration that can be readily employed under field conditions to recover microbes from water. The hydraulics of DEUF and microbe recovery for a new DEUF method were investigated using 100-liter tap water samples. Pressure, flow rate, and temperature were investigated using four hollow-fiber ultrafilter types. Based on hydraulic performance, the Asahi Kasei REXEED 25S ultrafilter was selected for microbe recovery experiments. Microbe recovery experiments were performed using MS2 bacteriophage, Enterococcus faecalis, Clostridium perfringens spores, and Cryptosporidium parvum oocysts. Microbes were recovered from ultrafilters by backflushing using a surfactant solution. Average flow rates were 2.1 liters/min for 100-liter water samples having turbidities of 0.28 to 4.3 nephelometric turbidity units (NTU), and no evidence of appreciable filter clogging was observed. The DEUF average recovery efficiencies for each study analyte in tap water were as follows: for E. faecalis, 93% ± 16%; for MS2, 57% ± 7.7%; for C. perfringens spores, 94% ± 22%; and for C. parvum, 87% ± 18%. Average microbe recoveries for tap water amended with surface water (average turbidity = 4.3 NTU) were as follows: for E. faecalis, 78% ± 12%; for MS2, 73% ± 13%; for C. perfringens, 57% ± 21%; and for C. parvum, 83% ± 21%. These data demonstrate that DEUF is an effective method for recovering diverse microbes from water and should be a useful tool for field-based environmental investigations.
There are an estimated 4 million to 33 million cases of acute gastrointestinal illness each year in the United States due to drinking water (3, 11). From 2005 to 2006, 20 reports of waterborne disease and outbreaks (WBDOs) associated with drinking water were submitted to the national Waterborne Disease and Outbreak Surveillance System (19). In addition, a record number (78 reports) of WBDOs associated with recreational water were also submitted to the Waterborne Disease and Outbreak Surveillance System in 2005 and 2006 (20). Detecting the etiologic agents for WBDOs is challenging due to such factors as the time delay between case exposure and water sampling, microbial die-off, and water dilution or treatment prior to sampling. Because it is likely that pathogens will be present at low concentrations in water sampled for outbreak investigations, relatively large volumes of water (e.g., 40 to 100 liters) should be collected. In addition, sampling water for a diverse array of microbes is sometimes a goal when multiple etiologic agents are suspected for a WBDO (13) or during emergency responses when the contaminant has not been identified.
Hollow-fiber ultrafiltration (UF) has been shown to be an effective technique for collecting large-volume water samples for recovery of diverse microbes, including viruses, vegetative bacteria, bacterial spores, and parasites (5, 6, 10, 12, 14). However, most hollow-fiber UF techniques utilize a tangential-flow approach that requires comprehensive operator training and which is generally not conducive to rapid-response implementation for field sampling. While the tangential-flow (i.e., recirculating flow) UF technique has been shown to be effective for microbe recovery, it is a more complicated sampling technique than traditional direct-filtration techniques, such as membrane filtration for coliforms (1), microfiltration for Cryptosporidium and Giardia (oo)cysts (18), and adsorption-elution microfiltration for viruses (4). For emergency response, outbreak investigations, or other field investigations performed by personnel with limited training in water sample collection, a dead-end UF (DEUF) technique would be useful for capturing and recovering multiple microbe classes.
Relatively few studies have reported using hollow-fiber UF in a DEUF configuration. Kearns et al. (7) reported using an automated DEUF system to recover Bacillus atrophaeus spores from tap water, with reported recovery efficiencies of 23 to 40% in ∼100-liter samples with low-level seeding (330 to 1,000 CFU). These researchers performed filter backflushing using a phosphate buffer containing either Tween 20 or sodium polyphosphate. Kearns et al. also reported suspected ultrafilter fouling based on measured reductions in flow rates for ∼100-liter samples (7). The Kearns et al. observations indicate that the ultrafilter pore size and filter area are important hydraulic performance variables for the DEUF technique. Leskinen and Lim (9) reported using hollow-fiber DEUF for recovery of enterococci from 100-liter samples of beach water. These researchers used a urea-lysine solution to elute (instead of backflush) enterococci from ultrafilters. Leskinen and Lim reported a wide range of recovery efficiencies (4 to 708%; average = 251%) for their DEUF method but did not discuss whether water quality (other than a potential variability in microbe distribution in the 100-liter samples) could have contributed to ultrafilter fouling or variable method performance (9).
The present study was designed to investigate DEUF using different commercially available hollow-fiber ultrafilters having a range of pore sizes and filter medium sizes. The parameters tested included the effect of the water sample flow rate and temperature on system pressure and the effect of turbidity on the permeate flow rate and microbial recovery efficiencies. A suite of four distinctly different microbes (MS2 bacteriophage, Enterococcus faecalis, Clostridium perfringens spores, and Cryptosporidium parvum oocysts) was studied to determine the performance of the DEUF method for simultaneous recovery of diverse microbes.
MATERIALS AND METHODS
Water samples.
Experiments were performed using tap water and tap water amended with surface water to achieve target turbidity levels of <0.50 nephelometric turbidity units (NTU) (low-range turbidity), 1.0 NTU (midrange turbidity), and 4.0 NTU (high-range turbidity) relative to typical drinking-water turbidity levels. Tap water samples were collected from a faucet in the laboratory at the Centers for Disease Control and Prevention (CDC) in Atlanta, GA. Surface-water samples were collected from Murphey Candler Park (MCP) lake in Atlanta, GA. Investigators collected MCP lake water once a week by pumping the water with a peristaltic pump into plastic cubitainers and then transporting the samples back to the laboratory, where they were stored at 5°C when not being used for experiments. Tap water samples were collected by opening the cold-water faucet in the lab for at least 5 min prior to filling a 30-gallon high-density polyethylene tank that had been previously calibrated to 100 liters using 10-liter gradations. At the beginning of each experiment, the 30-gallon tank was filled with water to the 100-liter mark, which was mixed with a metal stir rod, and a 100-ml sample was collected for water quality testing. Water quality testing consisted of analyses for turbidity, temperature, and total organic carbon (TOC). The temperature of the in-tank bulk water samples was measured using an infrared/type K noncontact thermometer (catalog no. 15-077-57; Fisher Scientific). Turbidity was measured with a model 2100N laboratory turbidimeter (Hach Company). TOC was measured using a low-range TOC reagent set and a DR/2400 portable spectrophotometer (Hach Company).
Lake water was volumetrically added to tap water based on the turbidity of the lake water sample to achieve target mid- and high-range turbidity levels. Final mid- and high-range-turbidity sample volumes were ∼102 liters. The sample water was dechlorinated with 50 ml of 10% sodium thiosulfate and then tested using N,N-diethyl-p-phenylenediamine-free chlorine reagents (Hach Company) to ensure no residual chlorine was present.
Dead-end UF setup.
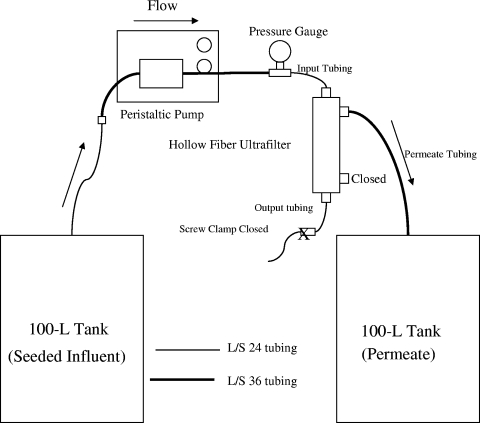
The DEUF setup is shown in Fig. 1. Each 30-gallon tank was sterilized before each experiment with a 1:10 dilution of bleach followed by a 3% solution of hydrogen peroxide and then rinsed three times with deionized (DI) water and sprayed with a 1% sodium thiosulfate solution. The tank was then rinsed two more times with DI water. Contact time with each disinfectant was at least 30 min. A Cole-Parmer Masterflex L/S peristaltic pump (model 7550-30) and pump heads were used with L/S 36 and L/S 24 silicone tubing. New tubing was used for each of the microbe-seeding experiments. The ultrafilters were set up with the input port on the top, connected with L/S 24 tubing to a 30-lb/in2 liquid-filled pressure gauge and clamped with plastic tubing clamps to prevent leakage. The pressure gauge was then fastened with L/S 36 tubing, which extended into the pump head. The L/S 36 tubing was used so that a maximum pump rate of 2,900 ml/min could be achieved, if desired. After extension out of the pump head, the L/S 36 tubing was connected to L/S 24 tubing using a polypropylene reducing connector and the L/S 24 tubing connected to the ultrafilter using an autoclaveable DIN adapter (a female fitting with threading for dialyzers on one end and a male barb on the other end) (special order from Molded Products, Inc., Harlan, IA; similar to the item under catalog no. MPC-855). Filtered water exited the ultrafilter through the permeate port (top-side port in Fig. 1) and was collected in a second 30-gallon “permeate” tank. For these DEUF experiments, the bottom output port was attached to a small piece of L/S 24 tubing to avoid leakage from the manufacturer's cap, and the tubing was clamped shut with a tubing clamp. All tubing connectors and clamps were autoclaved, and the brass fitting of the pressure gauge was sanitized with a 3% solution of hydrogen peroxide and a 1:10 dilution of bleach and then washed thoroughly with DI water prior to use in the filtration setup.
FIG. 1.
Schematic of the dead-end UF setup for filtering water samples.
Hydraulic performance of DEUF method with different ultrafilters.
Using nonamended tap water, the DEUF method was investigated with four ultrafilter types to evaluate the hydraulic characteristics (permeate flow rate versus pressure) of each filter type. Pressure changes were monitored versus different flow rates at two water temperature levels (room/tap temperature and ∼5°C). Permeate flow rates were measured using a 2-liter graduated cylinder and a stopwatch. The test ultrafilters included the following: Fresenius Optiflux F200NR (polysulfone, 2.0-m2 filter area, ∼30-kDa pore size), Baxter Exeltra Plus 210 (cellulose triacetate, 2.1-m2 filter area, 70-kDa pore size), Asahi Kasei REXEED 21S (polysulfone, 2.1-m2 filter area, ∼30-kDa pore size), and Asahi Kasei REXEED 25S (polysulfone, 2.5-m2 filter area, ∼30-kDa pore size). The pump speed was changed (and corresponding permeate flow rate and pressure recorded) every 5 min. Pump speeds were set at nominal flow rates of 500, 700, 900, 1,100, 1,300, 1,500, 1,700, 1,900, and 2,100 ml/min. Three replicate experiments were performed for each ultrafilter.
Additional experiments were performed with REXEED 25S filters to investigate the relationship between hydraulic performance (system pressure and permeate flow rate) for this filter and turbidity. One-hundred-liter samples of tap water (0.26 NTU, low-range turbidity) and tap water amended with surface water (to achieve a midrange target turbidity level of 1.0 NTU) were used. The system pressure and the permeate flow rate were measured every 5 min. Three replicate experiments were performed for each turbidity range.
Microbe recovery with DEUF and backflushing.
The recovery efficiency for a suite of microbes was assessed using the REXEED 25S filter and low-range-, midrange-, and high-range-turbidity water samples. After the filtration apparatus was set up, the filter was flushed with ∼1 liter of nonamended tap water to flush out the storage liquid that the vendor uses to fill REXEED 25S ultrafilters. After the sample of amended tap water was prepared for each experiment in the 30-gallon tank, the following samples were collected from the tank: a 100-ml water quality sample, a 2-liter “background” sample, and a 100-ml control sample. The background sample was tested to quantify study microbes that were present in the water sample prior to microbe seeding for an experiment. The control sample was subsequently seeded with microbes to quantify the number of microbes seeded into the 100-liter DEUF microbe recovery experiment.
The suite of microbes used for microbe recovery experiments consisted of C. perfringens spores (10,000 CFU BioBalls; BTF Pty. Ltd.), MS2 bacteriophage (ATCC 15597-B1), E. faecalis (ATCC 19433), and Iowa strain C. parvum oocysts (from Mike Arrowood, CDC). MS2 and E. faecalis were diluted in a diluent containing 0.01 M phosphate-buffered saline (Dulbecco's modification; pH 7.40), 0.01% (vol/vol) Tween 80 (Fisher), and 0.001% (vol/vol) Y-30 antifoam emulsion (Sigma). The C. perfringens spore Bioballs were added to 10 ml diluent (same as above) and placed in a Pall laboratory shaker for 30 min at 960 oscillations/min to suspend and disaggregate the spores. The C. parvum oocysts were diluted in DI water and heat inactivated (30 min in a heat block at 50°C). MS2 bacteriophage, E. faecalis, and C. perfringens spores were prefiltered using 0.1-μm, 5-μm, and 3-μm filters, respectively, to reduce the presence of microbe aggregates in the seed stocks added to water samples. The suite of microbes was then seeded into both the ∼100-liter input water sample and the 100-ml control sample. MS2 bacteriophage was seeded at 3.9 × 104 to 1.0 × 105 PFU, E. faecalis was seeded at 900 to 1,000 CFU, C. perfringens spores were seeded at 1,500 to 3,760 CFU, and C. parvum oocysts were seeded at 9 × 105 to 1.4 × 106 oocysts. The nominal filtration pump speed (2,100 ml/min) was constant throughout each experiment, and the permeate flow rate, system pressure, and temperature were monitored. At the 40- and 80-liter cumulative sample filtration time points, a permeate sample was collected and tested to evaluate filter integrity (i.e., microbial breakthrough).
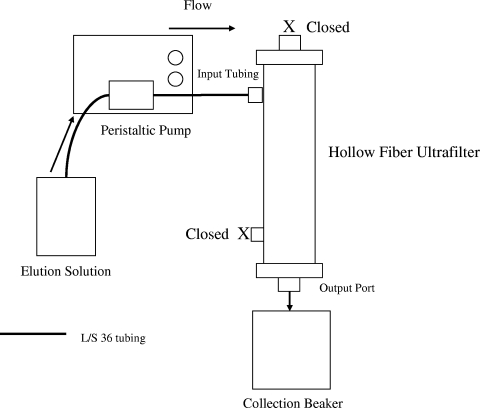
After the seeded sample was filtered, the filtration setup was adjusted for backflushing (Fig. 2). The input tubing was replaced with a filter port cap. The permeate port tubing was replaced by a clean piece of L/S 36 tubing that was used as the new input tubing (threaded through the pump head), and the output port tubing was removed. While the output tubing was being carefully unscrewed, a beaker was placed under the port so that there was no liquid loss from the filter. Five hundred milliliters of backflush solution (0.5% Tween 80, 0.01% sodium polyphosphate [Sigma-Aldrich, catalog no. 305553], and 0.001% Y-30 antifoam emulsion) was pumped through the permeate port and collected in a beaker positioned under the output port (Fig. 2). The pump rate was set at a constant 650 ml/min. The volume of the recovered backflush sample was measured with a graduated cylinder, and all of the samples were stored at 5°C until assaying or secondary processing took place. Final backflush sample volumes were an average of 533 ml.
FIG. 2.
Schematic of the setup for backflushing hollow-fiber ultrafilters.
Secondary processing of DEUF backflush samples was completed the same day as the DEUF procedure. The DEUF backflush sample and control were assayed by immunofluorescence assay microscopy per EPA method 1623 for C. parvum (18). The DEUF backflush, control, background, and permeate samples were analyzed for C. perfringens spores, E. faecalis, and MS2. MS2 was assayed by single-agar-layer plaque assay using EPA method 1602 with an E. coli Famp host (ATCC C-3000) (17) using 1 ml of control sample, 2 ml of UF concentrate, and 100 ml of permeate. E. faecalis was assayed by membrane filtration and mEI agar culture using EPA method 1600 (16) using 5 ml of control sample, 50 ml of UF concentrate, and 1.5 liters of permeate. C. perfringens was assayed by membrane filtration and mCP agar culture (2) using 10 ml of control sample, 50 ml of UF concentrate, and 1.5 liters of permeate.
Data analysis and statistical testing.
Percent recovery efficiencies were calculated by dividing the total number of each microbe measured in a backflush sample by the total number of each microbe measured in the input sample (microbes seeded plus microbes present as “background”) for the experiment and multiplying the fraction by 100. Tukey's studentized range (HSD) test was used to test for significant differences between mean recovery efficiencies for each microbe at each turbidity level (i.e., low-range, midrange, and high-range). An alpha value of 0.05 was used for all statistical tests.
RESULTS
Flow rate-versus-temperature experiments.
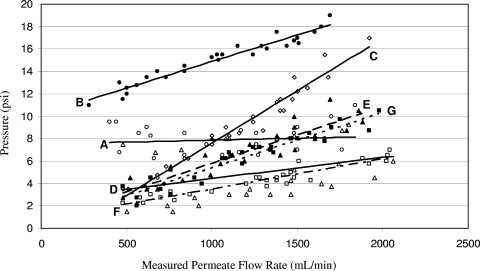
Experiments were performed to test the hydraulic characteristics of four ultrafilters (Fresenius Optiflux F200NR, Baxter Exeltra Plus 210, REXEED 21S, and REXEED 25S) at different nominal flow rates and two water temperatures. The average water temperatures and turbidities for these experiments, representing moderate and cold water conditions, were 18°C and 0.17 NTU and 4.8°C and 0.33 NTU, respectively (Table 1). For the 18°C experiments, the F200NR filters had the highest system pressures when permeate flow rates were above approximately 1,000 ml/min. F200NR filters also exhibited the highest rate of pressure increase versus permeate flow rate. REXEED 25S filters (which had the largest filter medium area of the four filters investigated) had the lowest overall pressures, ranging from 2 to 6 lb/in2 when water was approximately 18°C (Fig. 3). When the DEUF procedure was performed using cold water (averaging 4.8°C), more force (measured as pressure) was needed to achieve the same permeate flow rates achieved with warmer (18°C) water (Fig. 3). While Exeltra Plus 210 filters exhibited little pressure increase versus permeate flow rate for 18°C water (∼1.0- to 1.5-lb/in2 increase between flow rates of 440 and 1,800 ml/min), for 4.8°C, water pressures increased from approximately 12 lb/in2 at permeate flow rates of 500 ml/min to approximately 18 lb/in2 when permeate flow rates were 1,600 ml/min. The REXEED 21S and 25S filters generally exhibited the lowest pressures of the four ultrafilter types studied. As anticipated, the REXEED 25S filters required less pressure to achieve the same permeate flow rates as the REXEED 21S, likely because the REXEED 25S had 2.5 m2 of filter medium versus 2.1 m2 for the REXEED 21S filters.
TABLE 1.
Water quality data for the DEUF studya
| Expt set | Water temp (°C) | Turbidity (NTU) | TOC (mg/liter) |
|---|---|---|---|
| Flow rate vs temp | 18 ± 1.2 | 0.17 ± 0.09 | 2.5 ± 0.31 |
| Flow rate vs temp | 4.8 ± 1.4 | 0.33 ± 0.39 | 1.6 ± 0.74 |
| Low-range turbidity vs permeate rate | 17 ± 0.21 | 0.26 ± 0.04 | 2.2 ± 0.40 |
| Midrange turbidity vs permeate rate | 18 ± 0.82 | 1.1 ± 0.06 | 3.7 ± 0.21 |
| Microbe recovery (low-range turbidity) | 17 ± 0.62 | 0.29 ± 0.16 | 2.1 ± 0.83 |
| Microbe recovery (midrange turbidity) | 15 ± 1.0 | 1.5 ± 0.29 | 3.1 ± 0.62 |
| Microbe recovery (high-range turbidity) | 16 ± 0.28 | 4.3 ± 0.51 | 2.7 ± 0.94 |
Values are averages ± standard deviations.
FIG. 3.
Pressure and flow rates for the following ultrafilters when filtering tap water at 18°C (open symbols) or 4.8°C (shaded symbols): Exeltra Plus 210 (A and B) (○ and •, respectively), F200NR (C) (⋄), REXEED 21S (D and E) (▵ and ▴, respectively), and REXEED 25S (F and G) (□ and ▪, respectively).
Turbidity-versus-permeate flow rate experiments.
Based on hydraulic performance versus water temperature, the Rexeed 25S ultrafilters were used to examine the effect of turbidity on the hydraulic performance of ultrafilters used in the DEUF configuration. The hydraulic performances of the REXEED 25S filters were tested at two water turbidity levels: low-range turbidity (0.26 NTU) and midrange turbidity (1.1 NTU). The average water temperatures for the low-range- and midrange-turbidity experiments were similar (17 to 18°C), but the average TOC for the low-range-turbidity water samples (2.2 mg/liter) was significantly lower than that for the midrange-turbidity water samples (3.7 mg/liter) (Table 1). At a nominal flow rate of 2,100 ml/min, the average permeate flow rates at the beginning of the 100-liter low-range- and midrange-turbidity UF experiments were 1,920 ml/min and 1,970 ml/min, respectively. At the end of the low-range- and midrange-turbidity UF experiments (when nearly 100 liters had passed through each filter), the permeate flow rates were an average of 14% and 9% lower, respectively (data not shown). For these experiments, no attempt was made to increase the pump speed to increase the flow rate through the filters. Pressures at the end of the low-range- and midrange-turbidity experiments were approximately 19% lower (average final pressure, 6.2 lb/in2) and 34% lower (average final pressure, 6.8 lb/in2), respectively, than at the beginning of the experiments. For all the low-range- and midrange-turbidity hydraulic performance experiments, 100-liter samples were filtered within 60 min.
Microbe recovery experiments.
Using REXEED 25S ultrafilters, seeded 100-liter water samples were used to investigate the recovery efficiencies of the DEUF method for low-range-, midrange-, and high-range-turbidity water samples. For these experiments, the average turbidity values for the low-range-, midrange-, and high-range-turbidity water samples were substantially different: 0.29, 1.5, and 4.3 NTU, respectively (Table 1). As observed for previous hydraulic performance experiments, permeate flow rates for the 100-liter microbial recovery experiments did not change substantially between the beginning and the end of the experiments. Average permeate flow rates for low-range-, midrange-, and high-range-turbidity water experiments were initially 2,230 ml/min, 2,050 ml/min, and 2,080 ml/min, respectively, and were 10%, 5%, and 3% lower, respectively, at the end of the experiments (data not shown).
Average microbial recovery efficiencies for each of the study microbes were greater than 50% for water samples at each of the turbidity levels and were generally higher for low-range-turbidity water samples (Table 2). The average percent recovery for E. faecalis was highest for the low-range-turbidity water (93% ± 16%), with significant differences found compared to average E. faecalis recoveries in midrange-turbidity water (71% ± 11%) but not compared to recoveries from high-range-turbidity water. Recovery efficiencies for C. perfringens spores were significantly higher for the low-range-turbidity water (94% ± 22%) than for midrange- and high-range-turbidity water samples. As observed for E. faecalis and C. perfringens spores, the average recovery efficiency for C. parvum oocysts was higher for low-range-turbidity water (87% ± 18%), but no significant differences were found between low-range-turbidity recovery efficiencies and recovery efficiencies from midrange- and high-range-turbidity water samples. MS2 bacteriophage was the only study microbe for which recovery efficiencies were lower in low-range-turbidity water samples (57% ± 7.7%) than in midrange- and high-range-turbidity water samples, but the differences were significant only between low-range- and midrange-turbidity conditions (and not between low-range and high-range conditions). No microbial breakthrough of the study ultrafilters was observed.
TABLE 2.
Recovery efficiency of DEUF for microbes seeded into 100 liters of water at different turbidity levels
| Exptl condition (na) | Avg recovery efficiency ± SDb
|
|||
|---|---|---|---|---|
| E. faecalis | C. perf. | MS2 | Crypto. | |
| Low-range turbidity [0.29 NTU] (4-5) | 93 ± 16 | 94 ± 22 | 57 ± 7.7 | 87 ± 18 |
| Midrange turbidity [1.5 NTU] (5-6) | 71 ± 11 | 60 ± 8.1 | 82 ± 14 | 63 ± 21 |
| High-range turbidity [4.3 NTU] (6) | 78 ± 12 | 57 ± 21 | 73 ± 13 | 83 ± 21 |
n, no. of experiments.
C. perf., C. perfringens spores; Crypto., C. parvum oocysts.
DISCUSSION
The results from this study show that commercially available disposable hollow-fiber ultrafilters can be effectively used in a DEUF configuration for recovering diverse microbes from water samples of moderate turbidity. Hydraulic data indicated that there were appreciable differences in permeate flow rate-versus-pressure relationships between hollow-fiber ultrafilters having approximately the same filter area. As expected, higher system pressure was needed to achieve higher permeate flow rates, and the relationship was found to be more pronounced for colder water samples (Fig. 3). Although permeate rates were, in general, lower at the end of a 100-liter DEUF procedure, the reductions were generally modest (e.g., below 10%). For water samples having turbidities between 0.29 and 4.3 NTU, permeate flow rates through REXEED 25S filters were approximately 2 liters/min after a 100-liter volume was filtered. The 2-liter sample processing rate was higher than has been reported for some recirculating (tangential-flow) hollow-fiber UF methods (5, 12) using approximately the same-size ultrafilters and approximately the same permeate flow rate as that reported by Simmons et al. (15). The system pressures associated with a 2-liter/min DEUF permeate flow rate in the present study were generally less than 15 lb/in2, which is lower than the 25-lb/in2 system pressure used by Simmons et al. (15) to achieve the same general permeate flow rate. Although the upper limit for system pressure in a commercial ultrafilter (generally sold for medical uses, e.g., dialysis and hemoconcentration) has not been established, the data from this study suggest that permeate flow rates of greater than 2 liters/min are possible for the DEUF configuration without risking system pressures of greater than 25 lb/in2.
Overall, microbial recovery efficiencies ranged from 57% to 94% and were generally highest for the low-range-turbidity experiments and lowest for the midrange-turbidity experiments (Table 2). While recovery efficiencies were generally higher for the low-turbidity water samples, no consistent trends were observed between microbial recovery efficiency and water sample turbidity. These results demonstrate that the DEUF method was effective for recovering microbes from 100-liter water samples having low to moderate turbidity. The recovery efficiencies measured for E. faecalis in the present study were consistently high (71% ± 11% to 93% ± 16%). While a previous DEUF study performed by Leskinen and Lim (9) did focus on recovery of enterococci from water samples, the recovery efficiencies reported for their method were 4% to 708%, a range that makes it difficult to compare method performances. It is possible that the >100% recovery efficiencies reported by Leskinen and Lim were due to undercounting the low levels of naturally occurring enterococci or were due to disaggregation of enterococcal clumps during UF. While no previous DEUF studies have focused on recovery of MS2, C. perfringens spores, or Cryptosporidium oocysts, the recoveries of these microbes reported for the DEUF method in the present study are similar to, or greater than, recovery efficiencies reported for tangential-flow hollow-fiber UF studies (5, 8, 10, 15). Although Hill et al. (5) reported generally higher recovery efficiencies for MS2, E. faecalis, and C. perfringens spores than those reported in the present study, these authors acknowledged that the high recovery efficiencies were likely due in part to disaggregation during UF of microbe aggregates present in seed stocks.
The data reported in this study demonstrate that a simple DEUF method can be effective for rapid sample collection and efficient recovery of microbes present in 100-liter water samples of low to moderate turbidity. For a rapid response to suspected water contamination events, the DEUF method can enable untrained field personnel to readily employ an UF technique to recover diverse target microbes or unidentified microbial agents from large-volume water samples. Instead of shipping tens of liters of water to an analytical laboratory, use of the DEUF technique should decrease the costs and effort needed to analyze large-volume water samples by enabling field personnel to filter in the field and ship ultrafilters (instead of bulk water samples) to the laboratory for final processing and analysis. Additional research is warranted to investigate whether the DEUF method can be effective for large-volume water samples of higher turbidity (e.g., river or lake water). Additional research should also focus on whether permeate flow rates higher than 2 liters/min can be achieved at moderate system pressures.
Acknowledgments
We thank Jacquelin Roberts (CDC/NCZVED/DPD) for performing statistical analyses for this project.
This publication was supported in part by funds made available through the Centers for Disease Control and Prevention, Coordinating Office for Terrorism Preparedness and Emergency Response.
The use of trade names and names of commercial sources is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention or the U.S. Department of Health and Human Services. The findings and conclusions in this presentation are those of the authors and do not necessarily represent those of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 26 June 2009.
REFERENCES
- 1.American Public Health Association, American Water Works Association, and Water Environment Federation. 2005. Standard methods for the examination of water and wastewater, 21st ed. American Public Health Association, Washington, DC.
- 2.Bisson, J. W., and V. J. Cabelli. 1979. Membrane filter enumeration method for Clostridium perfringens. Appl. Environ. Microbiol. 37:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colford, J. M., Jr., S. Roy, M. J. Beach, A. Hightower, S. E. Shaw, and T. J. Wade. 2006. A review of household drinking water intervention trials and an approach to the estimation of endemic waterborne gastroenteritis in the United States. J. Water Health 4(Suppl. 2):71-88. [DOI] [PubMed] [Google Scholar]
- 4.Dahling, D. R. 2002. An improved filter elution and cell culture assay procedure for evaluating public groundwater systems for culturable enteroviruses. Water Environ. Res. 74:564-568. [DOI] [PubMed] [Google Scholar]
- 5.Hill, V. R., A. M. Kahler, N. Jothikumar, T. B. Johnson, D. Hahn, and T. L. Cromeans. 2007. Multistate evaluation of an ultrafiltration-based procedure for simultaneous recovery of enteric microbes in 100-liter tap water samples. Appl. Environ. Microbiol. 73:4218-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill, V. R., A. L. Polaczyk, D. Hahn, J. Narayanan, T. L. Cromeans, J. M. Roberts, and J. E. Amburgey. 2005. Development of a rapid method for simultaneous recovery of diverse microbes in drinking water by ultrafiltration with sodium polyphosphate and surfactants. Appl. Environ. Microbiol. 71:6878-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kearns, E. A., S. Magana, and D. V. Lim. 2008. Automated concentration and recovery of micro-organisms from drinking water using dead-end ultrafiltration. J. Appl. Microbiol. 105:432-442. [DOI] [PubMed] [Google Scholar]
- 8.Kuhn, R. C., and K. H. Oshima. 2002. Hollow-fiber ultrafiltration of Cryptosporidium parvum oocysts from a wide variety of 10-L surface water samples. Can. J. Microbiol. 48:542-549. [DOI] [PubMed] [Google Scholar]
- 9.Leskinen, S. D., and D. V. Lim. 2008. Rapid ultrafiltration concentration and biosensor detection of enterococci from large volumes of Florida recreational water. Appl. Environ. Microbiol. 74:4792-4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindquist, H. D. A., S. Harris, S. Lucas, M. Hartzel, D. Riner, P. Rochele, and R. DeLeon. 2007. Using ultrafiltration to concentrate and detect Bacillus anthracis, Bacillus atrophaeus subspecies globigii, and Cryptosporidium parvum in 100-liter water samples. J. Microbiol. Methods 70:484-492. [DOI] [PubMed] [Google Scholar]
- 11.Messner, M., S. Shaw, S. Regli, K. Rotert, V. Blank, and J. Soller. 2006. An approach for developing a national estimate of waterborne disease due to drinking water and a national estimate model application. J. Water Health 4(Suppl. 2):201-240. [DOI] [PubMed] [Google Scholar]
- 12.Morales-Morales, H. A., G. Vidal, J. Olszewski, C. M. Rock, D. Dasgupta, K. H. Oshima, and G. B. Smith. 2003. Optimization of a reusable hollow-fiber ultrafilter for simultaneous concentration of enteric bacteria, protozoa, and viruses from water. Appl. Environ. Microbiol. 69:4098-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Reilly, C. E., A. B. Bowen, N. E. Perez, J. P. Sarisky, C. A. Shepherd, M. D. Miller, B. C. Hubbard, M. Herring, S. D. Buchanan, C. C. Fitzgerald, V. Hill, M. J. Arrowood, L. X. Xiao, R. M. Hoekstra, E. D. Mintz, and M. F. Lynch. 2007. A waterborne outbreak of gastroenteritis with multiple etiologies among resort island visitors and residents: Ohio, 2004. Clin. Infect. Dis. 44:506-512. [DOI] [PubMed] [Google Scholar]
- 14.Polaczyk, A. L., J. Narayanan, T. L. Cromeans, D. Hahn, J. M. Roberts, J. E. Amburgey, and V. R. Hill. 2008. Ultrafiltration-based techniques for rapid and simultaneous concentration of multiple microbe classes from 100-L tap water samples. J. Microbiol. Methods 73:92-99. [DOI] [PubMed] [Google Scholar]
- 15.Simmons, O. D., III, M. D. Sobsey, C. D. Heaney, F. W. Schaefer III, and D. S. Francy. 2001. Concentration and detection of Cryptosporidium oocysts in surface water samples by method 1622 using ultrafiltration and capsule filtration. Appl. Environ. Microbiol. 67:1123-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.U.S. Environmental Protection Agency. 2005. Method 1600: enterococci in water by membrane filtration using membrane-enterococcus indoxyl-β-d-glucoside agar (mEI). EPA-821-R-04-023. U.S. EPA Office of Water, Washington, DC.
- 17.U.S. Environmental Protection Agency. 2001. Method 1602: male-specific (F+) and somatic coliphage in water by single agar layer (SAL) procedure. EPA 821-R-01-029. U.S. EPA Office of Water, Washington, DC.
- 18.U.S. Environmental Protection Agency. 2001. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. EPA-821-R-01-025. U.S. EPA Office of Water, Washington, DC.
- 19.Yoder, J., V. Roberts, G. F. Craun, V. Hill, L. A. Hicks, N. T. Alexander, V. Radke, R. L. Calderon, M. C. Hlavsa, M. J. Beach, and S. L. Roy. 2008. Surveillance for waterborne disease and outbreaks associated with drinking water and water not intended for drinking—United States, 2005-2006. MMWR Surveill. Summ. 57:39-62. [PubMed] [Google Scholar]
- 20.Yoder, J. S., M. C. Hlavsa, G. F. Craun, V. Hill, V. Roberts, P. A. Yu, L. A. Hicks, N. T. Alexander, R. L. Calderon, S. L. Roy, and M. J. Beach. 2008. Surveillance for waterborne disease and outbreaks associated with recreational water use and other aquatic facility-associated health events—United States, 2005-2006. MMWR Surveill. Summ. 57:1-29. [PubMed] [Google Scholar]