Abstract
The effect of agricultural management practices on geochemical cycles in moderate ecosystems is by far better understood than in semiarid regions, where fertilizer availability and climatic conditions are less favorable. We studied the impact of different fertilizer regimens in an agricultural long-term observatory in Burkina Faso at three different plant development stages (early leaf development, flowering, and senescence) of sorghum cultivars. Using real-time PCR, we investigated functional microbial communities involved in key processes of the nitrogen cycle (nitrogen fixation, ammonia oxidation, and denitrification) in the rhizosphere. The results indicate that fertilizer treatments and plant development stages combined with environmental factors affected the abundance of the targeted functional genes in the rhizosphere. While nitrogen-fixing populations dominated the investigated communities when organic fertilizers (manure and straw) were applied, their numbers were comparatively reduced in urea-treated plots. In contrast, ammonia-oxidizing bacteria (AOB) increased not only in absolute numbers but also in relation to the other bacterial groups investigated in the urea-amended plots. Ammonia-oxidizing archaea exhibited higher numbers compared to AOB independent of fertilizer application. Similarly, denitrifiers were also more abundant in the urea-treated plots. Our data imply as well that, more than in moderate regions, water availability might shape microbial communities in the rhizosphere, since low gene abundance data were obtained for all tested genes at the flowering stage, when water availability was very limited.
Land degradation is one of the most serious threats to food production on the African continent. Soil erosion, nutrient depletion, low organic matter content, and unfavorable pH values are some of the reasons for a deficient soil fertility (30), mainly in Central African countries. Combined with high variability and irregular distribution of rainfall, these factors contribute to negative nutrient balances. For example, 4.4 million tons of nitrogen (N) are lost per year in African soils, but only 0.8 million tons are reapplied by fertilization (12, 34). Since nitrogen is a key nutrient determining the productivity of agroecosystems (7, 11, 43), it is of central importance to optimize the nitrogen balance in these countries, mainly by steering the genetic resources of soil microbes in a way that losses of applied nitrogen are minimized and biological nitrogen fixation is increased. The aim should be to obtain a highly efficient nitrogen turnover, with leaching of nitrate and losses of gaseous products such as nitrous oxide (N2O) or dinitrogen (N2) as low as possible.
Despite the importance of this issue, not much data are available on microbial community structure and function related to the nitrogen cycle in agroecosystems of Central Africa, and scenarios from moderate climatic regions cannot simply be transferred to tropical agroecosystems. Furthermore, the few studies published thus far only investigated effects of agricultural management on a single process of the nitrogen turnover (18, 20, 32) and ignore the fact that nitrogen turnover is a network of closely interlinked processes.
Therefore, we sought to investigate the effects of different fertilizer regimens on multiple transformation processes within the nitrogen cycle in agroecosystems from semiarid areas in Central Africa. We investigated nitrogen dynamics on a full-cycle approach, including the most important steps in well-aerated agricultural soils (nitrification, denitrification, and nitrogen fixation). We hypothesized that each fertilizer regimen results in typical abundance pattern of the functional populations. While in moderate agroecosystems the plant (respectively, the plant development stage and the plant performance) plays an important role in shaping microbial community structure and function in the rhizosphere, we further postulated that nitrogen turnover and the corresponding populations are also influenced by the availability of water in semiarid soils. To test these hypotheses on a molecular basis, we quantified bacterial genes encoding the nitrogenase reductase (nifH), ammonia monooxygenase (amoA), and nitrite reductase (nirK and nirS), as well as archaeal amoA genes, by real-time PCR and linked the data to soil properties and grain yield.
MATERIALS AND METHODS
Location.
The experimental site is located at the Saria Agricultural Research Station (Saria II, 12°16′N, 2°9′W, 300-m altitude) in Burkina Faso. This site represents the North-Sudanese zone, with tropical climate and an average rainfall of 800 mm per year. The climate in Burkina Faso is characterized by two seasons: the dry season from October to April and the rainy season from May to September. The soil is strongly weather-beaten, eroded, and classified as ferric lixisol (13).
Site description and sampling strategy.
A long-term fertilizer experiment was established in 1980, including six replicates of six treatments in a randomized plot design. The treatments unamended control, control plus urea (C+U; 60 kg ha−1), straw (sorghum; 8.3 tons ha−1), straw plus urea (S+U), cattle manure (10 tons ha−1), and cattle manure plus urea (M+U) were applied to a continuous sorghum cropping system [Sorghum bicolor (L.) Moench] 2 months before sowing. Rhizosphere samples were collected according to standard procedures (28) in August (EC30, early leaf development), September (EC60, flowering stage), and November 2006 (EC90, senescence stage) by mixing roots of three plants per plot to form one composite sample. The six replicate plots were used as independent replicates. Rhizosphere samples were filled into cryotubes, shock frozen in solid carbon dioxide right after sampling, and stored at −80°C for nucleic acid extraction.
Soil physical and chemical properties.
Physical and chemical analyses have been performed in all treatments in soil samples taken at the flowering stage (i.e., EC30). Soil pH values were measured in 2 M KCl suspensions at a soil/liquid ratio of 1:2.5. The total amount of nitrogen and carbon was quantified by using the combustion system ThermoFinnigan Flash EA 1112 (ThermoFinnigan, France), and the colorimetric determination of total and available phosphorus (P) was performed according to the method of Dabin (10). Corresponding data are listed in Table 1.
TABLE 1.
Soil physical and chemical properties, plant biomass and grain yields of the differently amended plots
| Sample | Mean ± SDa
|
|||||
|---|---|---|---|---|---|---|
| Amt (mg g−1)
|
pH (KCl) | Amt (kg ha−1)
|
||||
| Total C | Total N | Total P | Grain yield | Plant biomass | ||
| Control | 2.16 ± 0.33A | 0.15 ± 0.05A | 0.11 ± 0.0AB | 4.58 ± 0.33A | 167 ± 174A | 480 ± 426A |
| C+U | 2.22 ± 0.77A | 0.16 ± 0.05AB | 0.10 ± 0.03A | 4.09 ± 0.17B | 207 ± 124A | 627 ± 286A |
| Straw | 2.48 ± 0.38A | 0.15 ± 0.11A | 0.11 ± 0.02AB | 5.19 ± 0.24CD | 90 ± 60A | 300 ± 192A |
| S+U | 2.69 ± 0.42A | 0.17 ± 0.07AB | 0.11 ± 0.02AB | 4.76 ± 0.23AC | 233 ± 154A | 600 ± 390A |
| Manure | 3.57 ± 0.2B | 0.27 ± 0.12B | 0.15 ± 0.02B | 5.51 ± 0.30D | 730 ± 319B | 1,893 ± 779B |
| M+U | 3.56 ± 0.43B | 0.25 ± 0.05AB | 0.15 ± 0.02B | 5.14 ± 0.36CD | 737 ± 350B | 1,900 ± 806B |
Significant differences between treatments are indicated by different superscript letters (n = 6).
Determination of ammonium (NH4+) and nitrate (NO3−) in rhizosphere soil.
Prior to chemical analyses, 3 g of rhizosphere soil was overhead shaken for 30 min with 12 ml of 0.01 M CaCl2. Each extract was filtered through a Millex HV Millipore filter (pore size, 0.45 μm). Ammonium and nitrate measurements were performed on a Nanocolor 300D photometer from Macherey-Nagel (Germany) by using the commercial kits Nanocolor Ammonium 3 and Nitrate 50 according to the manufacturer's protocol (data are indicated in Table 2).
TABLE 2.
Nitrate and ammonium concentrations in rhizosphere soil in different treatments at three different plant development stages
| Sample | Mean concn (mg of N kg of soil−1) ± SDa
|
|||||
|---|---|---|---|---|---|---|
| NO3+-N
|
NH4+-N
|
|||||
| EC30c | EC60 | EC90 | EC30 | EC60 | EC90 | |
| Control | 6.4 ± 3.6A1 | 5.4 ± 1.5A1 | 68.7 ± 36.9B1 | 1.9 ± 1.6A1 | 3.9 ± 2.7A1 | 1.1 ± 0.7A1 |
| C+U | 18.4 ± 13.2A1 | 7.9 ± 3.4A1 | 84.6 ± 42.5B1 | 4.2 ± 3.5A1 | 3.8 ± 2.1A1 | 1.6 ± 1.1A1 |
| Straw | 5.3 ± 3.5A1 | 6.2 ± 1.7A1 | 82.5 ± 44.9B1 | 1.5 ± 1.0A1 | 5.1 ± 1.9B1 | 3.7 ± 6.1AB1 |
| S+U | 19.2 ± 13.8A1 | 8.9 ± 3.0A1 | 136.9 ± 70.6B1 | 1.7 ± 1.0A1 | 5.8 ± 1.9B1 | 5.7 ± 6.2AB1 |
| Manure | 17.4 ± 15.9A1 | 6.6 ± 2.9A1 | 97.3 ± 41.1B1 | 2.4 ± 0.6A1 | 6.5 ± 2.0B1 | 2.2 ± 1.9A1 |
| M+U | 36.1 ± 20.8A1 | (12.4) | 74.9 ± 34.1B1 | 3.1 ± 1.7A1 | (5.6) | 1.8 ± 2.0A1 |
n = 6; n = 1 for data in parentheses. EC30, young leaf development; EC60, flowering; EC90, senescence. Significant differences of one treatment over three different plant development stages are indicated by different superscript letters. Significant differences among treatments at one plant development stage are indicated by different superscript numbers.
Nucleic acid extraction.
DNA was extracted from 0.5 g of soil by using the method described by Griffiths et al. (15). Extraction was performed by using Precellys-Keramik kit lysing tubes (PEQLAB Biotechnologie GmbH, Germany) in combination with the Bertin Precellys 24 beat-beading system (Bertin Technologies, France). DNA yield and purity were measured by using a microvolume fluorospectrometer (NanoDrop Technologies, Delaware).
Real-time PCR assay.
Absolute quantification of all investigated genes was carried out in triplicate on the ABI Prism 7300 Cycler (Applied Biosystems, Germany). The following reagents were used for the real-time PCR assay: bovine serum albumin (Sigma-Aldrich, Germany), primers (5, 17, 21, 27, 31, 33, 37, 42) (Metabion, Germany), dimethyl sulfoxide (Sigma, Germany), and Power SYBR Green PCR master mix (Applied Biosystems). The composition of each reaction mix is given in Table 3. All PCR runs started with an initial enzyme activation step performed at 95°C for 10 min. The subsequent thermal profile was different for each gene, as indicated in Table 4. The specificity of the amplification products was confirmed by melting-curve analysis, and the expected sizes of the amplified fragments were checked in a 1.5% agarose gel stained with ethidium bromide.
TABLE 3.
Reaction components of a 25-μl Mastermix assay used for quantification of the functional target genes involved in nitrogen turnover
| Target metabolic group | Target gene | Amt (μl) of assay component
|
||||||
|---|---|---|---|---|---|---|---|---|
| 2× Power SYBR green | 3% BSA | Primer (10 pmol μl−1)
|
Dimethyl sulfoxide | DNA | PCR water | |||
| Forward | Reverse | |||||||
| Nitrogen-fixing bacteria | nifH | 12.5 | 0.5 | 0.30 | 0.30 | 2 | 9.4 | |
| AOB | amoA | 12.5 | 0.5 | 0.75 | 0.75 | 2 | 8.5 | |
| AOA | amoA | 12.5 | 0.5 | 0.50 | 0.50 | 2 | 9.0 | |
| Denitrifying bacteria | nirK | 12.5 | 0.5 | 0.50 | 0.50 | 0.625 | 2 | 8.375 |
| nirS | 12.5 | 0.5 | 0.50 | 0.50 | 0.625 | 2 | 8.375 | |
TABLE 4.
Primer sets and thermal profiles used for the absolute quantification of functional target genes involved in nitrogen turnover
| Target gene | Primer set | Reference | Thermal cycling profile | No. of cycles |
|---|---|---|---|---|
| nifH | nifH-F-Rösch | 30 | 95°C/45 s, 55°C/45 s, 72°C/45 s | 40 |
| nifH-R-Rösch | ||||
| amoA (AOB) | amoA-1F | 32 | 94°C/60 s, 60°C/60 s, 72°C/60 s | 39 |
| amoA-2R | ||||
| amoA (AOA) | 19F | 21 | 94°C/45 s, 50°C/45 s, 72°C/45 s | 40 |
| CrenamoA616r48x | 36 | |||
| nirK | nirK-876 | 17 | 95°C/15 s, 63°C-58°C/30 s, 72°C/30 s | 6a |
| nirK-5R | 4 | 40 | ||
| nirS | nirS-cd3af | 26 | 94°C/60 s, 57°C/60 s, 72°C/60 s | 39 |
| nirS-R3cd | 40 |
Touch down.
To test possible inhibitory effects on quantitative PCR amplification caused by coextracted humic substances, the optimal dilution for each DNA extract was determined by pre-experiments (data not shown). Dilution series of plasmid DNA targeting the bacterial nitrogen cycle genes (nifH, amoA, nirS, and nirK) and the fosmid clone 54d9 targeting archaeal amoA genes (referenced in Table 4) were used to generate a standard curve for each of the five target genes (standard dilutions used for creating a standard curve ranged from 101 to 106 gene copies/μl). The amplification efficiencies were calculated by using the formula Eff = [10(−1/slope) − 1].
Statistical analyses.
Data were analyzed by multifactorial analysis of variance (ANOVA; see Tables 1, 2, and 5) with the independent variables plant development stage, organic fertilizers (straw residues, manure) and urea treatment (3 × 3 × 2). Normal distribution of the residuals was checked by using histograms and the Kolmogorov-Smirnov test. If the requirement was not met, data were log transformed prior to analysis. The homogeneity of the variances was checked by the Levene test. For pairwise comparison of means, the Tukey's test was applied, and if the homogeneity of the variances was not given, a Games-Howell test was used. Due to the observed interactions between plant development stage and the different fertilizing treatments, a two-way ANOVA was carried out for each growth stage in order to explain the effects of the treatments (organic fertilizers and urea) and interactions on gene abundances for EC30, EC60, and EC90 separately (3 × 2 for each plant development stage). The statistical significance was set at P < 0.05. Statistical analysis was performed by using SPSS 13.0 (SPSS, Inc.).
TABLE 5.
Statistical evaluation of gene abundance by multifactorial ANOVA
| Factor |
Pa
|
||||
|---|---|---|---|---|---|
| nifH | amoA (AOB) | amoA (AOA) | nirS | nirK | |
| Total | |||||
| EC | 0.00016* | 0.00187* | 0.00000* | 0.00000* | 0.00000* |
| Organic | 0.00584* | 0.00010* | 0.00663* | 0.01226* | 0.00110* |
| Urea | 0.86490 | 0.00000* | 0.76170 | 0.03195* | 0.00897* |
| EC × organic | 0.01611* | 0.06726 | 0.75901 | 0.00226* | 0.02470* |
| EC × urea | 0.97056 | 0.01246* | 0.94151 | 0.77689 | 0.26311 |
| Organic × urea | 0.88557 | 0.91020 | 0.40479 | 0.48152 | 0.67149 |
| EC × organic × urea | 0.96940 | 0.34970 | 0.70371 | 0.11885 | 0.20102 |
| EC30 | |||||
| Organic | 0.21544 | 0.01357* | 0.01120* | 0.00461* | 0.00226* |
| Urea | 0.90946 | 0.00011* | 0.92578 | 0.08370 | 0.03994* |
| Organic × urea | 0.95764 | 0.50181 | 0.41531 | 0.54840 | 0.31779 |
| EC60 | |||||
| Organic | 0.00028* | 0.01036* | 0.44637 | 0.00033* | 0.12915 |
| Urea | 0.91401 | 0.00004* | 0.76804 | 0.44207 | 0.79858 |
| Organic × urea | 0.75057 | 0.43477 | 0.69985 | 0.98800 | 0.68548 |
| EC90 | |||||
| Organic | 0.78713 | 0.06200 | 0.12951 | 0.48282 | 0.72698 |
| Urea | 0.62794 | 0.000109* | 0.74827 | 0.19272 | 0.01398* |
| Organic × urea | 0.71996 | 0.30539 | 0.19268 | 0.08998 | 0.07509 |
The P values describe the impact of plant development stages and nitrogen fertilizing treatments on functional genes involved in nitrogen turnover. Asterisks indicate significant effects of plant development stages or fertilizing treatments, respectively, on the functional genes, after analysis by Tukey's test with SPSS 13.0.
RESULTS
Soil physical and chemical properties.
The pH values and total C, N, and P contents were measured in soil samples in all treatments at all plant development stages (Table 1). The pH values were low in general and revealed clear differences between the urea-treated plots (<4.76) and plots without urea application (<5.51). As expected, total C, N, and P contents were low in all plots compared to agro-ecosystems in moderate climate zones (38). However, a clear influence of fertilizer regimens was visible with the highest C, N, and P concentrations (Table 1) in the manure-amended plots (3.56 to 3.57 mg of C g of soil−1, 0.25 to 0.27 mg of N g of soil−1, and 0.15 mg of P g of soil−1). Since the C and N values responded similarly to the different treatments, the C/N ratios did not vary significantly between the treatments.
Ammonium and nitrate concentrations in the rhizosphere.
Ammonium concentrations in the rhizosphere of all treatments were low at the young leaf development (EC30) and the plant senescence (EC90) stages, with less than 5.7 mg kg of soil−1. The highest amounts of ammonium were found at the flowering stage (EC60) and reached 6.5 mg kg of soil−1 in the manure amendment. Despite enhanced ammonium values in the straw and manure treatments, significant responses to the treatments were not found (Table 2).
Nitrate concentrations were lowest at EC60, independent of the different treatments. The amounts of nitrate reached 8.9 mg kg of soil−1 at the mentioned sampling time point and were significantly increased at EC90 with up to 136.9 mg kg of soil−1. The nitrate values did not show significant responses to the treatments. However, the amounts of nitrate were always higher in the urea-treated plots (136.9 mg kg of soil−1) than in plots not receiving urea (97.3 mg kg of soil−1) (Table 2).
Plant biomass and grain yield.
Highest plant biomass yields were obtained from the manure-amended plots ranging between 1,893 kg ha−1 (manure) and 1,900 kg ha−1 (M+U). Interestingly, straw amendment reduced plant biomass yields (300 kg ha−1) compared to the untreated control (480 kg ha−1). A certain but not significant stimulatory effect of the urea application was visible (C+U, 627 kg ha−1; S+U, 600 kg ha−1). Grain yields followed the same trends described above for overall plant biomass yields (control, 167 kg ha−1; C+U, 207 kg ha−1; straw, 90 kg ha−1; S+U, 233 kg ha−1; manure, 730 kg ha−1; M+U, 737 kg ha−1). Regarding the fitness of the plant, we obtained not only higher plant and grain yields in manure-amended plots but also a more vigorous plant development compared to the other treatments, independent of plant development stage.
Abundance of functional genes involved in nitrogen cycling.
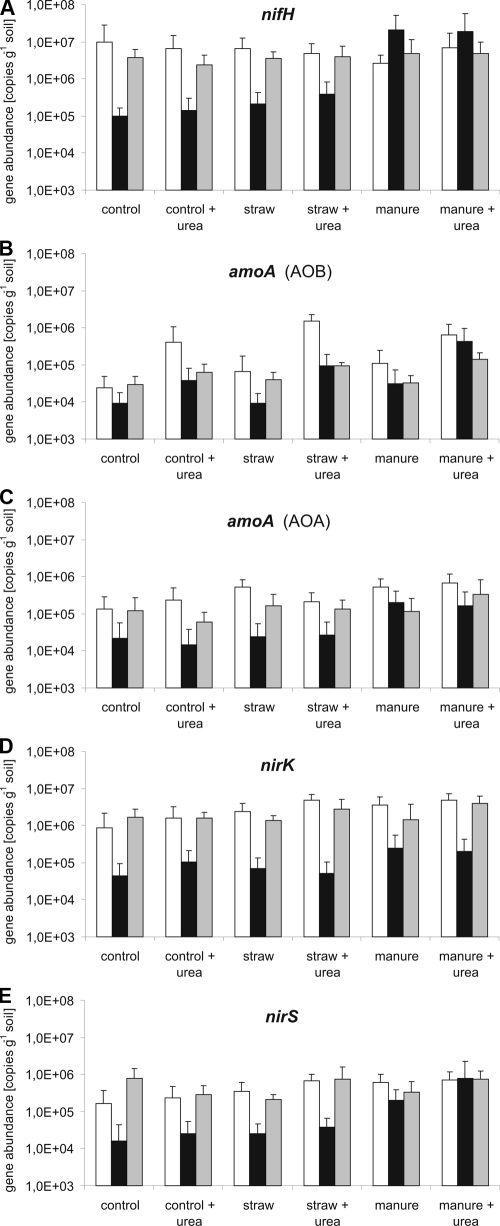
Real-time PCR was used to quantify the population size of nitrogen fixers (nifH), ammonia oxidizing bacteria (AOB-amoA) and ammonia-oxidizing archaea (AOA-amoA), and denitrifiers (nirK and nirS) of rhizosphere samples taken at three different plant development stages. All measured microbial genes showed a clear response to the different sampling time points, as well as fertilizer treatments (Table 5 and Fig. 1).
FIG. 1.
Copy numbers of functional genes involved in nitrogen cycling per gram of soil under different fertilizing regimens (unamended control, C+U, straw, S+U, manure, and M+U) at three different plant development stages (EC30, young leaf development [□]; EC60, flowering [▪]; and EC90, senescence [░⃞]). (A) nifH; (B) amoA (AOB); (C) amoA (AOA); (D) nirK; (E) nirS.
(i) Bacterial nifH genes.
Independent of treatment, similar nifH gene copy numbers were observed at EC30 and EC90, revealing 2.4 × 106 to 9.7 × 106 copies g of soil−1 (Fig. 1A). The copy numbers at EC60, however, differed distinctly from EC30 and EC90, with reduced numbers in control and straw-amended plots and enhanced numbers in manure-amended plots (see below). An influence of straw residues and urea on the nifH gene abundance was not apparent. Interestingly, nifH gene abundance was influenced by manure application. Both manure treatments (manure only and M+U) resulted in the highest nifH gene copy numbers, with up to 2.1 × 107 copies g of soil−1, but this was only the case at the flowering stage of the plant. For all other fertilizers, an opposite trend was observed, with low copy numbers measured at EC60 ranging from 9.9 × 104 to 4.0 × 105 gene copies g of soil−1.
(ii) Bacterial and archaeal amoA genes.
Overall, bacterial amoA gene abundance was highest at the young leaf development, revealing 1.5 × 106 copies g of soil−1, and decreased significantly in all treatments toward the flowering stage, amounting to <4.3 × 105 copies g of soil−1 (Fig. 1B and C). Among the different treatments, urea amendment had the strongest impact on the community size of bacterial ammonia oxidizers. Their number was at any plant development stage significantly increased compared to plots that did not receive urea. Even though organic treatments influenced the community size of bacterial ammonia oxidizers, this effect was less pronounced than the urea effect. The impact of straw was mainly apparent at the EC30 stage, while the manure effect was mainly apparent at EC30 and EC90.
The archaeal amoA gene copy numbers were significantly increased at EC30 and EC90 (6.0 × 104 to 6.6 × 105 copies g of soil−1) compared to EC60 (1.5 × 104 to 2.5 × 105 copies g of soil−1). Interestingly, whereas AOB were affected by urea, this was not true for AOA at any plant development stage. In contrast, organic treatments influenced the AOA mainly at the EC30 stage, resulting in higher gene copy numbers (5.3 × 105 copies g of soil−1) compared to the control plots (1.4 × 105 copies g of soil−1). Independent of the plant development stage, AOA outnumbered AOB when urea was not applied, with AOA/AOB ratios of >1 (control, < 5.6; straw, <7.9; manure, <4.8). In contrast, we obtained values of ≤1 for the AOA/AOB ratios in urea-amended plots (data not shown in detail), except for S+U (1.4) and M+U (2.4) at EC90.
(iii) Bacterial nirS and nirK genes.
To investigate the denitrification potential in the rhizosphere, we quantified two complementary genes encoding the nitrite reductase (nirS and nirK) (Fig. 1D and E). Gene abundances were clearly influenced by the plant development stage, with significantly increased gene copy numbers at the early plant development and the senescence stages. The nirK gene copy numbers ranged from 8.8 × 105 to 4.8 × 106 copies g of soil−1, and the nirS gene copy numbers ranged from 1.6 × 105 to 7.9 × 105 copies g of soil−1 at the mentioned sampling time points. The tendency of significantly reduced copy numbers at the flowering stage, as observed for all targeted microbial communities, remained the same for the denitrifiers, amounting to 4.3 × 104 to 2.5 × 105 nirK genes g of soil−1 and 1.6 × 104 to 7.7 × 105 nirS genes g of soil−1. This phenomenon could be observed in all treatments. However, nirK and nirS gene abundances were not only influenced by the plant development stage but also by fertilization. Although the abundance of nirK genes was enhanced by manure, straw, and urea mainly at EC30, nirS genes were increased by manure and straw at EC30 and only by manure at EC60. Overall, the two genes showed similar abundance patterns. However, lower nirS gene abundances were found compared to nirK in all treatments independent of the plant development stage.
DISCUSSION
The relevance of rhizosphere microorganisms for sustainable agricultural ecosystems was neglected for a long time. Only in the last decade, the impact of exudates on microbial communities in the rhizosphere and their nutrient supply to plants have been studied in more detail (2). However, semiarid ecosystems have yet to be examined with regard to the response of microbial communities in the rhizosphere to agricultural amendments and their role in agricultural sustainability.
Our results reveal that the abundances of all measured microbial groups involved in nitrogen turnover varied distinctly between the different plant development stages (P < 0.0002) (Fig. 1). Surprisingly, the abundances of all measured genes were significantly reduced at EC60 in all treatments, a finding which is in contrast to other studies from moderate regions. Most authors relate their results to enhanced root exudation during the flowering stage and high amounts of nutrients in the rhizosphere at that time point of plant development (6, 22), which are optimal conditions for nitrogen fixers (6) and denitrifiers (25, 26). However, in our experiment water availability in the soil was very low, since at the end of the rainy season (August and September 2006) precipitation fell short. Consequently, soil humidity (data not shown) was reduced in all treatments at the flowering stage. Nitrate concentrations at EC60 were also 3-fold reduced in urea-amended plots compared to those at EC30 and up to 15-fold reduced compared to those at EC90, which compromised the plant growth as well as the root exudation. Since the amount and composition of root exudates depend on environmental conditions (23), we suggest a combined effect of plant development stage and climatic conditions, which drive the abundance pattern of the genes under investigation. Since we performed a field experiment and both parameters are interlinked, it cannot be resolved which factor was more important for shaping microbial communities involved in nitrogen turnover. The different fertilization regimens clearly affected the community size of the functional groups as well, although to a lesser extent than the plant development stages.
nifH.
Although straw residues and urea did not induce any response from the nitrogen-fixing community, a strong effect of manure (P < 0.0003) on the nifH abundance became evident at the flowering stage (EC60). Likewise, the most vigorous plant growth was found in the manure-amended plots, where elevated concentrations of total carbon, nitrogen, and phosphorus have also been observed (Table 1), which might have been established over the years due to the continuous fertilizer application. The content of organic matter and the soil structure seem to be improved in a way that the decremental effects of the dry periods were reduced and the high abundance of nitrogen fixers, which can be explained by the increased ammonium concentrations (Table 2) in the manure-amended plots at EC30, might be based on a higher rhizodeposition. In contrast, low C and P concentrations were found in straw- and urea-amended plots. Furthermore, the same treatments yielded low nifH gene copy numbers at EC60, indicating that a lack of macronutrients not only limits the effectivity of biological nitrogen fixation (14, 35) by free-living diazotrophs but also reduces the population size of diazotrophs in semiarid ecosystems.
amoA-AOB.
AOB revealed a strong response (P < 0.0001) to urea application, as indicated by elevated amoA gene abundances in urea-amended plots and reduced gene abundances in plots that did not receive urea. Soil pH value seems to be an important factor for the abundance of bacterial ammonia oxidizers, as observed by Shen et al. (39). In contrast to our study, their experiments were conducted in an alkaline sandy loam with the application of NPK fertilizers in the form of urea. A decrease in the pH by 0.3 increased the bacterial amoA abundance 22.5-fold. Similar results were obtained in our study, with 23.1-fold-increased amoA gene copies in urea-treated plots, where pH values were reduced by 0.37 to 0.49. The phenomenon can be explained by the fact that, under acidic conditions, urea is assimilated preferentially and consequently nitrate accumulates (24). This is in line with our study, where elevated nitrate concentrations were obtained in urea-amended plots. The amounts of nitrate reached up to 136.9 mg of N kg of soil−1 and were thus almost three times higher than the nitrate concentrations in plots that did not receive urea.
amoA-AOA.
In contrast to AOB, the abundance of AOA was not affected by urea application. A recent study from Hatzenpichler et al. (16) showed that archaeal ammonia oxidizers were even inhibited by enhanced urea and ammonia concentrations. Determination of the AOB/AOA ratio indicated that AOB were predominant in urea-amended plots, ranging from 1.0 to 7.4, except in S+U-amended soil (0.7) and M+U-amended soil (0.4) at plant senescence. On the other hand, AOA were predominant in plots without urea application, with AOA/AOB ratios ranging from 2.4 to 7.9, independent of the plant development stage. Assuming 2.5 amoA gene copies per AOB cell and 1 amoA gene copy per AOA cell (21), the AOA/AOB ratio on a cell-based calculation was in the range of 5.9 to 19.7 independent of urea application and plant development stage. This finding is in accordance with recently published data from Chen et al. (9), who obtained ratios from 1.2 up to 69.3. The observation of AOA being predominant over AOB has been confirmed by several studies investigating different soils (1, 21) and in the rhizosphere of terrestrial plants (40, 41). However, even though few studies exhibited the opposite phenomenon, namely, that AOB outnumber AOA in some estuaries (4, 8, 36), it has been shown that AOA are more stable and do not respond as sensitively to environmental differences as their bacterial counterparts (36), as revealed by our study.
nirS and nirK.
Other than the two functionally redundant populations involved in the oxidation of ammonia (AOA and AOB), which were influenced differently by the investigated treatments, the two functionally redundant groups involved in nitrite reduction (harboring nirS and nirK as functional genes) showed similar response patterns in all treatments. However, the nirK copy numbers were larger in all cases compared to nirS. It is generally accepted that nitrogen fertilizers promote denitrification in agricultural soils (19, 29), which was confirmed by our study where organic fertilizers (manure and straw) increased the denitrifier population (nirS, P < 0.0046; nirK, P < 0.0023) significantly. However, the nirK gene abundance was even more increased when the organic fertilizers were applied in combination with urea. Similarly, Arcara et al. (3) reported low N2O-N losses from a maize-cropped soil when urea or pig slurry were applied; however, the combination of both fertilizers produced an increase in N2O emissions due to denitrification, which is in line with our increased gene abundance data.
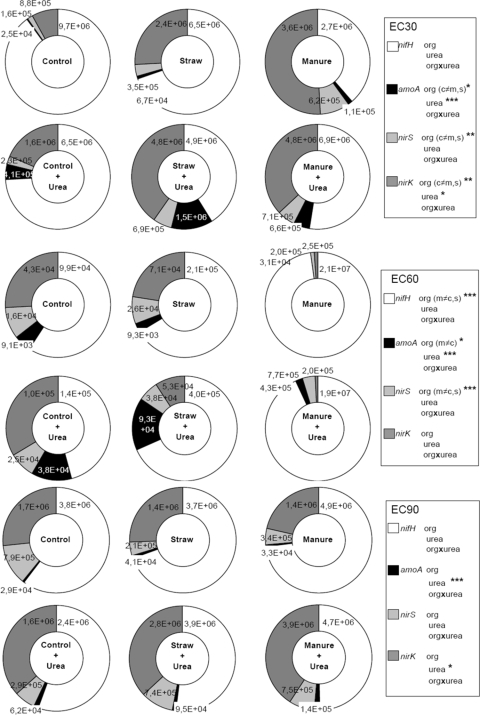
Our central hypothesis that each fertilizer treatment results in a typical ratio of nitrogen-fixing, ammonia-oxidizing, and denitrifying microbes is confirmed by Fig. 2, which shows clear alterations in the proportions of the different communities due to the fertilizer regimens. Interestingly, the most remarkable and obvious shifts were found in the manure-treated soils at the flowering stage, where the size of the nitrogen-fixing population was increased by a factor of 209. In contrast to the findings with the urea-treated plots, where an increase in nitrogen-fixing bacteria was accompanied by an increase in denitrifiers (C+U, 1.1; S+U, 4.4), in the manure-amended plots the ratio of nitrogen fixers and denitrifiers increased significantly (manure only, 46.2; M+U, 19.5). This might be a reason for the enormous plant growth and grain formation (enhanced plant biomass with 303 to 394%; enhanced grain yield with 354 to 436%) determined in manure-amended plots, since obviously the formed ammonium is not used by bacteria for nitrification and denitrification but by the plant for biomass production.
FIG. 2.
Effect of one particular fertilizing treatment on all bacterial populations measured in the present study (nifH, amoA [AOA and AOB], nirS, and nirK) at three different plant development stages (EC30, young leaf development; EC60, flowering; EC90, senescence). Significance (ANOVA): ***, P < 0.001; **, P < 0.001; *, P < 0.05. Abbreviations: org (organic) = manure + straw, m = manure, s = straw; e.g., c≠m = manure is significantly different from the control.
Summary and outlook.
Our data show the influence of different fertilizer treatments on the abundance of selected functional groups and the role of microbes in plant growth promotion in soils from semiarid regions. The presented data describe the sustainability of a particular treatment as population size of functional groups shape soil quality and turnover kinetics in the long term. As indicated by the ring diagrams (Fig. 2), plant and grain yields might benefit from reduced nitrogen losses by denitrification and enhanced biological nitrogen fixation in the manure treatments. However, the measured gene abundances only reflect a microbial potential for metabolizing nitrogen compounds and do not describe actual turnover rates in soils. Therefore, investigations of gene expression and enzyme activity and stability remain to be performed to compare the presence of functional groups with their activities and actual turnover rates. Furthermore, repeating our study in successive seasons and on additional sites would elucidate whether our data could be generalized for semiarid ecosystems. Finally, greenhouse experiments under controlled climatic conditions and using 13C-labeled carbon dioxide could clarify which factor was the main driver for the reduced gene abundances at the flowering stage: the plant development stage or the climate.
Acknowledgments
The Microbes Project was financially supported by the Agence Nationale de la Recherche.
We gratefully acknowledge Anton Hartmann for constructive comments on the manuscript; Sven Leininger for providing the AOA standard; and the anonymous reviewers, whose suggestions improved the manuscript substantially.
Footnotes
Published ahead of print on 5 June 2009.
REFERENCES
- 1.Adair, K., and E. Schwartz. 2008. Evidence that ammonia-oxidizing archaea are more abundant than ammonia-oxidizing bacteria in semiarid soils of northern Arizona, USA. Microb. Ecol. 56:420-426. [DOI] [PubMed] [Google Scholar]
- 2.Alami, Y., W. Achouak, C. Marol, and T. Heulin. 2000. Rhizosphere soil aggregation and plant growth promotion of sunflowers by an exopolysaccharide-producing Rhizobium sp. strain isolated from sunflower roots. Appl. Environ. Microbiol. 66:3393-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arcara, P. G., C. Gamba, D. Bidini, and R. Marchetti. 1999. The effect of urea and pig slurry fertilization on denitrification, direct nitrous oxide emission, volatile fatty acids, water-soluble carbon and anthrone-reactive carbon in maize-cropped soil from the Po plain (Modena, Italy). Biol. Fertil. Soils 29:270-276. [Google Scholar]
- 4.Boyle-Yarwood, S. A., P. J. Bottomley, and D. D. Myrold. 2008. Community composition of ammonia-oxidizing bacteria and archaea in soils under stands of red alder and Douglas fir in Oregon. Environ. Microbiol. 10:2956-2965. [DOI] [PubMed] [Google Scholar]
- 5.Braker, G., A. Fesefeldt, and K.-P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bürgmann, H., S. Meier, M. Bunge, F. Widmer, and J. Zeyer. 2005. Effects of model root exudates on structure and activity of a soil diazotroph community. Environ. Microbiol. 7:1711-1724. [DOI] [PubMed] [Google Scholar]
- 7.Cabello, P., M. D. Roldan, and C. Moreno-Vivian. 2004. Nitrate reduction and the nitrogen cycle in archaea. Microbiology 150:3527-3546. [DOI] [PubMed] [Google Scholar]
- 8.Caffrey, J. M., N. Bano, K. Kalanetra, and J. T. Hollibaugh. 2007. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J. 1:660-662. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X.-P., Y.-G. Zhu, Y. Xia, J.-P. Shen, and J.-Z. He. 2008. Ammonia-oxidizing archaea: important players in paddy rhizosphere soil? Environ. Microbiol. 10:1978-1987. [DOI] [PubMed] [Google Scholar]
- 10.Dabin, B. 1965. Application des dosages automatiques à l'analyse des sols. Cah. Orstom. Ser. Pedol. 3:335-366. [Google Scholar]
- 11.Dodds, W. K., M. A. Evans-White, N. M. Gerlanc, L. Gray, D. A. Gudder, M. J. Kemp, A. L. López, D. Stagliano, E. A. Strauss, J. L. Tank, M. R. Whiles, and W. M. Wollheim. 2000. Quantification of the nitrogen cycle in a prairie stream. Ecosystems 3:574-589. [Google Scholar]
- 12.FAO. 1995. FAO fertilizer yearbook 44. Food and Agriculture Organization of the United Nations, Rome, Italy.
- 13.FAO/UNESCO. 1994. Soil map of the world: ISRIC. Food and Agriculture Organization of the United Nations, Wageningen, The Netherlands.
- 14.Giller, K. E., and K. J. Wilson. 1991. Nitrogen fixation in tropical cropping systems. CABI, Wallingford, United Kingdom.
- 15.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatzenpichler, R., E. V. Lebecleva, E. Spieck, K. Stoecker, A. Richter, H. Daims, and M. Wagner. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. USA 105:2134-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry, S., E. Baudoin, J. C. Lopez-Gutierrez, F. Martin-Laurent, A. Baumann, and L. Philippot. 2004. Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J. Microbiol. Methods 59:327-335. [DOI] [PubMed] [Google Scholar]
- 18.Hermansson, A., and P.-E. Lindgren. 2001. Quantification of ammonia-oxidizing bacteria in arable soil by real-time PCR. Appl. Environ. Microbiol. 67:972-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser, E. A., K. Korths, K. Kucke, E. Schnug, J. C. Munch, and O. Heinemaeyer. 1998. Nitrous oxide release from cultivated soils: influence of different type of N-fertilizer. Biol. Fertil. Soils 28:36-43. [Google Scholar]
- 20.Kandeler, E., K. Deiglmayr, D. Tscherko, D. Bru, and L. Philippot. 2006. Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl. Environ. Microbiol. 72:5957-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, J. I. Prosser, S. C. Schuster, and C. Schleper. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 22.Lu, Y., R. Wassmann, H. U. Neue, and C. Huang. 1999. Impact of phosphorus supply on root exudation, aerenchyma formation, and methane emission of rice plants. Biogeochemistry 47:203-218. [Google Scholar]
- 23.Lynch, J. M., and J. M. Whipps. 1990. Substrate flow in the rhizosphere. Plant Soil 129:1-10. [Google Scholar]
- 24.Marsh, K. L., G. K. Sims, and R. L. Mulvaney. 2005. Availability of urea to autotrophic ammonia-oxidizing bacteria as related to the fate of 14C- and 15N-labeled urea added to soil. Biol. Fertil. Soils 42:137-145. [Google Scholar]
- 25.Mayer, J., F. Buegger, E. Jensen, M. Schloter, and J. Heß. 2004. Turnover of grain legume N rhizodeposits and effect of rhizodeposition on the turnover of crop residues. Biol. Fertil. Soils 39:153-164. [Google Scholar]
- 26.Mayer, J., F. Buegger, E. S. Jensen, M. Schloter, and J. Heß. 2003. Estimating N rhizodeposition of grain legumes using a 15N in situ stem labelling method. Soil Biol. Biochem. 35:21-28. [Google Scholar]
- 27.Michotey, V., V. Mejean, and P. Bonin. 2000. Comparison of methods for quantification of cytochrome cd1-denitrifying bacteria in environmental marine samples. Appl. Environ. Microbiol. 66:1564-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milling, A., K. Smalla, F. Maidl, M. Schloter, and J. Munch. 2004. Effects of transgenic potatoes with an altered starch composition on the diversity of soil and rhizosphere bacteria and fungi. Plant Soil 266:23-39. [Google Scholar]
- 29.Mulvaney, R. L., S. A. Khan, and C. S. Mulvaney. 1997. Nitrogen fertilizers promote denitrification. Biol. Fertil. Soils 24:211-220. [Google Scholar]
- 30.Murwira, H. K. 2003. Managing Africa's soils: approaches and challenges, p. 293-306. In M. P. Gichuru, A. Bationo, M. A. Bekunda, H. C. Goma, P. L. Mafangoya, D. N. Mugendi, H. M. Murwira, S. M. Nandwa, P. Nyathi, and M. Swift (ed.), Soil fertility management in Africa: a regional perspective. Academy Science Publishers, Nairobi, Kenya.
- 31.Roesch, C., A. Mergel, and H. Bothe. 2002. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68:3818-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roesch, L., F. Olivares, L. Pereira Passaglia, P. Selbach, E. de Sá, and F. de Camargo. 2006. Characterization of diazotrophic bacteria associated with maize: effect of plant genotype, ontogeny and nitrogen-supply. World J. Microbiol. Biotechnol. 22:967-974. [Google Scholar]
- 33.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez, P. A., K. D. Shepherd, M. J. Soule, F. M. Place, R. J. Buresh, A. N. Izac, A. U. Mokwunye, F. R. Kwesiga, C. G. Ndiritu, and P. L. Woomer. 1997. Soil fertility replenishment in Africa: an investment in natural resource capital: replenishing soil fertility in Africa. Soil Science Society of America, Madison, WI.
- 35.Sanginga, N., B. Vanlauwe, and S. K. A. Danso. 1995. Management of biological N2 fixation in alley cropping systems: estimation and contribution to N balance. Plant Soil 174:119-141. [Google Scholar]
- 36.Santoro, A. E., C. A. Francis, N. R. de Sieyes, and A. B. Boehm. 2008. Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ. Microbiol. 10:1068-1079. [DOI] [PubMed] [Google Scholar]
- 37.Schauss, K., A. Focks, S. Leininger, A. Kotzerke, H. Heuer, and S. Thiele-Bruhn. 2008. Dynamics and functional relevance of ammonia-oxidizing archaea in agricultural soil. Environ. Microbiol. doi: 10.1111/j.1462-2920.2008.01783.x. [DOI] [PubMed]
- 38.Scheffer, F., and P. Schachtschabel. 2002. Lehrbuch der Bodenkunde, p. 295-314. Spektrum, Heidelberg, Germany.
- 39.Shen, J.-P., L.-M. Zhang, Y.-G. Zhu, J.-B. Zhang, and J.-Z. He. 2008. Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ. Microbiol. 10:1601-1611. [DOI] [PubMed] [Google Scholar]
- 40.Simon, H. M., J. A. Dodsworth, and R. M. Goodman. 2000. Crenarchaeota colonize terrestrial plant roots. Environ. Microbiol. 2:495-505. [DOI] [PubMed] [Google Scholar]
- 41.Simon, H. M., C. E. Jahn, L. T. Bergerud, M. K. Sliwinski, P. J. Weimer, D. K. Willis, and R. M. Goodman. 2005. Cultivation of mesophilic soil crenarchaeotes in enrichment cultures from plant roots. Appl. Environ. Microbiol. 71:4751-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Throbäck, I. N., K. Enwall, A. Jarvis, and S. Hallin. 2004. Reassessing PCR primers targeting nirS, nirK, and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49:401-417. [DOI] [PubMed] [Google Scholar]
- 43.Zehr, J. P., and B. B. Ward. 2002. Nitrogen cycling in the ocean: new perspectives on processes and paradigms. Appl. Environ. Microbiol. 68:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]