Abstract
Cinnamic acids (i.e., ferulic and caffeic acids) that are esterified to the vegetable cell walls should be enzymatically released to be absorbed in a mammal's intestines. A low dosage of ferulic acid in rodent diets stimulates insulin production and alleviates symptoms caused by diabetes (M. Sri Balasubashini, R. Rukkumani, and V. P. Menon, Acta Diabetol. 40:118-122, 2003). Several lactic acid bacteria are able to display ferulic acid esterase (FAE) activity, suggesting that their probiotic activity could be, in part, mediated by the slow release of ferulic acid. In the present work, we describe the isolation of one strain identified as being Lactobacillus johnsonii that displayed strong FAE activity in stool samples from diabetes-resistant biobreeding rats. These animals are genetically susceptible to becoming diabetic but do not develop the disease. By using genomic analysis coupled to protein purification and catalytic screening, we were able to purify two proteins with FAE activity. The enzymes displayed 42% sequence identity and a broad range of substrate preferences. High affinities and catalytic efficiencies toward aromatic compounds such as ethyl ferulate (Km = 20 to 60 μM) and chlorogenic acid (Km = 10 to 50 μM) were observed. The strain isolated herein as well as the enzymes studied could be potentially useful for the formulation of probiotics to ameliorate diabetes symptoms.
According to the Centers for Disease Control and Prevention (CDC), approximately 23.6 million Americans suffer from diabetes. In the year 2000, this metabolic disease was directly or indirectly responsible for 200,000 deaths. On average, diabetes costs more than 170 billion dollars annually to the American economy. Diets abundant in carbohydrates, increasing rates of obesity, and sedentary lifestyles in combination with genetic predisposition are key factors associated with the development of diabetes. In addition to these environmental factors, notable changes in the composition of the gut microbial ecosystem of diabetic patients were recently described (36).
A clever analysis utilized to measure microbially diverse populations based on genomic principles reported previously by Furrie (17) was used by Roesch and coworkers (30) to prove that rodents with diabetes have a specifically associated gut microflora. A deep characterization of the intestinal microflora of biobreeding diabetes-resistant (BB-DR) rats showed that the presence of Lactobacillus reuteri, Lactobacillus johnsonii, and Bifidobacterium species is predominant, compared with the bacterial species observed in biobreeding diabetes-prone (BB-DP) rats (30). In agreement with the large presence of probiotic-like bacteria, it was previously reported that the oral administration of lactic acid bacteria (LAB) can help to reduce blood glucose levels (25-27, 40) by stimulating insulin secretion via changes in autonomic neurotransmission (34). However, it is not clear how LAB species are able to induce these changes. Neither genetic nor physiological characteristics of the strains utilized have been extensively evaluated so far.
LAB are traditional fermentation agents used in food ripening, preservation, an flavor enhancement and as probiotics (28, 37). They are relatively easy to culture and are frequently used in many industrial processes. LAB can be massively produced in fermentation tanks and administered to patients as a dairy food supplement (10). The enhancement of insulin secretion using LAB is a promising alternative since it is natural and inexpensive. Nevertheless, before suggesting the use of LAB to alleviate or even prevent diabetes, it is necessary to understand the mechanisms that stimulate insulin secretion and the genetic characteristics of the strains to be used.
A low dosage of cinnamic acids (especially ferulic acid) has been related with the stimulation of insulin secretion (1, 6, 32), prevention of oxidative stress, lipid peroxidation (33), and inhibition of diabetic nephropathy progression (16). The enzymatic release of cinnamic acid esters from fibers present in food takes place in the intestines of mammalian digestive systems. Although ferulic acid esterase (FAE) activity was detected in small intestine epithelial cells, a higher level of activity was detected in samples obtained from the intestinal lumen (3, 4). This suggests that the FAE activity produced by the intestinal microflora is a key factor in releasing and assimilating the previously esterified phenolic compounds (11, 21).
In this study, we focused our attention on FAE-producing LAB strains that we isolated from frozen samples studied previously by Roesch et al. (30) and that were shown to be predominant in BB-DR rats or decreased in BB-DP rats. Our working hypothesis was that the strains that are highly associated with BB-DR rats could be good producers of FAEs. Here, we describe the isolation and identification of a Lactobacillus johnsonii strain isolated from stool samples of BB-DR rats as well as the biochemical characteristics of two new FAEs produced by this strain.
MATERIALS AND METHODS
All materials were purchased from Sigma unless specified otherwise.
Strain isolation.
The strains were isolated by plating out aliquots of BB-DP and BB-DR rat stool samples directly onto selective Rogosa agar plates (31) grown at 37°C under anaerobic conditions in a 5% carbon dioxide atmosphere. The isolated strains were conserved in glycerol deeps at −86°C in 96-well plates.
Feruloyl esterase screening assay.
The isolated LAB were screened on Man, Rogosa, and Sharpe (MRS) agar plates with glucose omitted and that contained 0.1% ethyl ferulate (Apin Biochem) (MRS-F) (14). The ferulate assay plates were inoculated with cells obtained from individual cultures grown overnight on MRS broth and incubated at 37°C for a maximum of 3 days. Ferulate esterase activity was evidenced by the formation of a clearing zone (halo) around the colonies. The strains with the largest zones of activity were selected for further analysis.
Strain identification.
The selected strains were identified by the sequencing of internal 16S rRNA gene fragment from genomic DNA. LAB genomic DNA was isolated using a Qiagen DNeasy kit, and the 231-bp fragments were PCR amplified with primers LactoF and LactoR (9). The purified amplification product was sequenced, and the sequence was blasted against the MedLine Database to identify the donor species.
Cloning and purification of enzyme.
The genes of interest were PCR amplified from genomic DNA isolated as described above. The fragments were cloned into pET-15b-TV according to methods described previously by Lorca et al. (23). The expression of His6-tagged proteins was carried out in Escherichia coli BL21(DE3) cells (Stratagene) by using IPTG (isopropyl-β-d-thiogalactopyranoside) (1 mM) as an inducer. The cells were collected by centrifugation, resuspended in binding buffer (5 mM imidazole, 500 mM NaCl, 20 mM Tris-HCl [pH 7.9]), and disrupted by use of a French press. The His6-tagged proteins were purified by affinity chromatography as previously described (18). The purified proteins were dialyzed at 4°C against a solution containing 50 mM Tris-HCl buffer (pH 8.00), 500 mM NaCl, and 1 mM dithiothreitol. After dialysis, the samples were flash-frozen and preserved at −86°C in small aliquots until use.
Enzymatic assays: determination of enzyme substrate preference.
The enzymatic substrate profile of each purified protein was determined at 25°C using the ester library described previously by Liu et al. (22). The purified enzymes were diluted (800 μg/ml) and redialyzed against 5 mM BES [N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid] buffer (pH 7.2), and the enzymatic activity was determined using a colorimetric method described previously by Janes et al. (20). The final concentration of each component of the reaction mixture was as follows: 1 mM ester substrate, 0.44 mM p-nitrophenol (as a proton acceptor), 4.39 mM BES (pH 7.2), 7.1% acetonitrile, and 30 to 35 μg per ml of enzyme. The enzymatic assays were performed with 105 μl in 96-well plates in a Synergy HT Biotek reader. The decrease in absorbance was continuously monitored during 30 min at 412 nm, and the concentration of p-nitrophenol was estimated by using the extinction coefficient (ɛ = 16,300 M−1 cm−1). All assays and controls were performed in triplicate, and the average activities were quantified. Results are shown as means ± standard deviations.
Saturation assays.
Classical model substrates (p-nitrophenyl and naphthyl esters) were used to determine the biochemical parameters of the purified enzymes Lj0536 and Lj1228. The assays were conducted at the optimal conditions (pH and temperature) determined for each purified enzyme according to protocols described previously by Gonzalez et al. (18). Lj0536 reactions were done using 20 mM HEPES buffer (pH 7.8) at 25°C, and the assays with Lj1228 were performed using 20 mM MES (morpholineethanesulfonic acid) buffer (pH 6.7) at 30°C. The effect of pH was done using overlapping buffers (pH 4 to pH 11); the divalent cations assayed were used in the chloride form. The effects of bile salts such as sodium glycocholate and taurocholic, cholic, and deoxycholic acids were assayed at a range of concentrations (2 to 10 mM) using p-nitrophenyl butyrate as an enzymatic model substrate. All kinetic parameters were determined from the average of data from triplicate assays.
Assays using ethyl ferulate and chlorogenic acid as enzyme substrates.
Enzymatic hydrolysis of ethyl ferulate and chlorogenic acid was done by the continuous reading of reaction mixture absorbance at 324 nm (maximal absorbance of ethyl ferulate and chlorogenic acid). The typical reaction mixture contained 20 mM buffer, 0.01 to 0.20 mM substrate, and 0.05 to 0.1 μg/ml of purified enzyme. The decrease in absorbance at 324 nm was monitored at 25°C during 10 min in a 96-well UV plate in a microplate reader (Synergy HT; Biotek). The ethyl ferulate extinction coefficient (ɛ = 15,390 M−1 cm−1) and the chlorogenic acid extinction coefficient (ɛ = 26,322 M−1 cm−1) were determined experimentally and used to calculate the amount of substrate hydrolyzed. The kinetic parameters were determined from the averages of data from triplicate assays with a correlation coefficient higher than 0.98 after nonlinear regression analysis using Origin 8 software.
HPLC analysis.
A high-performance liquid chromatography (HPLC) assay was performed according to methods described previously by Mastihuba et al. (24), using a Phenomenex Synergy 4-μm Hydro RP-80A column, HP1090 Series II liquid chromatography with a UV detector, and an HP1047A RI detector. The assay was based on the measurement of acid derivative compounds released by enzymatic action from the substrate utilized in the reaction mixture. A typical reaction mixture contained 15 μg/ml of enzyme, 20 mM buffer, and 5 mM of substrate.
RESULTS AND DISCUSSION
Strain isolation and screening.
LAB strains used in this work were isolated from stool samples obtained from BB-DR and BB-DP rats. More than 300 colonies were picked from each sample and transferred into the screening medium (MRS-F). LAB colonies that displayed the best FAE activity produced clear zones of 0.8 to 0.9 cm resulting from the enzymatic release of ferulic acid. Approximately 80% ± 5% (mean ± standard deviation) of the colonies isolated from BB-DR rats were positive for FAE; interestingly, only 41% ± 7% of the strains isolated from BB-DP rats were able to demonstrate good hydrolytic abilities. Among all of the colonies isolated from stool samples of BB-DP and BB-DR rats, five colonies derived from BB-DR rats showed the largest halos in MRS-F plates and were selected for identification by sequencing methods.
Identification of the best producer strain.
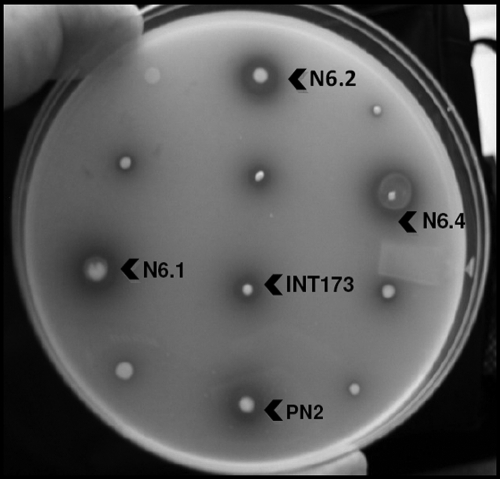
The 16S rRNA gene sequence amplified from the selected colonies belongs to three different LAB. The first sequence analyzed was closely related to L. reuteri (identity 99%); the second sequence showed 96% identity with Lactobacillus helveticus. The last three belong to the same species; they showed 100% identity with L. johnsonii 16S rRNA genes sequence (Fig. 1). The colonies identified as being L. johnsonii, in particular strain N6.2 in our primary analysis, demonstrated that it could hydrolyze the ethyl ferulate better than L. reuteri or L. helveticus-like colonies when they were plated together in the same MRS-F plate (Fig. 2). Consequently, strain N6.2 was selected as the DNA donor to clone genes encoding esterases that are potentially active against feruloyl esters.
FIG. 1.
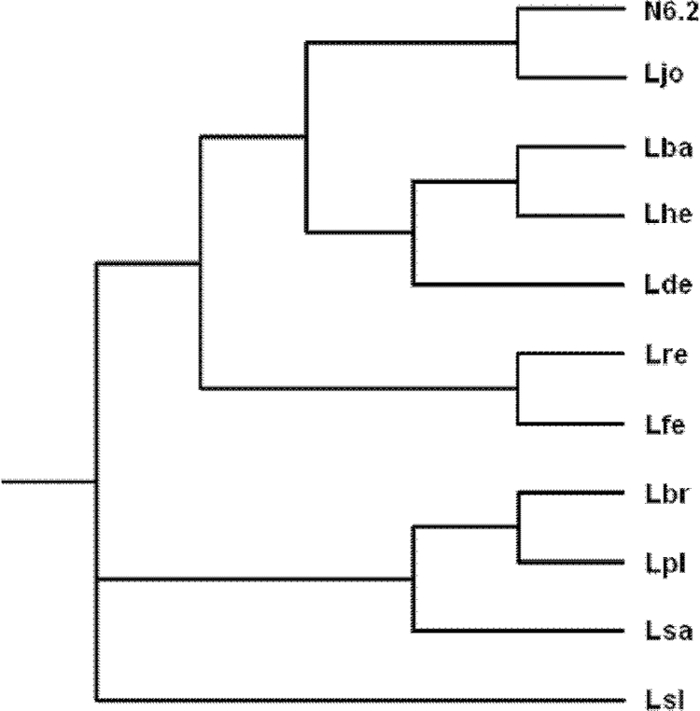
Phylogenetic tree of LAB 16S rRNA genes gene sequences and strain N6.2 isolated from stool samples from BB-DR rats. N6.2, isolated strain (best FAE producer colony); Ljo, Lactobacillus johnsonii NCC 533 (locus tag, LjR007); Lba, Lactobacillus acidophilus NCFM (locus tag, LBA2001); Lhe, Lactobacillus helveticus DPC 4571 (locus tag, lhv_3101); Lde, Lactobacillus delbrueckii subsp. bulgaricus ATCC BAA-365 (locus tag, LBUL_r0045); Lre, Lactobacillus reuteri F275 (locus tag, LAR_16SrRNA01); Lfe, Lactobacillus fermentum IFO 3956 (locus tag, LAF_16SrRNA01); Lbr, Lactobacillus brevis ATCC 367 (locus tag, LVIS_r0082); Lpl, Lactobacillus plantarum WCFS1 (locus tag, lp_rRNA01); Lsa, Lactobacillus sakei 23K (locus tag, LSAr01); Lsl, Lactobacillus salivarius UCC118 (locus tag, LSL_RNA001). Alignment was done using ClustalX, and the phylogenetic tree was constructed using the neighbor-joining method and visualized with Treeview.
FIG. 2.
Comparative feruloyl esterase activities displayed by isolated LAB on solid MRS-0.1% (vol/vol) ethyl ferulate culture medium. Hydrolysis is visualized as clear zones around the individual colonies. The plates were incubated overnight at 37°C during 72 h. N6.1, N6.2, and N6.4 are L. johnsonii colonies. INT173 is an L. reuteri-like colony. PN2 is an L. helveticus-like colony.
In silico selection of targets and enzymatic screening.
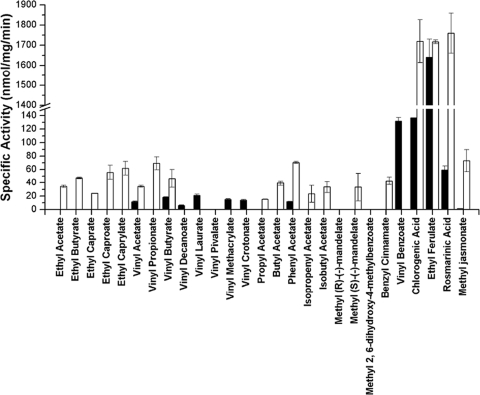
The precise identification of L. johnsonii in the stool samples allowed the use of comparative genomics to select our targets in silico. We selected five open reading frames (ORFs) encoding proteins that displayed the characteristic motif described previously for esterases (8, 12). The target genes were selected from a group of 346 ORFs encoding hypothetical (306 ORFs) or putative (40 ORFs) proteins as they are annotated in the genome used as a reference, strain L. johnsonii NCC 533 (http://cmr.jcvi.org/tigr-scripts). Based on the genome sequence of L. johnsonii NCC 533 (http://cmr.jcvi.org/tigr-scripts), primers were designed, and L. johnsonii N6.2 chromosomal DNA was used as a template for gene cloning and protein purification. The enzyme activity profile of the purified proteins was evaluated in parallel against a panel of 27 different substrates. Several aliphatic esters were included in the substrate array for comparative purposes. Two enzymes (Lj0536 and Lj1228) showed high preference and good enzymatic activity using aromatic esters as substrates. Lj1228 demonstrated similar hydrolytic abilities toward ethyl ferulate, chlorogenic acid, and rosmarinic acid. Instead, ethyl ferulate was the best substrate determined for Lj0536 (Fig. 3). Despite the fact that the stability of the enzymes can be affected during the necessary exhaustive dialysis in BES buffer and the conditions were not previously optimized, the technique used was helpful for selecting the enzymes with the best activity toward the substrates of interest.
FIG. 3.
Esterase activity (substrate profile) of Lj0536 (black) and Ljo1228 (white) toward a general ester library. The assays conditions and enzyme activities were carried out according to the protocols described previously by Janes and coworkers (20), using p-nitrophenol as a proton trapper. The reaction mixtures were formulated with 1 mM substrate and 30 to 35 μg ml−1 of purified enzymes. The selected enzymes were demonstrated to be active against a broad range of substrates, but they showed the highest activity toward aromatic esters such as ethyl ferulate and chlorogenic and rosmarinic acids. The figure displays the mean specific activities obtained (bars) toward different substrates, and lines on top of each bar represent the standard deviations estimated from three independent assays.
Bioinformatic analysis of selected targets.
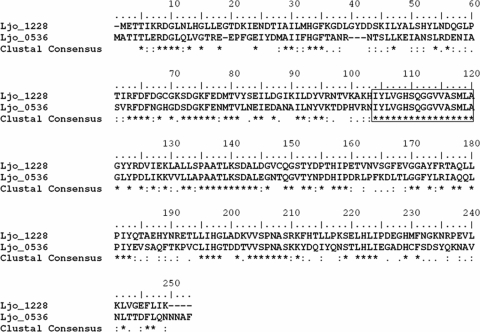
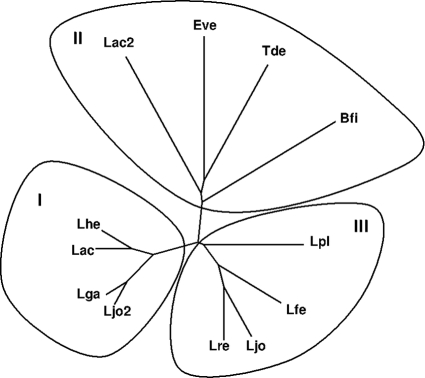
The genes encoding the proteins herein studied are located in two different and poorly characterized regions of the L. johnsonii chromosome. The flanking genes, described as being hypothetical proteins, of the Lj0536 ORF are transcribed in opposite directions. Lj1228 is flanked by two intergenic regions of ∼800 bp. The genomic data suggest that both genes are transcribed as independent transcription units. The proteins encoded by this genes share 42% identity based on the analysis of their amino acid sequences (Fig. 4). It is possible to identify five well-preserved clusters along the alignment of these protein sequences. A higher conservation was observed in the central cluster, which displays the sequence G108HSQGGVVASMLA120 (Fig. 4), containing the classical serine nucleophilic motif (GxSxG, in boldface type) described previously for some carboxyl esterases (8, 12). This sequence is a unique fully conserved cluster in all the homologous proteins analyzed and is included in the phylogenetic analysis performed in this work. The phylogenetic tree depicted in Fig. 5 was constructed with the closest sequences obtained after a Blast search against each of the L. johnsonii paralogs. The proteins grouped into cluster I are Lj1228-homologous proteins (Fig. 5). They are highly conserved (>80% identity) and are present only in homofermentative LAB (Lactobacillus gasseri, L. helveticus, and L. acidophilus). Lj0536 (Fig. 5) has a high level of identity with an L. reuteri (74%) homolog and is clustered together with proteins encoded only in the chromosome of heterofermentative strains (cluster III) (Fig. 5). These results and the absence of paralogs in the genome of L. gasseri suggested that Lj1228 could be originated by an intrachromosomal duplication and further diversification of Lj0536. Cluster II is integrated by proteins that share only 22% identity with each of the L. johnsonii FAEs studied herein. However, they were included because Butyrivibrio fibrisolvens CinI, annotated as cinnamoyl esterase, is the closest related protein previously purified and characterized (13). The homology to B. fibrisolvens CinI was used to automatically annotate other homologs within cluster II when their genomes were sequenced.
FIG. 4.
Binary alignment of two L. johnsonii enzymes (Lj0536 and Lj1228). The proteins showed 42% sequence identity, with the highest conservation in the central cluster (box including I104 to G121), where it is possible to distinguish the classical serine esterase catalytic motif G108XS110XG112.
FIG. 5.
Phylogenetic relationships of purified L. johnsonii FAE. Shown are L. johnsonii (Ljo [Lj0536] [GI 42519153] and Ljo2 [Lj1228] [GI 42519789]), L. acidophilus (Lac2 [GI 58337623] and Lac [GI 58338090]), Eubacterium ventriosum (Eve) (GI 154484090), Treponema denticola (Tde) (GI 41815924), Butyrivibrio fibrisolvens (Bfi) (GI 1622732), L. plantarum (Lpl) (GI 28379396), L. fermentum (Lfe) (GI 184155794), L. reuteri (Lre) (GI 194467183), L. gasseri (Lga) (GI 116630316), and L. helveticus (Lhe) (GI 161508065).
Biochemical properties of Lj0536 and Lj1228.
The purity of His-tagged FAEs was tested by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by PhastGelBlue R-350 staining. Both proteins purify as a single band with an apparent molecular mass of 31 kDa. The molecular mass determined was consistent with the theoretical protein mass predicted (27 kDa), in addition to the TV cleavage site and the His6 tag encoded in the plasmid that was fused to the native protein. Lj0536 exhibited its maximal activity at 20°C at pH 7.8 in 20 mM HEPES buffer, whereas the optimal parameters for Lj1228 were 30°C at pH 6.7 in 20 mM MES buffer. The optimal temperature seems to be low for proteins purified from bacteria living in rat intestines, but they demonstrated more than 70% residual activity in a wide range of temperature (15°C to 38°C). When the activity of Lj0536 was evaluated in the presence of divalent cations or iron chloride, only Cu2+ (1 mM) inhibited the activity by 90%. Lj1228 enzymatic activity was arrested with 1 mM of Zn2+, Fe3+, or Cu2+. Lj1228 was five times more sensitive to Fe3+ than the enzyme purified from L. acidophilus (38). The addition of EDTA to the reaction mixtures did not affect the enzymatic activity of these enzymes. Both enzymes were fully inhibited by phenylmethanesulfonyl fluoride and resistant to N-ethylmaleimide and iodoacetate. These results confirm the presence of serine as the nucleophilic residue in the active center, which is suggested by data from the bioinformatic analysis.
These bacterial proteins should be active in the gastrointestinal tract, where emulsifying compounds such as bile salts can affect their activity. Consequently, we evaluated their activity in the presence of conjugated and deconjugated bile salts. Lj1228 was not affected by any of the salts assayed, but surprisingly, in the presence of a low concentration of sodium glycocholate (2 mM), the activity of Lj0536 increased by 50% with respect to the control reaction. A further increase in the concentration of sodium glycocholate, up to 10 mM, increased the enzyme activity up to 2.5 times.
The enzymatic parameters obtained by steady-state saturation kinetics using a variety of esters as substrates are summarized in Table 1. As was determined by the screening method, both enzymes were active against a wide range of substrates, but they showed a high affinity toward aromatic esters. The best catalytic efficiency (kcat/Km) was obtained against ethyl ferulate and chlorogenic acid. Lj0536 also demonstrated a high affinity for p-nitrophenyl butyrate, but its catalytic efficiency was lower than those observed for phenolic esters. The hydrolysis of chlorogenic acid was also analyzed by HPLC by detecting the appearance of free caffeic acid in the reaction mixture. Chlorogenic acid is another important component of the human diet (present in coffee), but its absorption occurs only after microbial enzymatic degradation (29). Chlorogenic acid esterase activity was detected in several bacterial species, including E. coli (11). However, there are no records of purified bacterial enzymes with efficient chlorogenic acid esterase activity. The affinities of both L. johnsonii enzymes for chlorogenic acid are comparable to the Km described previously for Aspergillus niger (5).
TABLE 1.
Kinetic parameters of Lj0536 and Lj1228 using several esters as enzymatic substrates
| ORF and substrate |
Km (mM)
|
Vmax (μmol min−1 mg−1)
|
kcat (s−1) | kcat/Km (M−1 s−1) | ||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| Lj0536 | ||||||
| 1-Naphthyl-acetate | 0.298 | ±0.03 | 21.2 | ±1.23 | 9.75 | 3.27E+04 |
| 1-Naphthyl propionate | 0.162 | ±0.01 | 14 | ±0.05 | 6.43 | 3.97E+04 |
| 1-Naphthyl butyrate | 0.150 | ±0.01 | 12.7 | ±0.24 | 5.82 | 3.87E+04 |
| 2-Naphthyl acetate | 0.897 | ±0.22 | 1.83 | ±0.25 | 0.841 | 9.37E+02 |
| 2-Naphthyl propionate | 0.225 | ±0.02 | 0.533 | ±0.05 | 0.245 | 1.09E+03 |
| 2-Naphthyl butyrate | 0.222 | ±0.01 | 0.247 | ±0.01 | 0.114 | 5.10E+02 |
| p-Nitrophenyl acetate | 0.469 | ±0.14 | 8.4 | ±1.03 | 3.86 | 8.23E+03 |
| p-Nitrophenyl butyrate | 0.040 | ±0.00 | 3.77 | ±0.18 | 1.73 | 4.30E+04 |
| p-Nitrophenyl caprylate | 0.198 | ±0.00 | 0.268 | ±0.01 | 0.123 | 6.20E+02 |
| Ethyl ferulate | 0.020 | ±0.01 | 17.2 | ±3.24 | 7.89 | 3.93E+05 |
| Chlorogenic acid | 0.053 | ±0.01 | 61.2 | ±2.75 | 28.1 | 5.32E+05 |
| Lj1228 | ||||||
| 1-Naphthyl-acetate | 0.737 | ±0.08 | 2.97 | ±0.17 | 1.36 | 1.85E+03 |
| 1-Naphthyl propionate | 0.395 | ±0.07 | 2.25 | ±0.15 | 1.03 | 2.61E+03 |
| 1-Naphthyl butyrate | 0.186 | ±0.03 | 0.846 | ±0.05 | 0.39 | 2.10E+03 |
| 2-Naphthyl propionate | 0.317 | ±0.06 | 0.25 | ±0.02 | 0.11 | 3.65E+02 |
| 2-Naphthyl butyrate | 0.115 | ±0.01 | 0.10 | ±0.00 | 0.04 | 3.87E+02 |
| p-Nitrophenyl acetate | 0.946 | ±0.22 | 0.64 | ±0.09 | 0.29 | 3.11E+02 |
| p-Nitrophenyl butyrate | 0.225 | ±0.02 | 0.56 | ±0.01 | 0.26 | 1.14E+03 |
| p-Nitrophenyl caprylate | 0.258 | ±0.01 | 0.15 | ±0.01 | 0.06 | 2.58E+02 |
| Ethyl ferulate | 0.063 | ±0.03 | 1.11 | ±0.28 | 0.50 | 7.80E+04 |
| Chlorogenic acid | 0.010 | ±0.00 | 8.68 | ±0.49 | 3.97 | 3.69E+05 |
The biological importance of LAB as probiotics is extensively documented and discussed (37). The beneficial effects of LAB in ameliorating certain diabetes symptoms were recently described. However, there is still no satisfactory explanation for this observation (25-27, 40). The present work does not answer that question but joins important elements to enrich the discussion in pursuing our understanding of the bacterium-diabetic host relationship. Based on our microflora analysis of BB-DP and BB-DR rats, LAB are one of the groups of bacteria that are naturally enriched in the gut of a nondiabetic host (30). We also know that an important amount of the cinnamoyl esterase activity is provided by the enzymes produced by the gut microflora (29, 39) and that ferulic acid can stimulate insulin secretion (1, 6, 16, 32). These three important elements together suggest that the ability of LAB to produce FAEs could play a role in releasing ferulic acid from the diet in the digestive tract to overcome diabetes symptoms of genetically predisposed diabetic hosts. Further in vivo evidence using knockout FAE mutants will be necessary to discuss this observation in depth.
As was discussed in recent reviews, most FAEs have been isolated from phytopathogenic fungi (15, 35). Thus, the second important contribution of this work is related exclusively with the biochemistry of two new bacterial enzymes that display FAE activity. Lactobacillus esterases are enzymes that play an important role in the modification of the texture and flavor of fermented food.
Even though FAE activity has been detected in homo- and heterofermentative LAB species including L. helveticus, L. acidophilus, L. fermentum, L. casei, L. rhamnosus, L. reuteri, and L. farciminis (7, 14, 19), we found only one report describing the biochemical characteristics of a purified enzyme (38). In that work, those authors described the purification of a 36-kDa L. acidophilus FAE, and the N-terminal sequence of the purified protein was determined. A year later, the genome sequence of L. acidophilus became available (2). With the exception of l-lactate dehydrogenase, there is no protein in L. acidophilus that matches the sequence described previously by Wang and coworkers (38). This indicates that they did not identify the correct protein responsible for the activity described in their work. As a consequence, this is the first publication associating the sequence with the biochemical characteristics of two new proteins with feruloyl esterase activity cloned from LAB.
The in vivo regulation and expression of L. johnsonii N6.2 FAE-encoding genes in rat intestines as well as the potential use of the isolated strain in the formulation of probiotics are subjects of current investigation in our laboratory.
Acknowledgments
This work was supported by the Research Innovation Award, Institute of Food and Agricultural Sciences, University of Florida.
We thank J. Neu and D. Schatz from the Department of Pediatrics, M. Atkinson and C. Wasserfall from the Department of Pathology, Immunology, and Laboratory Medicine, and E. Triplett and J. Larkin from Department of Microbiology and Cell Science, University of Florida, for their invaluable support and scientific discussions. We thank Beverly Driver for her valuable technical assistance, Andy Burrion for his support with enzyme production and purification, and Ricardo Valladares for proofreading of the manuscript.
Footnotes
Published ahead of print on 5 June 2009.
REFERENCES
- 1.Adisakwattana, S., P. Moonsan, and S. Yibchok-Anun. 2008. Insulin-releasing properties of a series of cinnamic acid derivatives in vitro and in vivo. J. Agric. Food Chem. 56:7838-7844. [DOI] [PubMed] [Google Scholar]
- 2.Altermann, E., W. Russell, M. Azcarate-Peril, R. Barrangou, B. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreasen, M., P. Kroon, G. Williamson, and M. Garcia-Conesa. 2001. Esterase activity able to hydrolyze dietary antioxidant hydroxycinnamates is distributed along the intestine of mammals. J. Agric. Food Chem. 49:5679-5684. [DOI] [PubMed] [Google Scholar]
- 4.Andreasen, M., P. Kroon, G. Williamson, and M. Garcia-Conesa. 2001. Intestinal release and uptake of phenolic antioxidant diferulic acids. Free Radic. Biol. Med. 31:304-314. [DOI] [PubMed] [Google Scholar]
- 5.Asther, M., M. I. Estrada Alvarado, M. Haon, D. Navarro, M. Asther, L. Lesage-Meessen, and E. Record. 2005. Purification and characterization of a chlorogenic acid hydrolase from Aspergillus niger catalysing the hydrolysis of chlorogenic acid. J. Biotechnol. 115:47-56. [DOI] [PubMed] [Google Scholar]
- 6.Balasubashini, M., R. Rukkumani, P. Viswanathan, and V. Menon. 2004. Ferulic acid alleviates lipid peroxidation in diabetic rats. Phytother. Res. 18:310-314. [DOI] [PubMed] [Google Scholar]
- 7.Bhathena, J., A. Kulamarva, A. Urbanska, C. Martoni, and S. Prakash. 2007. Microencapsulated bacterial cells can be used to produce the enzyme feruloyl esterase: preparation and in-vitro analysis. Appl. Microbiol. Biotechnol. 75:1023-1029. [DOI] [PubMed] [Google Scholar]
- 8.Brenner, S. 1988. The molecular evolution of genes and proteins: a tale of two serines. Nature 334:528-530. [DOI] [PubMed] [Google Scholar]
- 9.Byun, R., M. Nadkarni, K. Chhour, F. Martin, N. Jacques, and N. Hunter. 2004. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J. Clin. Microbiol. 42:3128-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cogan, T., T. Beresford, J. Steele, J. Broadbent, N. Shah, and Z. Ustunol. 2007. Advances in starter cultures and cultured foods. J. Dairy Sci. 90:4005-4021. [DOI] [PubMed] [Google Scholar]
- 11.Couteau, D., A. McCartney, G. Gibson, G. Williamson, and C. Faulds. 2001. Isolation and characterization of human colonic bacteria able to hydrolyse chlorogenic acid. J. Appl. Microbiol. 90:873-881. [DOI] [PubMed] [Google Scholar]
- 12.Cygler, M., J. Schrag, J. Sussman, M. Harel, I. Silman, M. Gentry, and B. Doctor. 1993. Relationship between sequence conservation and three-dimensional structure in a large family of esterases, lipases, and related proteins. Protein Sci. 2:366-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalrymple, B., Y. Swadling, D. Cybinski, and G. Xue. 1996. Cloning of a gene encoding cinnamoyl ester hydrolase from the ruminal bacterium Butyrivibrio fibrisolvens E14 by a novel method. FEMS Microbiol. Lett. 143:115-120. [DOI] [PubMed] [Google Scholar]
- 14.Donaghy, J., P. Kelly, and A. McKay. 1998. Detection of ferulic acid esterase production by Bacillus spp. and lactobacilli. Appl. Microbiol. Biotechnol. 50:257-260. [DOI] [PubMed] [Google Scholar]
- 15.Fazary, A., and Y. Ju. 2007. Feruloyl esterases as biotechnological tools: current and future perspectives. Acta Biochim. Biophys. Sin. (Shanghai) 39:811-828. [DOI] [PubMed] [Google Scholar]
- 16.Fujita, A., H. Sasaki, A. Doi, K. Okamoto, S. Matsuno, H. Furuta, M. Nishi, T. Nakao, T. Tsuno, H. Taniguchi, and K. Nanjo. 2008. Ferulic acid prevents pathological and functional abnormalities of the kidney in Otsuka Long-Evans Tokushima fatty diabetic rats. Diabetes Res. Clin. Pract. 79:11-17. [DOI] [PubMed] [Google Scholar]
- 17.Furrie, E. 2006. A molecular revolution in the study of intestinal microflora. Gut 55:141-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez, C., M. Proudfoot, G. Brown, Y. Korniyenko, H. Mori, A. Savchenko, and A. Yakunin. 2006. Molecular basis of formaldehyde detoxification. Characterization of two S-formylglutathione hydrolases from Escherichia coli, FrmB and YeiG. J. Biol. Chem. 281:14514-14522. [DOI] [PubMed] [Google Scholar]
- 19.Guglielmetti, S., I. De Noni, F. Caracciolo, F. Molinari, C. Parini, and D. Mora. 2008. Bacterial cinnamoyl esterase activity screening for the production of a novel functional food product. Appl. Environ. Microbiol. 74:1284-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janes, L. E., A. C. Löwendahl, and R. J. Kazlauskas. 1998. Quantitative screening of hydrolase libraries using pH indicators: identifying active and enantioselective hydrolases. Eur. J. 4:2324-2331. [Google Scholar]
- 21.Kroon, P. A., C. B. Faulds, P. Ryden, J. A. Robertson, and G. Williamson. 1997. Release of covalently bound ferulic acid from fiber in the human colon. J. Agric. Food Chem. 45:661-667. [Google Scholar]
- 22.Liu, A. M. F., N. A. Somers, R. J. Kazlauskas, T. S. Brush, F. Zocher, M. M. Enzelberger, U. T. Bornscheuer, G. P. Horsman, A. Mezzetti, C. Schmidt-Dannert, and R. D. Schmid. 2001. Mapping the substrate selectivity of new hydrolases using colorimetric screening: lipases from Bacillus thermocatenulatus and Ophiostoma piliferum, esterases from Pseudomonas fluorescens and Streptomyces diastatochromogenes. Tetrahedron Asymmetry 12:545-556. [Google Scholar]
- 23.Lorca, G., A. Ezersky, V. Lunin, J. Walker, S. Altamentova, E. Evdokimova, M. Vedadi, A. Bochkarev, and A. Savchenko. 2007. Glyoxylate and pyruvate are antagonistic effectors of the Escherichia coli IclR transcriptional regulator. J. Biol. Chem. 282:16476-16491. [DOI] [PubMed] [Google Scholar]
- 24.Mastihuba, V., L. Kremnický, M. Mastihubová, J. Willett, and G. Côté. 2002. A spectrophotometric assay for feruloyl esterases. Anal. Biochem. 309:96-101. [DOI] [PubMed] [Google Scholar]
- 25.Matsuzaki, T., Y. Nagata, S. Kado, K. Uchida, S. Hashimoto, and T. Yokokura. 1997. Effect of oral administration of Lactobacillus casei on alloxan-induced diabetes in mice. APMIS 105:637-642. [PubMed] [Google Scholar]
- 26.Matsuzaki, T., Y. Nagata, S. Kado, K. Uchida, I. Kato, S. Hashimoto, and T. Yokokura. 1997. Prevention of onset in an insulin-dependent diabetes mellitus model, NOD mice, by oral feeding of Lactobacillus casei. APMIS 105:643-649. [DOI] [PubMed] [Google Scholar]
- 27.Matsuzaki, T., R. Yamazaki, S. Hashimoto, and T. Yokokura. 1997. Antidiabetic effects of an oral administration of Lactobacillus casei in a non-insulin-dependent diabetes mellitus (NIDDM) model using KK-Ay mice. Endocr. J. 44:357-365. [DOI] [PubMed] [Google Scholar]
- 28.Oliszewski, R., R. B. Medina, S. N. Gonzalez, and A. B. Perez Chaia. 2007. Esterase activities of indigenous lactic acid bacteria from Argentinean goats’ milk and cheeses. Food Chem. 101:1446-1450. [Google Scholar]
- 29.Plumb, G. W., M. T. Garcia-Conesa, P. A. Kroon, M. Rhodes, S. Ridley, and G. Williamson. 1999. Metabolism of chlorogenic acid by human plasma, liver, intestine and gut microflora. J. Sci. Food Agric. 79:390-392. [Google Scholar]
- 30.Roesch, L., G. Lorca, G. Casella, A. Giongo, A. Naranjo, A. Pionzio, N. Li, V. Mai, C. Wasserfall, D. Schatz, M. Atkinson, J. Neu, and E. Triplett. 19 February 2009. Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J. [Epub ahead of print.] doi: 10.1038/ismej.2009.5. [DOI] [PMC free article] [PubMed]
- 31.Rogosa, M., J. Mitchell, and R. Wiseman. 1951. A selective medium for the isolation and enumeration of oral lactobacilli. J. Dent. Res. 30:682-689. [DOI] [PubMed] [Google Scholar]
- 32.Sri Balasubashini, M., R. Rukkumani, and V. P. Menon. 2003. Protective effects of ferulic acid on hyperlipidemic diabetic rats. Acta Diabetol. 40:118-122. [DOI] [PubMed] [Google Scholar]
- 33.Srinivasan, M., A. Sudheer, and V. Menon. 2007. Ferulic acid: therapeutic potential through its antioxidant property. J. Clin. Biochem. Nutr. 40:92-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanida, M., T. Yamano, K. Maeda, N. Okumura, Y. Fukushima, and K. Nagai. 2005. Effects of intraduodenal injection of Lactobacillus johnsonii La1 on renal sympathetic nerve activity and blood pressure in urethane-anesthetized rats. Neurosci. Lett. 389:109-114. [DOI] [PubMed] [Google Scholar]
- 35.Topakas, E., C. Vafiadi, and P. Christakopoulos. 2007. Microbial production, characterization and applications of feruloyl esterases. Proc. Biochem. 42:497-509. [Google Scholar]
- 36.Vaarala, O., M. Atkinson, and J. Neu. 2008. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes 57:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter, J. 2008. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl. Environ. Microbiol. 74:4985-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, X., X. Geng, Y. Egashira, and H. Sanada. 2004. Purification and characterization of a feruloyl esterase from the intestinal bacterium Lactobacillus acidophilus. Appl. Environ. Microbiol. 70:2367-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williamson, G., A. Day, G. Plumb, and D. Couteau. 2000. Human metabolic pathways of dietary flavonoids and cinnamates. Biochem. Soc. Trans. 28:16-22. [DOI] [PubMed] [Google Scholar]
- 40.Yamano, T., M. Tanida, A. Niijima, K. Maeda, N. Okumura, Y. Fukushima, and K. Nagai. 2006. Effects of the probiotic strain Lactobacillus johnsonii strain La1 on autonomic nerves and blood glucose in rats. Life Sci. 79:1963-1967. [DOI] [PubMed] [Google Scholar]