Abstract
We report for the first time a quantitative mlrA gene-directed TaqMan PCR assay for the rapid detection of microcystin-degrading bacteria. This was applied, in combination with 16S ribosomal DNA-directed quantitative PCR and denaturing gradient gel electrophoresis, to study virgin sand filter column biofilm development and to correlate mlrA gene abundance with microcystin removal efficiency.
It has been predicted that the changing climatic conditions around the world are likely to increase both the occurrence and the intensity of blue-green algal (cyanobacterial) blooms (16). Of particular concern to the water industry are the blooms of Microcystis, Anabaena, Nostoc, and Planktothrix species, which are capable of producing microcystin toxins within surface water storages used for potable water supply (4, 5, 12). In dissolved (extracellular) form, microcystins are not efficiently removed by conventional water treatment processes (6), and more advanced treatment options, such as activated carbon application or ozonation, are usually employed. However, these are expensive alternatives, and removal efficiencies are often compromised by the presence of natural organic matter (15). Biological filtration of microcystins is now recognized as an alternative treatment barrier (1, 7, 8, 14) and is favored by water utilities, as the process is generally low technology, chemical free, and requires little maintenance, where retrofitting of the process into existing water treatment plant (WTP) infrastructure is often feasible.
To date, 10 different microcystin-degrading bacteria have been isolated from rivers, lakes, and biofilters (7), and the mlr gene cluster has been demonstrated to encode proteins involved in the initial steps of microcystin biodegradation by such organisms (2, 3). The MlrA protein is responsible for the initial hydrolytic cleavage of the cyclic microcystin structure, and conventional mlrA gene-directed PCR has been employed for qualitative detection of microcystin-degrading bacteria from lakes (18) and within the biofilm of biofilters (1, 7, 8). However, these conventional PCR assays do not allow for accurate quantitation of mlrA gene abundance and have not been designed with degenerate primer sequences to allow for variations that exist between different mlrA homologues. In this study, we report for the first time a quantitative mlrA gene-directed TaqMan PCR assay, including degenerate oligonucleotides targeting conserved DNA regions, for the rapid detection of microcystin-degrading bacteria.
Using all available mlrA nucleotide sequences to date (GenBank accession numbers DQ112243, AF411068, AB114203, AB161685, and AB114202), primers qmlrAf (5′-AGCCCKGGCCCRCTGC-3′) and qmlrAr (5′-ATGCCARGCCCACCACAT-3′) and the TaqMan probe qmlrA-tm, which was labeled with 6-carboxyfluorescein (FAM) at the 5′ end and labeled with black hole quencher 1 (BHQ1) at the 3′ end (5′-FAM-TGCCSCAGCTSCTCAAGAAGTTTG-BHQ1-3′), were designed to target highly conserved regions of the mlrA gene for quantitative TaqMan PCR. Reactions resulted in the amplification of a 120-bp product and were carried out in quadruplicate on a Rotor Gene 6000 (Corbett Research, New South Wales, Australia) thermal cycling system. Each 25-μl reaction mixture contained 200 μM of each deoxynucleoside triphosphate, 2.0 mM of MgCl2, 1× PCR buffer, 0.5 μM of primers qmlrAf and qmlrAr, 0.25 μM of TaqMan probe qmlrA-tm, 400 μg/ml bovine serum albumin, 0.5 U of Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA), and 2.5 μl of either a DNA standard or sample template. Thermal cycling conditions consisted of an initial denaturation at 95°C for 2 min followed by 45 cycles of denaturation at 95°C for 5 s and annealing/extension at 62°C for 25 s. Data were collected in the FAM channel (gain set to 6) at the end of each annealing/extension step.
Quantitation was performed using DNA standard curves constructed from a serial dilution, in Milli-Q water, of an 807-bp mlrA gene fragment (18) from the two microcystin-degrading bacteria Sphingomonas sp. strain ACM-3962 (2) and Sphingopyxis sp. strain LH21 (7). Results were linear over the range of 1 × 101 to 1 × 109 mlrA gene copies/μl with linear coefficient values (R2) for ACM-3962 and LH21 of 0.999 and 0.998 and reaction efficiencies of 0.97 and 0.99, respectively. Specificity of the mlrA TaqMan assay was verified by the absence of signals for genomic DNA from the non-microcystin-degrading bacteria Aeromonas hydrophila (ATCC 7966), Bacillus subtilis (ATCC 10145), Escherichia coli (ATCC 11775), Pseudomonas aeruginosa (ATCC 10145), and Staphylococcus epidermidis (ATCC 12228).
Determination of the DNA extraction efficiency, linear range, and the limit of detection of the TaqMan PCR assay under environmentally relevant conditions was achieved by the spiking of known amounts of Sphingopyxis sp. strain LH21 into Myponga reservoir water and Morgan WTP filter sand (containing a biofilm) in log10-fold increments ranging from 0 to 1 × 108 cells/ml or g, respectively. DNA from each water (0.5 ml) and sand preparation (0.5g) was then extracted in triplicate using the UltraClean soil DNA isolation kit (MoBio Laboratories Inc., Carlsbad, CA). For reservoir water, the TaqMan assay was linear over 6 log10 orders of magnitude, with an R2 of 0.998 and a limit of detection equivalent to 102 mlrA copies/ml. For sand filter medium, the TaqMan assay was linear over 5 log10 orders of magnitude, with an R2 equivalent to 0.998 and a limit of detection equivalent to 103 mlrA copies/g. These results are comparable to other quantitative PCR (qPCR) data cited in the literature, where the detection limit was 2 × 102 cells/ml for Methylocystis sp. from water (13) and approximately 103 cells/g for Escherichia coli O157:H7 (11) and Rhodococcus sp. (17) cells from soil.
The mlrA TaqMan assay was then applied, in conjunction with 16S rDNA qPCR (10) and 16S rDNA PCR-denaturing gradient gel electrophoresis (DGGE) cluster analysis (9), to investigate the attachment and subsequent biofilm formation upon virgin sand particles within a laboratory scale sand filter and also to investigate the previously unknown relationship between mlrA gene copy abundance and microcystin removal through biofiltration processes. Virgin sand (effective particle size, 1.25 mm; uniformity coefficient, 1.4; particle density, 1.62 g/cm3) (Riversands Pty Ltd, Carbrook, Queensland, Australia) was washed and sterilized by autoclaving. Sand was then packed into a glass column (length, 30 cm; internal diameter, 2.5 cm) at a bed height of 15 cm, and the column was continually fed with Myponga reservoir water (empty bed contact time, 15 min; UV254/cm, 0.412; dissolved organic carbon, 11.8 mg/liter; specific UV absorbance, 3.5 liters/mg-m; and pH, 6.7; South Australia) and spiked daily with microcystin-LR at a target concentration of 5 μg/liter. Full details of the sand column apparatus are described by Ho et al. (8). Influent and effluent water samples were taken at regular intervals for microcystin-LR determination by high-performance liquid chromatography (8). Sand samples (0.5 g) were sampled in triplicate from the top surface of the sand bed, and the biofilm DNA samples were extracted using the UltraClean soil DNA isolation kit (Mo Bio).
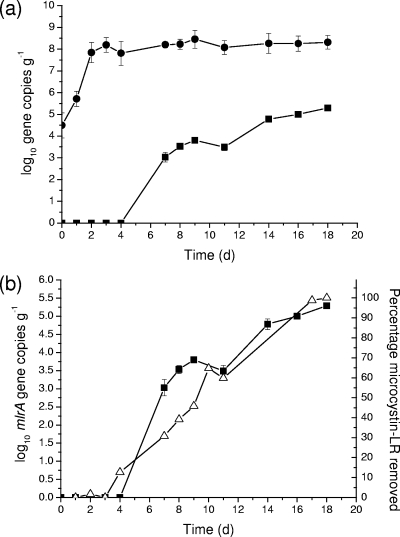
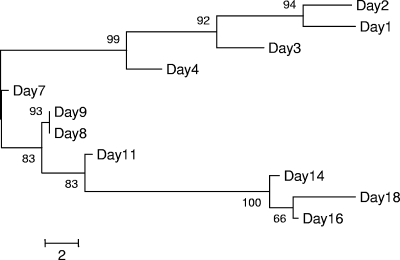
As shown in Fig. 1a, bacterial biofilm development (determined by 16S rDNA qPCR) appeared to begin within the first 24 h of start-up, and following day 3, the 16S bacterial abundance remained steady at approximately 1 × 108 copies/g. Despite the limited change in total bacterial abundance following day 3, 16S rDNA PCR-DGGE cluster analysis revealed a dynamic shift in the overall bacterial community composition throughout the 18-day study (Fig. 2). There was also a 3-day period before removals of microcystin were observed through the column (Fig. 1b). At day 4, there was a 12.6% removal of microcystin, although the abundance of mlrA-containing bacteria on day 4 remained below the limit of detection of the TaqMan assay (Fig. 1b). By day 7, the removal of microcystin had increased to 30.8% and was accompanied by the detection of 1.17 × 103 mlrA gene copies/g of sand. For the remainder of the study, there was a close association between the trend of microcystin removal and the abundance of mlrA gene copies/g of sand (Fig. 1b). These data suggested that the efficiency of microcystin removals by biofiltration processes was directly related to the abundance of microcystin-degrading bacteria within the sand filter biofilm. Within biofilters that have had no preexposure to microcystins, lag periods have been reported to be as short as 2 to 4 days (1, 8), but they have been reported to be up to 211 days for a filter containing virgin sand (19). In most cases, 100% removal of microcystin through biofilters usually occurs rapidly within several days following the lag period (8, 19), although complete removal of microcystin in this study was achieved 15 days following the 3-day lag period (Fig. 1b).
FIG. 1.
(a) Abundance of 16S rDNA (closed circles) and mlrA gene copies (closed squares) within the sand filter column biofilm. (b) Abundance of mlrA gene copies (closed squares) within the sand filter column biofilm and the percentage of microcystin-LR removed (open triangles) through the column. Error bars represent standard deviations of the results for triplicate analyses.
FIG. 2.
Cluster analysis of DGGE band profiles for the sand filter column study. Scale bar represents two band differences.
A clone library was then constructed to investigate the diversity of mlrA homologues detected by the mlrA TaqMan PCR assay within the sand filter column on day 18. Following sequence analysis of 50 cloned DNA fragments, three unique mlrA gene sequences, MC-A, MC-B, and MC-C, were obtained. Each sequence had greater than 98% similarity to previously described mlrA gene sequences, where MC-A, MC-B, and MC-C represented 22, 72, and 6% of the clone library, respectively.
In summary, this study has demonstrated the development of an mlrA gene-directed TaqMan PCR assay for the assessment of microcystin-degrading bacteria within biologically active sand filter biofilm. The data presented here revealed that during the early phase of operation of a sand filter column, the removal of microcystin was directly related to the abundance of microcystin-degrading bacteria within the biofilm, although removals began only once an adequate number of microcystin-degrading bacteria were established. It is envisaged that the TaqMan PCR assay will be valuable for WTP operators who wish to investigate the abundance of microcystin-degrading bacteria within their biofilters and, in turn, assess the capacity of the biofilters for removing microcystin toxins—especially as such episodes are particularly transient in nature.
Nucleotide sequence accession numbers.
The three unique mlrA gene sequences, MC-A, MC-B, and MC-C, obtained in this study have been deposited in GenBank under accession numbers FJ438525, FJ438526, and FJ438527, respectively.
Acknowledgments
We thank J. P. Rasmussen for his assistance with the design of the primers and probe reported in this study.
This work was funded by the Industry Cooperative Innovation Program (ICIP) of AusIndustry. We thank the Australian Water Quality Centre and the South Australian Water Corporation for supporting this work.
Footnotes
Published ahead of print on 5 June 2009.
REFERENCES
- 1.Bourne, D. G., R. L. Blakeley, P. Riddles, and G. J. Jones. 2006. Biodegradation of the cyanobacterial toxin microcystin LR in natural water and biologically active slow sand filters. Water Res. 40:1294-1302. [DOI] [PubMed] [Google Scholar]
- 2.Bourne, D. G., G. J. Jones, R. L. Blakeley, A. Jones, A. P. Negri, and P. Riddles. 1996. Enzymatic pathway for the bacterial degradation of the cyanobacterial cyclic peptide toxin microcystin LR. Appl. Environ. Microbiol. 62:4086-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourne, D. G., P. Riddles, G. J. Jones, W. Smith, and R. L. Blakeley. 2001. Characterisation of a gene cluster involved in bacterial degradation of the cyanobacterial toxin microcystin LR. Environ. Toxicol. 16:523-534. [PubMed] [Google Scholar]
- 4.Carmichael, W. W. 1992. Cyanobacteria secondary metabolites—the cyanotoxins. J. Appl. Bacteriol. 72:445-459. [DOI] [PubMed] [Google Scholar]
- 5.Codd, G. A. 1995. Cyanobacterial toxins: occurrence, properties and biological significance. Water Sci. Technol. 32:149-159. [Google Scholar]
- 6.Himberg, K., A. M. Keijola, L. Hiisvirta, H. Pyysalo, and K. Sivonen. 1989. The effect of water treatment processes on the removal of hepatotoxins from Microcystis and Oscillatoria cyanobacteria. A laboratory study. Water Res. 23:979-984. [Google Scholar]
- 7.Ho, L., D. Hoefel, C. P. Saint, and G. Newcombe. 2007. Isolation and identification of a novel microcystin-degrading bacterium from a biological sand filter. Water Res. 41:4685-4695. [DOI] [PubMed] [Google Scholar]
- 8.Ho, L., T. Meyn, A. Keegan, D. Hoefel, J. Brookes, C. P. Saint, and G. Newcombe. 2006. Bacterial degradation of microcystin toxins within a biologically active sand filter. Water Res. 40:768-774. [DOI] [PubMed] [Google Scholar]
- 9.Hoefel, D., P. T. Monis, W. L. Grooby, S. Andrews, et al. 2005. Profiling bacterial survival through a water treatment process and subsequent distribution system. J. Appl. Microbiol. 99:175-186. [DOI] [PubMed] [Google Scholar]
- 10.Hoefel, D., P. T. Monis, W. L. Grooby, S. Andrews, and C. P. Saint. 2005. Culture-independent techniques for rapid detection of bacteria associated with loss of chloramine residual in a drinking water system. Appl. Environ. Microbiol. 71:6479-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibekwe, A. M., P. M. Watt, P. J. Shouse, and C. M. Grieve. 2004. Fate of Escherichia coli O157:H7 in irrigation water on soils and plants as validated by culture method and real-time PCR. Can. J. Microbiol. 50:1007-1014. [DOI] [PubMed] [Google Scholar]
- 12.Keil, C., A. Forchert, J. Fastner, U. Szewzyk, W. Rotard, I. Chorus, and R. Kratke. 2002. Toxicity and microcystin content of extracts from a Planktothrix bloom and two laboratory strains. Water Res. 36:2133-2139. [DOI] [PubMed] [Google Scholar]
- 13.Kikuchi, T., K. Iwasaki, H. Nishihara, Y. Takamura, and O. Yagi. 2002. Quantitative and rapid detection of the trichloroethylene-degrading bacterium Methylocystis sp. M in groundwater by real-time PCR. Appl. Microbiol. Biotechnol. 59:731-736. [DOI] [PubMed] [Google Scholar]
- 14.Lee, Y. J., J. M. Jung, M. H. Jang, K. Ha, and G. J. Joo. 2006. Degradation of microcystins by adsorbed bacteria on a granular active carbon (GAC) filter during the water treatment process. J. Environ. Biol. 27:317-322. [PubMed] [Google Scholar]
- 15.Newcombe, G., D. Cook, S. Brooke, L. Ho, and N. Slyman. 2003. Treatment options for microcystin toxins: similarities and differences between variants. Environ. Toxicol. 24:299-308. [DOI] [PubMed] [Google Scholar]
- 16.Paerl, H. W., and J. Huisman. 2008. Climate. Blooms like it hot. Science 320:57-58. [DOI] [PubMed] [Google Scholar]
- 17.Rodrigues, J. L. M., M. R. Aiello, J. W. Urbance, T. V. Tsoi, and J. M. Tiedje. 2002. Use of both 16S rRNA and engineered functional genes with real-time PCR to quantify and engineered, PCB-degrading Rhodococcus in soil. J. Microbiol. Methods 51:181-189. [DOI] [PubMed] [Google Scholar]
- 18.Saito, T., K. Okana, H.-D. Park, T. Itayama, Y. Inamori, B. A. Neilan, B. P. Burns, and N. Sugiura. 2003. Detection and sequencing of the microcystin LR-degrading gene, mlrA, from new bacteria isolated from Japanese lakes. FEMS Microbiol. Lett. 229:271-276. [DOI] [PubMed] [Google Scholar]
- 19.Wang, H., L. Ho, D. M. Lewis, J. D. Brookes, and G. Newcombe. 2007. Discriminating and assessing adsorption and biodegradation removal mechanisms during granular activated carbon filtration of microcystin toxins. Water Res. 41:4262-4270. [DOI] [PubMed] [Google Scholar]