Abstract
The objective of this study was to identify a microbial marker for pig manure contamination. We quantified the persistence of four dominant bacterial groups from the pig intestinal tract throughout manure handling at 10 livestock operations (including aerobic digestion) by using molecular typing. The partial 16S rRNA genes of Bacteroides-Prevotella, Eubacterium-Clostridiaceae, Bacillus-Streptococcus-Lactobacillus (BSL), and Bifidobacterium group isolates were amplified and analyzed by capillary electrophoresis single-strand conformation polymorphism. The most dominant bacterial populations were identified by cloning and sequencing their 16S rRNA genes. The results showed that Bifidobacterium spp. and, to a lesser extent, members of the BSL group, were less affected by the aerobic treatment than either Eubacterium-Clostridiaceae or Bacteroides-Prevotella. Two Bifidobacterium species found in raw manure were still present in manure during land application, suggesting that they can survive outside the pig intestinal tract and also survive aerobic treatment. The 16S-23S rRNA internal transcribed spacer of one species, Bifidobacterium thermacidophilum subsp. porcinum, was sequenced, and a specific pair of primers was designed for its detection in the environment. With this nested PCR assay, this potential marker was not detected in samples from 30 bovine, 30 poultry, and 28 human fecal samples or in 15 urban wastewater effluents. As it was detected in runoff waters after spreading of pig manure, we propose this marker as a suitable microbial indicator of pig manure contamination.
Brittany represents only 7% of France but is the main pig production area and hosts approximately 14 million fatteners per year. This high concentration of confined pig feeding has led to the overapplication of manure to soil, which contributes to water pollution. Physical and biological manure treatment processes have been developed to limit nitrogen and phosphorus pollution (5). As these treatments were not designed to eliminate microbial pollution, even treated manure can contain pathogenic microorganisms (27) and agricultural soils and water systems can thus potentially still be contaminated through surface runoff and seepage. As manure application can increase the number of pathogens in the soil (18), pig feces may represent a significant risk to human health in Brittany. Currently, the monitoring of bacteria to assess fecal contamination (Escherichia coli, fecal coliforms, and enterococci) does not differentiate contamination from pig slurry from pollution by other animals or humans. It is thus important to develop analytic tools to specifically detect this source of pollution.
Many studies have already proposed potential markers for the detection of host-specific fecal pollution (2, 3, 8, 12-15, 20, 37, 38, 48, 49). Much of this research has concentrated on distinguishing human and animal sources of contamination (3, 8, 20, 30, 38). Some studies have focused on identifying individual sources of animal pollution and have described molecular markers for feces from ducks (13), chickens (37), bovines (2, 3, 49), or cervids (6). Biomarkers have been proposed for porcine fecal contamination but rarely for porcine manure, the bacterial composition of which differs from that of porcine feces (9). Molecular markers have been developed to target the 16S rRNA gene sequences of dominant Eubacteria (2, 14, 43, 48) or methanogenic Archaebacteria (54) of the pig intestinal tract, whereas Khatib et al. (29) targeted the STII toxin gene from enterotoxigenic E. coli. Among the dominant groups of pig fecal Eubacteria, which include Bacteroides-Prevotella, Eubacterium-Clostridiacea, Lactobacillus-Streptococcus (34, 45, 51, 58), and to a lesser extent Bifidobacterium (40), the Bacteroides-Prevotella group has been particularly well studied (14, 22, 44). This marker of pig feces was described by Okabe et al. (44), but their work was based on feces sampled from only two farms and the number of clones analyzed was low. Gourmelon et al. (22) also detected the presence of a specific marker of pig feces belonging to the Bacteroides-Prevotella group in five stored manure samples. Although these studies revealed the presence of specific markers in fecal samples and in the subsequent pig manure samples, they did not address the possible disappearance of these anaerobic bacteria during the storage or biological treatment of the manure.
Due to the lack of data concerning the bacterial flora of manure, the aims of this study were (i) to compare the monitoring of the Bacteroides-Prevotella group with that of Eubacterium-Clostridiaceae, Bacillus-Streptococcus-Lactobacillus (BSL), and Bifidobacterium throughout the biological manure treatment process and (ii) to search for a molecular marker among these groups of bacteria that was consistently present in the manure intended for land application. In the first part of this study, the persistence of the dominant bacteria throughout treatment was studied by using molecular typing, capillary electrophoresis-single-strand conformation polymorphism (CE-SSCP) (45) based on the analysis of the 16S rRNA genes. CE-SSCP is a fingerprinting technique in which single-stranded DNA fragments of the same length are separated based on the conformation of their secondary structure (23). The major advantages of this technique are its reproducibility between runs and its high resolution power with fewer false results than with denaturing gradient gel electrophoresis (25, 26).
The second part of this article describes the relevance of the potential marker of pig manure (Bifidobacterium thermacidophilum subsp. porcinum) selected according to the results of the CE-SSCP profiles and the subsequent identification of dominant peaks of the CE-SSCP profiles. The specificity of this pig marker was then tested by assessing the host distribution in a selection of fecal, manure, and wastewater samples.
MATERIALS AND METHODS
Sample collection. (i) Manure samples.
Manure was collected from 17 piggeries located across Brittany. At these farms, raw manure was stored for 2 to 8 weeks in a primary anaerobic tank, followed by aeration treatment for 3 to 4 weeks before final anaerobic storage for 3 to 9 months. The chemical characteristics of the manure were similar on all of the farms. The mean pHs of the raw and treated manure samples were 7.5 and 7.8, respectively. The corresponding dry-matter contents were 4.3 and 5.1% (wt/wt); the total Kjeldahl nitrogen contents were 4.3 and 2.0 g liter−1, and the soluble chemical organic demands were 9.7 and 2.4 g O2 liter−1, respectively. All of the manure stored in tanks was homogenized by mixing with a propeller agitator for at least 30 min before sampling. A volume of 30 liters of manure was removed and transferred to the laboratory. The samples were then remixed with a propeller homogenizer. One liter of homogenized manure was transferred to a flask. The manure was then centrifuged at 16,000 × g to form a pellet of approximately 250 mg (wet weight). The pellets were stored at −20°C.
(ii) Fecal samples.
A total of 90 samples of animal feces (30 bovine, 30 pig, and 30 poultry fecal samples) were collected from 62 farms across Brittany. Twenty-eight samples of human feces from healthy people were obtained from two French research institutes (IFREMER [Brest] and INRA [Jouy-en-Josas]). Approximately 250 mg (wet weight) of each feces was transferred into a microtube and stored at −20°C.
(iii) Water samples.
Fifteen urban wastewater samples (5 raw and 10 treated effluent samples) were collected from locations across Brittany. Six independent samples of field runoff water (R1 to R6) were collected 40 to 50 min after six rainfall simulations on an experimental agricultural plot previously spread with either pig (samples R1 to R3) or bovine (samples R4 to R6) manure. The samples were collected and poured into 2-liter flasks. Two samples were taken from two lagoons which receive treated liquid manure from piggeries. The retention time for the storage lagoons was between 5 days (L1) and 9 months (L2).
Volumes of approximately 200 ml of water were centrifuged at 4,000 × g for 30 min, and pellets were transferred into microtubes for storage at −20°C.
Enumeration of E. coli bacteria.
E. coli bacteria were enumerated in all of the water samples by using 3M Petrifilm E. coli to estimate the level of fecal contamination. Tenfold serial dilutions were performed with peptone water up to 10−4. The gel of the Petrifilm was rehydrated with 1 ml of water (diluted or undiluted) and incubated at 44°C for 24 h. Blue colonies (glucuronidase positive) were counted to determine the concentration of E. coli, which was expressed as the number of CFU per 100 ml. All enumerations were performed in triplicate.
Collection Bifidobacterium strains.
The strains used in this study were B. animalis subsp. animalis DSM 20104T, B. boum DSM 20432T, B. longum subsp. suis DSM 20211T, B. merycicum DSM 6492T, B. pseudolongum subsp. globosum DSM 20092T, B. ruminantium DSM 6489T, B. thermacidophilum subsp. porcinum DSM 17755T, and B. thermophilum DSM 20210T. All strains were cultured on the medium described by Beerens (1) and incubated at 37°C in a jar under anaerobic conditions. One milliliter of an overnight culture of each strain was centrifuged at 17,000 × g for 10 min. The pellets were stored at −20°C.
Extraction of DNA.
DNA was extracted from the pellets stored at −20°C with a QIAamp DNA stool kit (Qiagen) in accordance with the manufacturer's instructions. The elution volume was 50 μl.
Bacterial group PCRs.
PCRs for each bacterial group were performed with a general bacterial forward primer (W18) and a group-specific reverse primer targeting BSL, Eubacterium-Clostridiaceae, or Bacteroides-Prevotella (GE08, GE09, or rBacPre, respectively) and with a group-specific primer pair for the Bifidobacterium group (g-BIFID-F and g-BIFID-R) (Table 1). The reaction mixture comprised deoxynucleoside triphosphates (dNTPs) at 0.2 mM, primers at 350 nM each, 1× AccuPrime Taq DNA polymerase buffer II, 2.5 U of AccuPrime Taq DNA polymerase (Invitrogen), and 1 μl of manure DNA diluted five times in water. The final reaction volume was 20 μl. The annealing temperatures were 61, 55, 55, and 53°C for the BSL, Eubacterium-Clostridiaceae, Bacteroides-Prevotella, and Bifidobacterium groups, respectively. After a denaturation step of 94°C for 2 min, the reactions were carried out by 30 cycles of 94°C for 30 s, the annealing temperature for 90 s, and 68°C for 90 s. No final elongation was performed, as recommended by the supplier (Invitrogen). The reaction was stopped by cooling the mixture to 10°C.
TABLE 1.
Sequences and target positions of the primers used in this study
| Primer | Sequence (5′-3′)a | E. coli position | 16S rRNA target(s) | Reference |
|---|---|---|---|---|
| W18 | GAGTTTGATCMTGGCTCAG | 9 | Bacteria | 21 |
| W34 | ACGGTCCAGACTCCTACGGG | 330 | V3 bacteria | 11 |
| W49 | 6FAM-TTACCGCGGCTGCTGGCACb | 500 | V3 universal, Bacillus spp., Lactobacillus spp. | 11 |
| GE08 | ATTYCACCGCTACACATG | 679 | Pediococcus spp., Leuconostoc spp., Weissella spp., Streptococcus spp. | 24 |
| GE09 | CCCTTTACACCCAGTAA | 561 | Clostridiacea | 55 |
| rBacPre | TCACCGTTGCCGGCGTACTC | 887 | Prevotella, Bacteroides | 59 |
| g-BIFID-F | CTCCTGGAAACGGGTGG | 153 | Bifidobacterium | 39 |
| g-BIFID-R | GGTGTTCTTCCCGATATCTACA | 699 | Bifidobacterium | 39 |
| ITSF | GTCGTAACAAGGTAGCCGTA | Total ITS (universal primer) | 7 | |
| ITSR | GCCAAGGCATCCACC | Total ITS (universal primer) | 7 | |
| GE35 | ATGGTATCGCGGGGGTCGTC | B. thermacidophilum subsp. porcinum ITS | This study | |
| GE36 | GAACACCCGGGAAGGAA | B. thermacidophilum subsp. porcinum ITS | This study |
M = A/C, N = A/T/C/G, Y = C/T.
6FAM, 6-carboxyfluorescein primer label.
The sizes of the amplification products were confirmed by agarose gel electrophoresis (1× Tris-borate-EDTA and 0.7 or 1.5% [wt/vol] agarose for total bacteria and bacterial groups, respectively). The PCR products were visualized under UV light after gel staining with ethidium bromide.
A volume of 1 μl of each PCR product was used as the template for further PCR and CE-SSCP analyses.
Analysis by CE-SSCP PCR.
We used a nested PCR in which the first PCR (described above) was done with the group-specific primers to target the microbial groups of interest. As the amplified DNA fragments are larger than the V3 region, each group-specific PCR product was amplified again in a second PCR using the bacterial W34-W49 primers to target the V3 region and label the DNA fragment with the fluorescent dye present on primer W49. These two primers were used specifically for SSCP since they target the 16S rRNA gene V3 region that is the right length (200 bp) and has the necessary diversity for SSCP analysis of microbial communities. This approach facilitates the PCRs and enables careful comparison of the different patterns generated with the same primers.
The reaction mixture comprised dNTPs at 0.2 mM, primers at 390 nM, 1× PfuTurbo buffer, 0.625 U of PfuTurbo polymerase (Stratagene), and 1 μl of the PCR products amplified previously. The final reaction volume was 20 μl. The amplification conditions were 1 cycle of 94°C for 2 min, followed by 25 cycles of 30 s at 94°C, 30 s at 61°C, and 30 s at 72°C and then a final elongation step of 10 min at 72°C. The resulting PCR products were then separated by SSCP capillary electrophoresis with an ABI 310 genetic analyzer (Applied Biosystems) as described by Delbès et al. (10) but with a 5.58% conformation analysis polymer-10% glycerol polymer (Applied Biosystems).
Cloning and sequencing.
For each bacterial group, cloning was performed on a mixture of two PCR products selected according to their SSCP profiles (with the most numerous and highest peaks). The mixed PCR products were cloned and transformed into competent E. coli cells with the StrataClone PCR cloning kit (Stratagene, La Jolla, CA) by following the manufacturer's instructions, except for the ligation time, which was increased from 5 to 15 min.
A total of 275 clones were further analyzed: 96 for the Eubacterium-Clostridiaceae group (48 from raw manure and 48 from treated manure), 35 for the BSL group (11 from raw manure and 24 from treated manure), 72 for the Bacteroides-Prevotella group (48 from raw manure and 24 from treated manure), and 72 for the Bifidobacterium group (24 from raw manure and 48 from treated manure).
The clones were randomly picked, and their inserts were screened by nested PCR and CE-SSCP as follows. In the first step, plasmid inserts were amplified by PCR with plasmid-targeted primers T7 (5′-TAATACGACTCACTATAGGG-3′) and P13 (5′-GACCATGATTACGCCA-3′) (Stratagene, La Jolla, CA). The reaction mixture was 0.2 mM dNTPs, 700 nM each primer, 1× RedTaq buffer, 2.5 U RedTaq polymerase, and deionized water to bring the volume to 25 μl. The amplification conditions were 10 min at 94°C, followed by 25 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C and a final elongation step of 72°C for 10 min. One microliter of these PCR products was used to perform a CE-SSCP PCR as described above. Inserts yielding a peak that comigrated with distinguishable peaks from the manure CE-SSCP profiles were sequenced for peak identification.
A total of 139 clones were sequenced. Sequence reactions were performed at the Ouest Genopole Sequencing Facility (CNRS, Roscoff, France) with primer T7. DNA sequences were identified by comparison with their closest relatives available in databases by using BLAST from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) and Ribosomal Database Project II (http://rdp.cme.msu.edu/).
Specific amplification of the Bifidobacterium thermacidophilum subsp. porcinum ITS.
The total internal transcribed spacer (ITS) sequence of B. thermacidophilum subsp. porcinum DSM 17755T was amplified by PCR with the primer set ITSF/ITSR, designed by Cardinale et al. (7). The reaction mixture was 1× RedTaq buffer, 5 U RedTaq polymerase, 0.2 mM dNTPs, 700 nM primers, and deionized water to bring the volume to 20 μl. The amplification program was as described by Cardinale et al. (7), except for the elongation temperature (72°C), which was adapted to RedTaq polymerase. The PCR product was sequenced by the Ouest Genopole Sequencing Facility (CNRS, Roscoff, France). The sequence obtained was aligned, by using the ClustalW2 software (52), with the seven ITS sequences of Bifidobacterium strains present in GenBank (B. breve, B. adolescentis, B. longum, B. choerinum, B. animalis, B. thermophilum, and B. pseudolongum) and to the ITS sequence of B. longum biotype suis, which was obtained in this study as described above. Based on the comparison of these sequences, a pair of primers specific to B. thermacidophilum subsp. porcinum was designed (GE35/GE36) (Table 1).
Specific detection of B. thermacidophilum subsp. porcinum was then performed with a nested PCR. All Bifidobacterium ITSs were first amplified with the primer pair ITSF/ITSR as described above. The resulting PCR products were diluted 10 times, and 1 μl was used as the template for a second PCR with the primer pair GE35/GE36. The GE35/GE36 PCR mixture comprised 1× AccuPrime Taq DNA polymerase buffer II, 2.5 U of AccuPrime Taq polymerase (Invitrogen), 350 nM each primer, and deionized water to bring the total volume to 20 μl. The PCR was performed under the following conditions: 1 cycle of 94°C for 2 min and 30 cycles of 94°C for 30 s, 59°C for 30 s, and 68°C for 1.5 min.
Nucleotide sequences accession numbers.
Sequences were deposited in the EMBL database under accession numbers AM991308 to AM991325.
RESULTS
Comparison of the dominant microbial groups of raw and treated manure.
For each bacterial group, the CE-SSCP profiles obtained from the 10 raw and treated manure samples were aligned and compared (see Fig. 1 to 4). The Eubacterium-Clostridiaceae profiles provided the lowest resolution with a high background level below the peaks, underlining the complexity of this bacterial group (Fig. 1). The raw manure profiles had 9 to 11 comigrating peaks in common and a similar number of distinct peaks before and after treatment. However, in most cases, the peaks of raw manure did not comigrate with the peaks of treated manure.
FIG. 1.
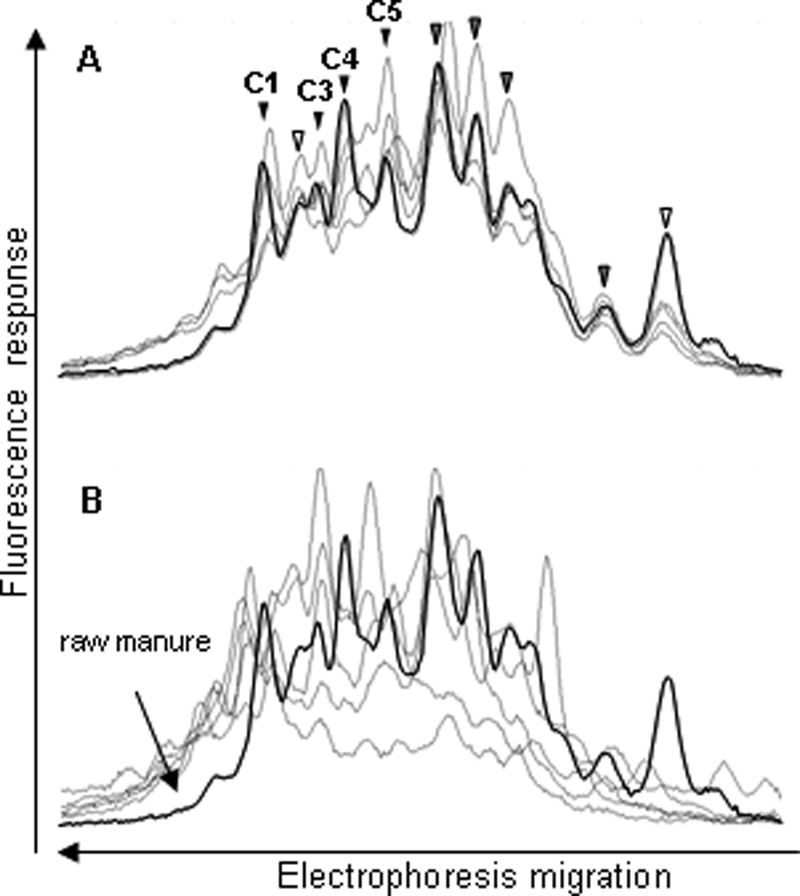
Comparison of the Eubacterium-Clostridiaceae group CE-SSCP profiles from five raw manure samples (A) and five treated manure samples (B). One raw manure profile (in bold) is also shown in panel B for comparison. The peaks corresponding to the dominant bacterial populations are indicated by arrowheads. The white arrows correspond to unidentified peaks, gray arrows to peaks identified by one sequence only, and black arrows to peaks identified by at least two sequences. Peaks that could be identified are designated C1 to C5, as in Table 3.
FIG. 4.
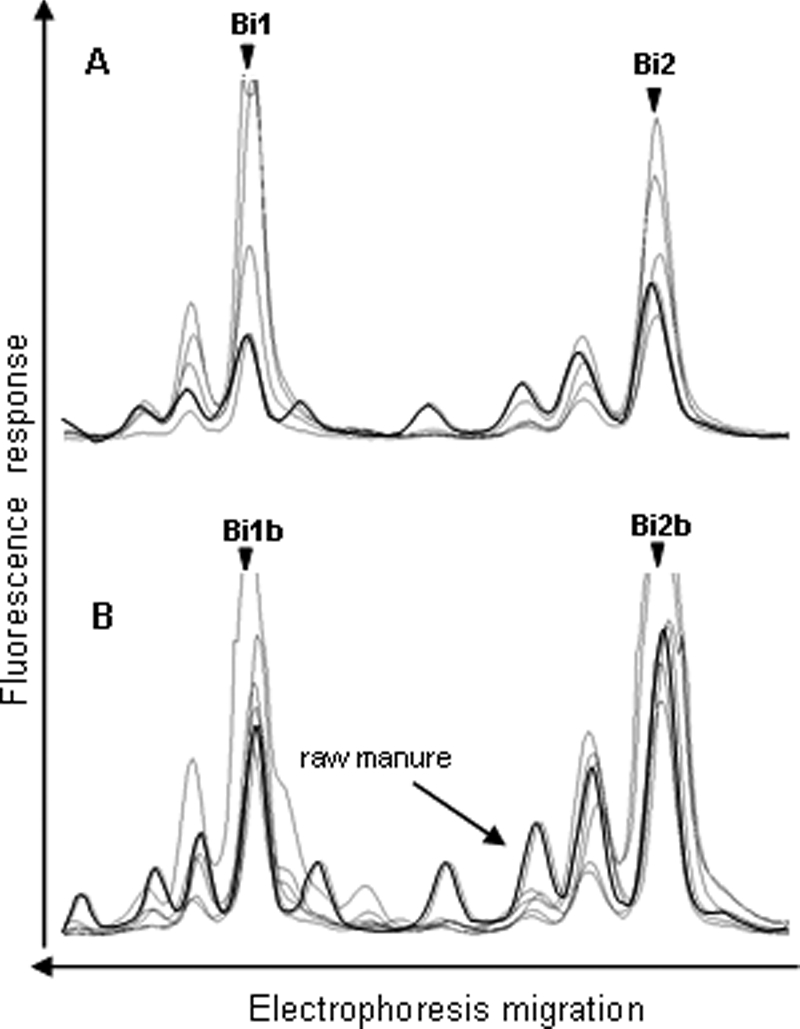
Comparison of the Bifidobacterium SSCP profiles of five raw manure samples (A) and five treated manure samples (B). The symbols are the same as those in Fig. 1. Peaks that could be identified are designated Bi1, Bi2, Bi1b, and Bi2b, as shown in Table 3.
The BSL group profiles provided a lower background signal than that observed for the Eubacterium-Clostridiaceae group (Fig. 2). The profiles of raw and treated manure consisted of 10 and 12 peaks, respectively. After aerobic treatment, seven peaks from the treated manure profiles comigrated with peaks from the raw manure profiles.
FIG. 2.
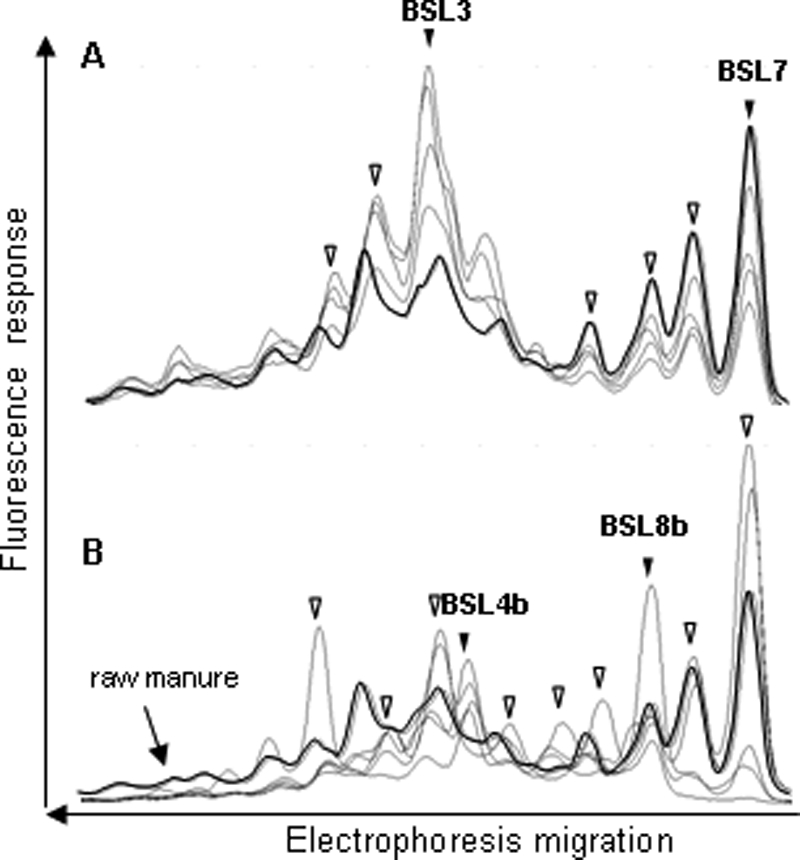
Comparison of the BSL group CE-SSCP profiles from five raw manure samples (A) and five treated manure samples (B). The symbols are the same as those in Fig. 1. Peaks that could be identified are designated BSL3, BSL7, BSL4b, and BSL8b, as in Table 3.
The CE-SSCP profiles of the Bacteroides-Prevotella and Bifidobacterium groups differed from the BSL and Eubacterium-Clostridiaceae group profiles by the absence of background and the small number of peaks detected (Fig. 3 and 4). These profiles yielded three and two dominant peaks, respectively, consistently preceded by smaller artifactual peaks which were also visible with purified clones (data not shown). These artifactual peaks were probably produced either during migration in capillary electrophoresis or during PCR amplification. In the latter, they would represent a small proportion of PCR fragments that have ended prematurely. The three peaks from the Bacteroides-Prevotella group detected in all of the raw manure were not detected in treated manure, which contained two other distinguishable peaks (Fig. 3B). The first peak (BA3) was common to all of the treated manure samples, whereas the position of the second peak (BA4) differed from one sample to another. The profiles of the Bifidobacterium group were characterized by two peaks which were detected in all of the raw and treated manure samples (Fig. 4).
FIG. 3.
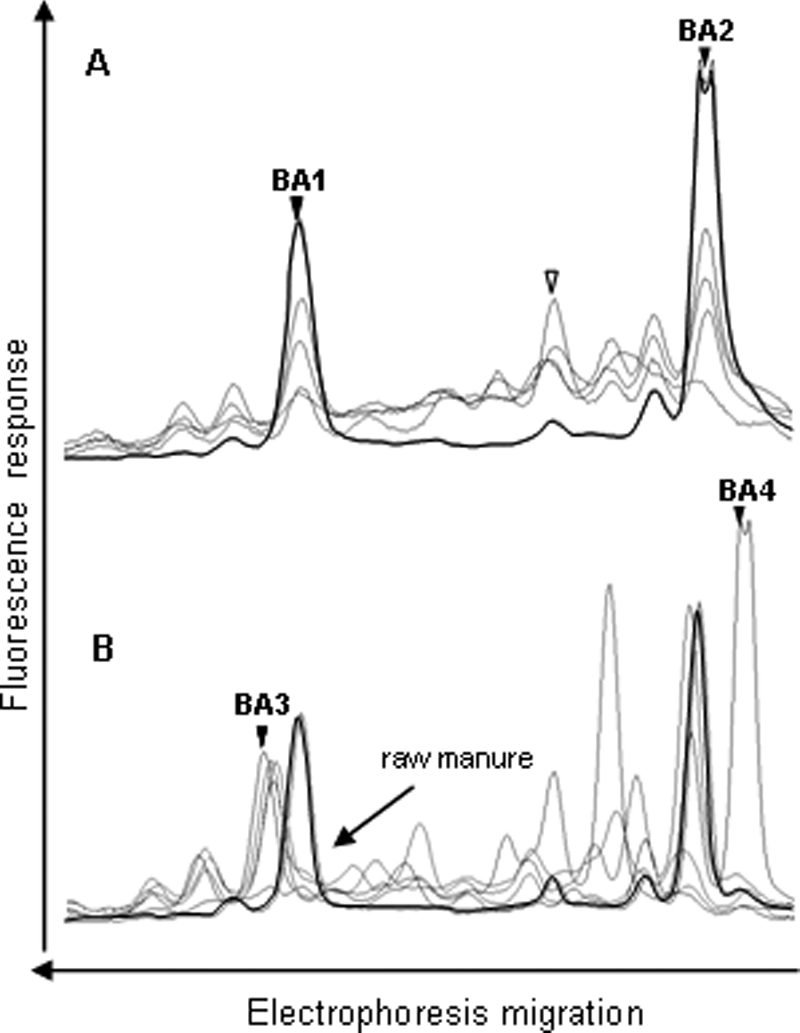
Comparison of the Bacteroides-Prevotella group SSCP profiles from five raw manure samples (A) and five treated manure samples (B). The symbols are the same as those in Fig. 1. Peaks that could be identified are designated BA1, BA2, BA3, and BA4 (further characterized in Table 3). Because of their strong dominance over the profiles, peaks BA2 and BA4 saturated the fluorescence detector when other peaks were detectable.
Identification of the major peaks of each group.
The dominant peaks were identified by cloning and sequencing of the corresponding 16S rRNA gene fragments. A total of 275 clones were screened by CE-SSCP, and 139 were sequenced. The phylogenetic affiliation of the clones corresponding to the major peaks of the CE-SSCP profiles is presented in Table 2. Only 37.5% of the Eubacterium-Clostridiaceae 16S rRNA gene sequences found in raw manure demonstrated more than 97% similarity to sequences in databases. Four of the dominant peaks in the Eubacterium-Clostridiacea raw manure profiles were identified, but no identity could be assigned to peaks obtained from the treated manure profiles. The closest relatives of the four sequences identified were sequences from uncultured bacteria from various sources, including the effluent treatment plant, the solid waste digester, and the pig manure storage pit.
TABLE 2.
Phylogenetic affiliations of 16S rRNA gene sequences
| Peak designationa | Sequence length (bp) | Closest relative
|
Reference | |||
|---|---|---|---|---|---|---|
| Name (accession no.) | Affiliation group | % Similarity | Source | |||
| C1 | 525 | Clone B-87 (AY676487) | Clostridiaceae | 97 | Bovine teat canal | 19 |
| C3 | 530 | Clone M75 (DQ640962) | Clostridiaceae | 88 | Effluent treatment plant | V. C. Kalia et al. (unpublished, 2007) |
| C4 | 524 | Clone A35 D28 L B B12 (EF559222) | Clostridiaceae | 99 | Mesophilic solid waste digester | T. Li et al. (unpublished, 2007) |
| C5 | 525 | Clone P316 (AF261803) | Clostridiaceae | 98 | Manure storage pit | T. R. Whitehead and M. A. Cotta (unpublished, 2004) |
| BA1 | 707 | Clone BRC82 (EF436368) | Bacteroidetes | 92 | Buffalo rumen water | H. Mao et al. (unpublished, 2007) |
| BA2 | 844 | Clone SRRT42 (AB240481) | Bacteroidetes | 92 | Rhizosphere biofilm of phragmites | Y. Nakamura et al. (unpublished, 2005) |
| BA3 | 662 | Clone Z144 (EU029356) | Bacteroidetes | 94 | Raw milk | D. Raats and M. Halpern (unpublished, 2007) |
| BA4 | 405 | Clone oca46 (AY491639) | Bacteroidetes | 94 | Wastewater | 46 |
| BSL3 | 674 | Clone WTB_Y48 (EU009859) | Mollicutes | 91 | Turkey intestinal tract | E. Bent et al. (unpublished, 2007) |
| BSL7 | 674 | L. sobrius (AY700063) | Lactobacillus | 100 | Piglet intestinal tract | 32 |
| BSL4b | 645 | Clone R8C-A3 (AY678482) | Mollicutes | 88 | Estuarine sediment | 42 |
| BSL8b | 647 | Clone R8C-A3 (AY678482) | Firmicutes | 86 | Estuarine sediment | 42 |
| Bi1 | 513 | B. thermacidophilum subsp. porcinum (AY148470) | Bifidobacterium | 99 | Piglet intestinal tract | 60 |
| Bi2 | 522 | B. pseudolongum subsp. pseudolongum (AY174109) | Bifidobacterium | 100 | Porcine cecum | 50 |
| Bi1b | 513 | B. thermacidophilum subsp. porcinum (AY148470) | Bifidobacterium | 100 | Piglet intestinal tract | 60 |
| Bi2b | 514 | B. pseudolongum subsp. pseudolongum (AY174109) | Bifidobacterium | 98 | Porcine cecum | 50 |
Sequences from this study have been deposited in EMBL under accession numbers AM991308 to AM991325.
Two of the three dominant peaks of the Bacteroides-Prevotella raw manure profiles and peak BA3 of the treated manure profiles were identified. The closest relative of the Bacteroides-Prevotella sequences was found in various sources but not in pig feces or manure (Table 2). As mentioned above, a specific Bacteroides-Prevotella peak was found to be present in each treated manure profile. One of them (BA4) was cloned and sequenced. Its closest relative was a Bacteroidetes identified in microbial fuel cells fed with wastewater (46).
Two peaks of the BSL profiles of raw manure were assigned (BSL3 and BSL7). BSL3 was 91% similar to its closest relative, a turkey intestinal tract microorganism. The sequence of peak BSL7 was 100% similar to Lactobacillus sobrius isolated from piglet feces (31). The two BSL peaks identified in treated manure were only about 88% similar to cloned DNA from an estuarine sediment.
The sequences of the two peaks of the Bifidobacterium profiles, obtained from either raw or treated manure, were 99 to 100% similar to B. thermacidophilum subsp. porcinum isolated from piglet feces (peaks Bi1 and Bi1b) (60) and 98 to 100% similar to B. pseudolongum subsp. pseudolongum isolated from a porcine cecum (peaks Bi2 and Bi2b) (50).
Specificity of GE35/GE36 primers.
Among the four groups of bacteria analyzed in this study, only two species, B. thermacidophilum subsp. porcinum and B. pseudolongum subsp. pseudolongum, comigrated with a peak that was systematically detected in all of the raw and treated manure CE-SSCP profiles. Given that B. pseudolongum subsp. pseudolongum has previously been observed in the feces of various animals (4), the B. thermacidophilum subsp. porcinum strain was selected for further analyses. However, this species is genotypically too similar to B. thermophilum (57) to be differentiated at the 16S rRNA gene sequence level. The design of specific primers thus required targeting of the 16S-23S rRNA ITS region. A specific pair of primers (GE35/GE36) was designed and tested on B. thermacidophilum subsp. porcinum DSM 17755T and on seven other Bifidobacterium type strains representative of taxa of animal origin as previously described by Ventura et al. (56). The test showed that the primer set produced species-specific amplicons from B. thermacidophilum subsp. porcinum DSM 17755T and did not amplify any PCR products from the other seven strains (Table 3).
TABLE 3.
Specificity of PCR product formation with primer set GE035/GE036 tested on Bifidobacterium collection strains
| Bifidobacterium straina | PCR product formationb |
|---|---|
| B. boum DSM 20432T | − |
| B. thermophilum DSM 20210T | − |
| B. thermacidophilum subsp. porcinum DSM 17755T | + |
| B. merycicum DSM 6492T | − |
| B. pseudolongum subsp. globosum DSM 20092T | − |
| B. ruminantium DSM 6489T | − |
| B. animalis subsp. animalis DSM 20104T | − |
| B. longum subsp. suis DSM 20211T | − |
Specificity was tested with chromosomal DNA from Bifidobacterium strains previously detected in animal feces.
+, product formed; −, no product formed.
The host specificity of the species was then examined with the set of primers based on DNA originating from human, pig, bovine, and poultry feces (Table 4). All of the fecal samples gave a positive signal in the first universal ITS-targeted PCR, but the B. thermacidophilum subsp. porcinum marker was only found in pig feces when nested PCR and the GE35/GE36 primers were used.
TABLE 4.
Results of B. thermacidophilum subsp. porcinum PCR tested on DNA from human and animal feces
| Origin of feces | Total no. of samples | No. of positive samples |
|---|---|---|
| Pig | 30 | 30 |
| Bovine | 30 | 0 |
| Poultry | 30 | 0 |
| Human | 28 | 0 |
B. thermacidophilum subsp. porcinum and the concentration of E. coli were observed in manure and in water samples with our nested PCR assay (Table 5). Regardless of the level of E. coli, B. thermacidophilum subsp. porcinum was not detected in urban effluents of human origin or in runoff water impacted by bovine manure contamination in spite of the presence of E. coli. In the case of runoff water obtained after the application of pig manure, the three samples showed positive amplification. B. thermacidophilum subsp. porcinum was also found in raw and treated manure and in two types of lagoon supplied with treated liquid pig manure. In lagoon L1 (with a retention time of 5 days), the concentration of E. coli was 4.5 × 106 CFU/100 ml and positive amplification of the target bacteria was observed, whereas in lagoon L2 (with a retention time of 9 months), neither E. coli nor B. thermacidophilum subsp. porcinum was detected.
TABLE 5.
E. coli counts and detection of B. thermacidophilum subsp. porcinum in manure and in water impacted by human activity and contaminated with manure
| Type of sample | Origin of contamination | Mean E. coli count (CFU/100 ml) ± SD | No. of samples positive for target bacteria/total |
|---|---|---|---|
| Raw manure (pig) | 4.0 × 106 ± 4.2 × 106 | 17/17 | |
| Treated manure (pig) | 5.1 × 104 ± 3.3 × 104 | 10/10 | |
| Lagoon with retention time of 5 days (L1) (pig) | Treated liquid manure | 4.5 × 106 ± 4.1 × 105 | 1/1 |
| Lagoon with retention time of 9 mo (L2) (pig) | Treated liquid manure | Not detected | 0/1 |
| Runoff water (R1-R3) | Pig manure spread on field | 9.7 × 10 ± 3.3 × 103 | 3/3 |
| Runoff water (R4-R6) | Bovine manure spread on field | 7.5 × 103 ± 8 × 102 | 0/3 |
| Raw wastewater | Urban effluent (mainly human) | 1.8 × 106 ± 1.7 × 106 | 0/5 |
| Treated wastewater | Urban effluent (mainly human) | 3.3 × 103 ± 4.3 × 103 | 0/10 |
DISCUSSION
Although pig-specific genetic markers have been proposed to trace fecal pollution in the environment, their application has mainly focused on fecal samples (14, 22, 43, 44, 54) and data concerning manure intended for spreading are scarce (22, 29). Cotta et al. (9) reported a difference in composition between the bacterial communities of pig feces and stored manure. Furthermore, Peu et al. (45) observed differences in the bacterial communities in fresh manure located below the animals and manure stored in outdoor tanks. To be considered suitable, a microbial indicator of pig contamination must be abundant and found not only in feces but also in stored manure intended for land application.
Whereas studies concerning fecal markers have usually focused on a particular group of bacteria, we used a broader strategy (i.e., four groups instead of one) with the aim of identifying a potential microbial marker of pig contamination present in both raw and treated manure. The behavior of four pig fecal bacterial groups (34, 45, 53, 58) was monitored throughout pig manure biological treatment by molecular typing (CE-SSCP). These bacterial groups were selected either because they are dominant in manure microbial communities (Eubacterium-Clostridiacea, Bacteroides-Prevotella, BSL) or due to their known host specificity. Thus, phylogenetic groups of Bacteroides-Prevotella have been associated with pig feces (14, 22, 43, 44) and the genus Bifidobacterium consists of species of animal or human origin (17, 40).
SSCP profiles.
The 17 raw manure samples analyzed revealed the remarkable consistency of the SSCP profiles of the four bacterial groups (Fig. 1 to 4), regardless of the geographical location of the piggeries sampled and of the storage period of the manure. In practice, in Brittany piggeries, raw manure stores are rarely aerated and slurry tanks are not operated as closed batch reactors but are subject to regular additions of fresh manure. The major difference from one manure to another is thus the length of storage, which ranges from weeks to months, depending on the storage capacity of the tank. This consistency of the bacterial profile could be explained by the similarity of farm management practices (diet and the age of the animals) and manure storage conditions. Leung and Topp (35) and Peu et al. (45) obtained similar results by using molecular techniques to monitor pig manure microbial community dynamics during storage in a laboratory scale reactor and a manure storage tank for a period of 3 months, respectively. These data suggested that the dominant bacterial populations of manure stored under anoxic conditions are not strongly influenced by the length of storage.
Biological treatment of manure, comprising nitrification-denitrification by alternating periods of aerobic and anoxic conditions, caused changes in the composition of Eubacterium-Clostridium and of the Bacteroides-Prevotella groups. These results are in agreement with those of Leung and Topp (35), who observed significant changes in bacterial manure populations during aeration. It is interesting that the four bacterial groups targeted in this study, which are classified as anaerobes, presented different behaviors throughout treatment, suggesting different levels of oxygen tolerance. The composition of the Eubacterium-Clostridium and Bacteroides-Prevotella groups changed significantly, resulting in the disappearance of the dominant peaks found in raw manure, whereas new peaks appeared in treated manure. It has previously been reported that the presence of oxygen has significant effects on the survival ability of fecal Bacteroides spp. and Eubacterium-Clostridium groups (16, 47). In contrast, Bifidobacterium and, to a lesser extent, BSL appeared to be less sensitive to biological treatment because most of their peaks were detected in both raw and treated manure. The different behavior during treatment indicates that the BSL and Bifidobacterium groups are potentially more robust markers of manure contamination.
Identification of peaks of SSCP profiles.
Of the 16 peaks identified (Table 2), only 6 were identical or closely related to other sequences obtained specifically from pig feces or manure. The scarcity of available data on the bacterial populations of treated urban or animal effluents could explain the small number of sequence matches, particularly with the Eubacterium-Clostridium group. Peak C5 was closely related (98% similarity) to an uncultured Clostridium previously found in a manure storage pit (58), and peak BSL7 was identified as L. sobrius, which has previously been described in piglet (32) and pig feces (28). However, none of these peaks was found in treated manure whereas the two Bifidobacterium peaks were found in both raw and treated manure. These peaks presented 100% similarity to B. pseudolongum subsp. pseudolongum, which has been isolated from the feces of various animals (17), and with B. thermacidophilum subsp. porcinum, which has been recently described in pig and piglet feces (41, 60).
The absence of members of the Bacteroides-Prevotella group as a potential marker was surprising because several phylotypes of this group have previously been found in pig feces (14, 22, 34, 43, 58) and manure (35, 45, 58). This absence could be explained by the use of the CE-SSCP technique, which overrepresents the dominant bacterial populations when these populations make up more than 1% of the total community (36). The presence of two very dominant peaks in the raw and treated manure may have masked the diversity of less dominant species. These two peaks were not closely related to bacteria isolated from pig feces or manure and presented poor similarity (92%) to uncultured rumen and rhizosphere bacteria.
B. thermacidophilum subsp. porcinum targeting.
According to the results of the SSCP analyses, which highlighted the presence of B. thermacidophilum subsp. porcinum in manure, the host specificity of this genetic marker was then determined. As this species is closely related to B. thermophilum and B. boum (56), the 16S rRNA gene did not allow discrimination of the target bacteria. Nevertheless, the use of a nested PCR for the ITS region of the 16S and 23S rRNA genes led to differentiation of B. thermacidophilum subsp. porcinum from B. thermophilum and B. boum (Table 3). Lamendella et al. (33) reported that certain species of the genus Bifidobacterium were present in various environments whereas other species had a preferential host such as B. boum and B. thermophilum; these authors only detected the latter in pig feces (33). Our results also highlighted the host specificity of B. thermacidophilum subsp. porcinum, which was previously described in the pig intestinal tract (41, 60), as it was not detected in bovine, poultry, or human feces or in urban wastewaters containing domestic sewage. Our results show that by using a nested PCR, it is possible to detect B. thermacidophilum subsp. porcinum in water samples contaminated with manure. This is in agreement with the study of King et al. (30), who also used a nested PCR to detect B. adolescentis in samples of water impacted by human activities. As already reported by Lamendella et al. (33) and King et al. (30), our results confirm that certain species of Bifidobacterium might represent a good target population for assessing fecal contamination above a background level, for example, associated with heavy rainfall events.
Conclusions.
The comparison of dominant pig manure microbial communities throughout manure treatment by CE-SSCP allowed a large number of raw and treated manure samples to be screened. This demonstrated that bifidobacteria and, to a lesser extent, members of the BSL group were less affected by the handling and treatment of manure than were the Eubacterium-Clostridiaceae and Bacteroides-Prevotella groups. These data show that the Bifidobacterium species found in manure can persist outside the pig intestinal tract and that B. thermacidophilum subsp. porcinum can be used as an indicator of manure contamination in the environment.
Acknowledgments
We thank M. Gourmelon from IFREMER (Brest) and M. Leclerc from INRA (Jouy-en-Josas), who kindly provided bovine and human fecal samples for the study.
This research was financially supported by the Agence Française de Sécurité Sanitaire Environnementale et du Travail (AFSSET). Romain Marti is the recipient of a Cemagref-Ademe fellowship.
Footnotes
Published ahead of print on 12 June 2009.
REFERENCES
- 1.Beerens, H. 1991. Detection of bifidobacteria by using propionic acid as a selective agent. Appl. Environ. Microbiol. 57:2418-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhard, A. E., T. Goyard, M. T. Simonich, and K. G. Field. 2003. Application of a rapid method for identifying fecal pollution sources in a multi-use estuary. Water Res. 37:909-913. [DOI] [PubMed] [Google Scholar]
- 4.Biavati, B., M. Vescovo, S. Torriani, and V. Bottazzi. 2000. Bifidobacteria: history, ecology, physiology and applications. Ann. Microbiol. 50:117-131. [Google Scholar]
- 5.Burton, C. H. 1992. A review of the strategies in the aerobic treatment of pig slurry: purpose, theory and method. J. Agric. Eng. Res. 53:249-272. [Google Scholar]
- 6.Call, D. R., D. M. Satterwhite, and M. Soule. 2007. Using DNA suspension arrays to identify library-independent markers for bacterial source tracking. Water Res. 41:3740-3746. [DOI] [PubMed] [Google Scholar]
- 7.Cardinale, M., L. Brusetti, P. Quatrini, S. Borin, A. M. Puglia, A. Rizzi, E. Zanardini, C. Sorlini, C. Corselli, and D. Daffonchio. 2004. Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl. Environ. Microbiol. 70:6147-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carson, C. A., J. M. Christiansen, H. Yampara-Iquise, V. W. Benson, C. Baffaut, J. V. Davis, R. R. Broz, W. B. Kurtz, W. M. Rogers, and W. H. Fales. 2005. Specificity of a Bacteroides thetaiotaomicron marker for human feces. Appl. Environ. Microbiol. 71:4945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotta, M. A., T. R. Whitehead, and R. L. Zeltwanger. 2003. Isolation, characterization and comparison of bacteria from swine faeces and manure storage pits. Environ. Microbiol. 5:737-754. [DOI] [PubMed] [Google Scholar]
- 10.Delbès, C., R. Moletta, and J. J. Godon. 2001. Bacterial and archaeal 16S rDNA and 16S rRNA dynamics during an acetate crisis in an anaerobic digestor ecosystem. FEMS Microbiol. Ecol. 35:19-26. [DOI] [PubMed] [Google Scholar]
- 11.Delbès, C., R. Moletta, and J. J. Godon. 2000. Monitoring of activity dynamics of an anaerobic digester bacterial community using 16S rRNA polymerase chain reaction-single-strand conformation polymorphism analysis. Environ. Microbiol. 2:506-515. [DOI] [PubMed] [Google Scholar]
- 12.Delcenserie, V., N. Bechoux, T. Leonard, B. China, and G. Daube. 2004. Discrimination between Bifidobacterium species from human and animal origin by PCR-restriction fragment length polymorphism. J. Food Prot. 67:1284-1288. [DOI] [PubMed] [Google Scholar]
- 13.Devane, M. L., B. Robson, F. Nourozi, P. Scholes, and B. J. Gilpin. 2007. A PCR marker for detection in surface waters of faecal pollution derived from ducks. Water Res. 41:3553-3560. [DOI] [PubMed] [Google Scholar]
- 14.Dick, L. K., A. E. Bernhard, T. J. Brodeur, J. W. Santo Domingo, J. M. Simpson, S. P. Walters, and K. G. Field. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickerson, J. W., Jr., C. Hagedorn, and A. Hassall. 2007. Detection and remediation of human-origin pollution at two public beaches in Virginia using multiple source tracking methods. Water Res. 41:3758-3770. [DOI] [PubMed] [Google Scholar]
- 16.Flint, H. J., S. H. Duncan, K. P. Scott, and P. Louis. 2007. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ. Microbiol. 9:1101-1111. [DOI] [PubMed] [Google Scholar]
- 17.Gavini, F., V. Delcenserie, K. Kopeinig, S. Pollinger, H. Beerens, C. Bonaparte, and M. Upmann. 2006. Bifidobacterium species isolated from animal feces and from beef and pork meat. J. Food Prot. 69:871-877. [DOI] [PubMed] [Google Scholar]
- 18.Gessel, P. D., N. C. Hansen, J. F. Moncrief, and M. A. Schmitt. 2004. Rate of fall-applied liquid swine manure: effects on runoff transport of sediment and phosphorus. J. Environ. Qual. 33:1839-1844. [DOI] [PubMed] [Google Scholar]
- 19.Gill, J. J., P. M. Sabour, J. Gong, H. Yu, K. E. Leslie, and M. W. Griffiths. 2006. Characterization of bacterial populations recovered from the teat canals of lactating dairy and beef cattle by 16S rRNA gene sequence analysis. FEMS Microbiol. Ecol. 56:471-481. [DOI] [PubMed] [Google Scholar]
- 20.Gilpin, B., T. James, F. Nourozi, D. Saunders, P. Scholes, and M. Savill. 2003. The use of chemical and molecular microbial indicators for faecal source identification. Water Sci. Technol. 47:39-43. [PubMed] [Google Scholar]
- 21.Godon, J. J., E. Zumstein, P. Dabert, F. Habouzit, and R. Moletta. 1997. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gourmelon, M., M. P. Caprais, R. Segura, C. Le Mennec, S. Lozach, J. Y. Piriou, and A. Rince. 2007. Evaluation of two library-independent microbial source tracking methods to identify sources of fecal contamination in French estuaries. Appl. Environ. Microbiol. 73:4857-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hebenbrock, K., P. M. Williams, and B. L. Karger. 1995. Single-strand conformational polymorphism using capillary electrophoresis with two-dye laser-induced fluorescence detection. Electrophoresis 16:1429-1436. [DOI] [PubMed] [Google Scholar]
- 24.Heilig, H. G., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. Akkermans, and W. M. de Vos. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl. Environ. Microbiol. 68:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong, H., A. Pruden, and K. F. Reardon. 2007. Comparison of CE-SSCP and DGGE for monitoring a complex microbial community remediating mine drainage. J. Microbiol. Methods 69:52-64. [DOI] [PubMed] [Google Scholar]
- 26.Hori, T., S. Haruta, Y. Ueno, M. Ishii, and Y. Igarashi. 2006. Direct comparison of single-strand conformation polymorphism (SSCP) and denaturing gradient gel electrophoresis (DGGE) to characterize a microbial community on the basis of 16S rRNA gene fragments. J. Microbiol. Methods 66:165-169. [DOI] [PubMed] [Google Scholar]
- 27.Hutchison, M. L., L. D. Walters, A. Moore, and S. M. Avery. 2005. Declines of zoonotic agents in liquid livestock wastes stored in batches on-farm. J. Appl. Microbiol. 99:58-65. [DOI] [PubMed] [Google Scholar]
- 28.Jakava-Viljanen, M., and A. Palva. 2007. Isolation of surface (S) layer protein carrying Lactobacillus species from porcine intestine and faeces and characterization of their adhesion properties to different host tissues. Vet. Microbiol. 124:264-273. [DOI] [PubMed] [Google Scholar]
- 29.Khatib, L. A., Y. L. Tsai, and B. H. Olson. 2003. A biomarker for the identification of swine fecal pollution in water, using the STII toxin gene from enterotoxigenic Escherichia coli. Appl. Microbiol. Biotechnol. 63:231-238. [DOI] [PubMed] [Google Scholar]
- 30.King, E. L., D. S. Bachoon, and K. W. Gates. 2007. Rapid detection of human fecal contamination in estuarine environments by PCR targeting of Bifidobacterium adolescentis. J. Microbiol. Methods 68:76-81. [DOI] [PubMed] [Google Scholar]
- 31.Konstantinov, S. R., A. Awati, H. Smidt, B. A. Williams, A. D. Akkermans, and W. M. de Vos. 2004. Specific response of a novel and abundant Lactobacillus amylovorus-like phylotype to dietary prebiotics in the guts of weaning piglets. Appl. Environ. Microbiol. 70:3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konstantinov, S. R., E. Poznanski, S. Fuentes, A. D. Akkermans, H. Smidt, and W. M. de Vos. 2006. Lactobacillus sobrius sp. nov., abundant in the intestine of weaning piglets. Int. J. Syst. Evol. Microbiol. 56:29-32. [DOI] [PubMed] [Google Scholar]
- 33.Lamendella, R., J. W. Santo Domingo, C. Kelty, and D. B. Oerther. 2008. Bifidobacteria in feces and environmental waters. Appl. Environ. Microbiol. 74:575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Moller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung, K., and E. Topp. 2001. Bacterial community dynamics in liquid swine manure during storage: molecular analysis using DGGE/PCR of 16S rDNA. FEMS Microbiol. Ecol. 38:169-177. [Google Scholar]
- 36.Loisel, P., J. Harmand, O. Zemb, E. Latrille, C. Lobry, J. P. Delgenes, and J. J. Godon. 2006. Denaturing gradient electrophoresis (DGE) and single-strand conformation polymorphism (SSCP) molecular fingerprintings revisited by simulation and used as a tool to measure microbial diversity. Environ. Microbiol. 8:720-731. [DOI] [PubMed] [Google Scholar]
- 37.Lu, J., J. Santo Domingo, and O. C. Shanks. 2007. Identification of chicken-specific fecal microbial sequences using a metagenomic approach. Water Res. 41:3561-3574. [DOI] [PubMed] [Google Scholar]
- 38.Lynch, P. A., B. J. Gilpin, L. W. Sinton, and M. G. Savill. 2002. The detection of Bifidobacterium adolescentis by colony hybridization as an indicator of human faecal pollution. J. Appl. Microbiol. 92:526-533. [DOI] [PubMed] [Google Scholar]
- 39.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Kado, T. Takada, K. Matsumoto, and R. Tanaka. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 70:167-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayrhofer, S., K. J. Domig, E. Amtmann, A. H. Van Hoek, A. Petersson, C. Mair, H. K. Mayer, and W. Kneifel. 2007. Antibiotic susceptibility of Bifidobacterium thermophilum and Bifidobacterium pseudolongum isolates from animal sources. J. Food Prot. 70:119-124. [DOI] [PubMed] [Google Scholar]
- 41.Mølbak, L., L. E. Thomsen, T. K. Jensen, K. E. Bach Knudsen, and M. Boye. 2007. Increased amount of Bifidobacterium thermacidophilum and Megasphaera elsdenii in the colonic microbiota of pigs fed a swine dysentery preventive diet containing chicory roots and sweet lupine. J. Appl. Microbiol. 103:1853-1867. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen, J. L., A. Schramm, A. E. Bernhard, G. J. van den Engh, and D. A. Stahl. 2004. Flow cytometry-assisted cloning of specific sequence motifs from complex 16S rRNA gene libraries. Appl. Environ. Microbiol. 70:7550-7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okabe, S., N. Okayama, O. Savichtcheva, and T. Ito. 2007. Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl. Microbiol. Biotechnol. 74:890-901. [DOI] [PubMed] [Google Scholar]
- 44.Okabe, S., and Y. Shimazu. 2007. Persistence of host-specific Bacteroides-Prevotella 16S rRNA genetic markers in environmental waters: effects of temperature and salinity. Appl. Microbiol. Biotechnol. 76:935-944. [DOI] [PubMed] [Google Scholar]
- 45.Peu, P., H. Brugere, A. M. Pourcher, M. Kerouredan, J. J. Godon, J. P. Delgenes, and P. Dabert. 2006. Dynamics of a pig slurry microbial community during anaerobic storage and management. Appl. Environ. Microbiol. 72:3578-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phung, N. T., J. Lee, K. H. Kang, I. S. Chang, G. M. Gadd, and B. H. Kim. 2004. Analysis of microbial diversity in oligotrophic microbial fuel cells using 16S rDNA sequences. FEMS Microbiol. Lett. 233:77-82. [DOI] [PubMed] [Google Scholar]
- 47.Savichtcheva, O., N. Okayama, T. Ito, and S. Okabe. 2005. Application of a direct fluorescence-based live/dead staining combined with fluorescence in situ hybridization for assessment of survival rate of Bacteroides spp. in drinking water. Biotechnol. Bioeng. 92:356-363. [DOI] [PubMed] [Google Scholar]
- 48.Seurinck, S., M. Verdievel, W. Verstraete, and S. D. Siciliano. 2006. Identification of human fecal pollution sources in a coastal area: a case study at Oostende (Belgium). J. Water Health 4:167-175. [PubMed] [Google Scholar]
- 49.Shanks, O. C., J. W. Santo Domingo, R. Lamendella, C. A. Kelty, and J. E. Graham. 2006. Competitive metagenomic DNA hybridization identifies host-specific microbial genetic markers in cow fecal samples. Appl. Environ. Microbiol. 72:4054-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simpson, P. J., C. Stanton, G. F. Fitzgerald, and R. P. Ross. 2003. Genomic diversity and relatedness of bifidobacteria isolated from a porcine cecum. J. Bacteriol. 185:2571-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snell-Castro, R., J. J. Godon, J. P. Delgenes, and P. Dabert. 2005. Characterisation of the microbial diversity in a pig manure storage pit using small subunit rDNA sequence analysis. FEMS Microbiol. Ecol. 52:229-242. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tzortzis, G., A. K. Goulas, J. M. Gee, and G. R. Gibson. 2005. A novel galactooligosaccharide mixture increases the bifidobacterial population numbers in a continuous in vitro fermentation system and in the proximal colonic contents of pigs in vivo. J. Nutr. 135:1726-1731. [DOI] [PubMed] [Google Scholar]
- 54.Ufnar, J. A., D. F. Ufnar, S. Y. Wang, and R. D. Ellender. 2007. Development of a swine-specific fecal pollution marker based on host differences in methanogen mcrA genes. Appl. Environ. Microbiol. 73:5209-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Dyke, M. I., and A. J. McCarthy. 2002. Molecular biological detection and characterization of Clostridium populations in municipal landfill sites. Appl. Environ. Microbiol. 68:2049-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ventura, M., C. Canchaya, A. Del Casale, F. Dellaglio, E. Neviani, G. F. Fitzgerald, and D. van Sinderen. 2006. Analysis of bifidobacterial evolution using a multilocus approach. Int. J. Syst. Evol. Microbiol. 56:2783-2792. [DOI] [PubMed] [Google Scholar]
- 57.von Ah, U., V. Mozzetti, C. Lacroix, E. E. Kheadr, I. Fliss, and L. Meile. 2007. Classification of a moderately oxygen-tolerant isolate from baby faeces as Bifidobacterium thermophilum. BMC Microbiol. 7:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitehead, T. R., and M. A. Cotta. 2001. Characterisation and comparison of microbial populations in swine faeces and manure storage pits by 16S rDNA gene sequence analyses. Anaerobe 7:181-187. [Google Scholar]
- 59.Wood, J., K. P. Scott, G. Avgustin, C. J. Newbold, and H. J. Flint. 1998. Estimation of the relative abundance of different Bacteroides and Prevotella ribotypes in gut samples by restriction enzyme profiling of PCR-amplified 16S rRNA gene sequences. Appl. Environ. Microbiol. 64:3683-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu, L., W. Li, and X. Dong. 2003. Species identification of genus Bifidobacterium based on partial HSP60 gene sequences and proposal of Bifidobacterium thermacidophilum subsp. porcinum subsp. nov. Int. J. Syst. Evol. Microbiol. 53(Pt. 5):1619-1623. [DOI] [PubMed] [Google Scholar]


