Abstract
Microbial biotransformations have a major impact on environments contaminated with toxic elements, including arsenic, resulting in an increasing interest in strategies responsible for how bacteria cope with arsenic. In the present work, we investigated the metabolism of this metalloid in the bacterium Ochrobactrum tritici SCII24. This heterotrophic organism contains two different ars operons and is able to oxidize arsenite to arsenate. The presence of arsenite oxidase genes in this organism was evaluated, and sequence analysis revealed structural genes for an As(III) oxidase (aoxAB), a c-type cytochrome (cytC), and molybdopterin biosynthesis (moeA). Two other genes coding for a two-component signal transduction pair (aoxRS) were also identified upstream from the previous gene cluster. The involvement of aox genes in As(III) oxidation was confirmed by functionally expressing them into O. tritici 5bvl1, a non-As(III) oxidizer. Experiments showed that the As(III) oxidation process in O. tritici requires not only the enzyme arsenite oxidase but also the cytochrome c encoded in the operon. The fundamental role of this cytochrome c, reduced in the presence of arsenite in strain SCII24 but not in an O. tritici ΔaoxB mutant, is surprising, since to date this feature has not been found in other organisms. In this strain the presence of an aox system does not seem to confer an additional arsenite resistance capability; however, it might act as part of an As(III)-detoxifying strategy. Such mechanisms may have played a crucial role in the development of early stages of life on Earth and may one day be exploited as part of a potential bioremediation strategy in toxic environments.
Arsenic is naturally present in soil, water, and air, and arsenic contamination of drinking water constitutes an important public health problem in numerous countries throughout the world (33). Arsenic occurs in nature in the oxidation states +5 (arsenate), +3 (arsenite), 0 (elemental arsenic), and −3 (arsine). Although arsenic is most notorious as a poison threatening human health, recent studies suggest that arsenic species may have been involved in the ancestral taming of energy and played a crucial role in early stages in the development of life on Earth (reviewed in reference 34). The two soluble arsenic species, arsenate [As(V) as H2AsO4− and HAsO42−] and arsenite [As(III) as H3AsO30 and H2AsO3−] are the most common forms and exhibit different toxicities for living organisms. Several studies have documented the role of bacteria on speciation and mobilization of arsenic in the environment (23). Microorganisms are known to influence arsenic geochemistry by their metabolism, i.e., reduction, oxidation, and methylation (for reviews, see references 5, 19, and 22), affecting both the speciation and the toxicity of this element. Arsenate is less toxic than arsenite, but paradoxically, resistance to As(V) requires its reduction to As(III), which is then extruded by an active efflux pump.
Another well-documented arsenic transformation is the microbiological oxidation of arsenite to arsenate. This redox reaction is generally carried out by microorganisms either for detoxification or for energy generation to support cellular growth (23). The oxidation of As(III) by heterotrophic microorganisms is generally considered to be a detoxification strategy, since the microbes do not gain energy from this reaction (32). These heterotrophic As-oxidizing organisms include the most-studied Alcaligenes faecalis (3), Herminiimonas arsenoxidans (21), Thermus species (13, 14), Hydrogenophaga sp. strain NT-14 (35), and Agrobacterium tumefaciens (17). In contrast, other organisms have been described as autotrophic As(III) oxidizers able to use the energy gained from the oxidation reaction for growth. Autotrophic As(III) oxidation has been best studied in strain NT-26 (27, 28) but has also been reported for Thiomonas sp. (10), strain MLHE1 (24), and other environmental isolates (7, 16, 25, 26).
Of the arsenite-oxidizing bacteria, A. faecalis (3), NT-26 (27), and NT-14 (35) have been studied in detail and their arsenite oxidases purified and characterized. Moreover, a crystal structure of the A. faecalis arsenite oxidase has been elucidated (11). Genes encoding As(III) oxidases (aox) have also been identified and sequenced in several organisms, showing a common genetic organization, aoxA-aoxB, that encodes the small and large subunits, respectively. These aox operons usually contain additional genes, e.g., cytC, which encodes a cytochrome c, and moeA, which encodes an enzyme involved in molybdenum cofactor biosynthesis (32).
The genome exploration of the alphaproteobacterium Ochrobactrum tritici revealed that it possesses heretofore-unsuspected mechanisms for coping with arsenic. This work reports the identification of a locus involved in arsenic oxidation in a heterotrophic bacterium previously characterized as carrying two operons involved in arsenic resistance. One operon confers resistance to arsenite and antimonite, while the second one is responsible for resistance to arsenate.
MATERIALS AND METHODS
Media and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. O. tritici strains were grown aerobically at 30°C in a minimal defined medium (MDM) that consisted of MgSO4·7H2O at 1.0 g/liter, NH4Cl at 1.0 g/liter, Na2SO4 at 0.5 g/liter, K2HPO4 at 0.01 g/liter, CaCl2 at 0.067 g/liter, NaHCO3 at 0.08 g/liter, and 1% stock vitamin solution (31). The final pH of the medium was 7.2. Chemolithoautotrophic growth was achieved with arsenite as the electron donor and the heterotrophic growth medium was supplemented with 0.05% yeast extract with or without arsenite. Escherichia coli S17-1 was cultivated on Luria-Bertani medium (Difco). Gentamicin (15 μg/ml) and ampicillin (100 μg/ml) were included for selection of transformants when mobilizing plasmids into strain 5bvl1.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| Strains | ||
| O. tritici SCII24 | Type strain, As resistant, As(III) oxidizing | 6 |
| O. tritici 5bvl1 | As sensitive, not As(III) oxidizing | This study |
| 5pBBR | Strain 5bvl1 carrying vector pBBR1MCS-5 | This study |
| 5PaoxAB | Strain 5bvl1 carrying vector pBBR1MCS-5 containing the aox promoter and aoxA and aoxB genes | This study |
| 5PaoxABC | Strain 5bvl1 carrying vector pBBR1MCS-5 containing the aox promoter and aoxA, aoxB, and cytC genes | This study |
| O. tritici ΔaoxB | Mutant aoxB | This study |
| E. coli S17-1 | Pro− Mob+, conjugation donor | 9 |
| Plasmids | ||
| pK18mob | pUC18 derivative, lacZα Kanr, mob site, suicide plasmid | 30 |
| pBBR1MCS-5 | Broad-host-range cloning vector, mob site, InsP Gmr | 18 |
Amplification and sequencing of aox genes.
The amino acid sequences of some previously identified arsenite oxidase large subunits (aoxB) were aligned using ClustalW (15), and degenerated oligonucleotide primers were designed based on the conserved motifs aoxBf (5′-TGYCAYTWYTGYATCGTCGGCTG-3′) and aoxBr (5′-RWAGCCRAAMAGCATRAAGGT-3′), with Y indicating C/T, W indicating A/T, R indicating A/G, and M indicating A/C. PCRs were carried out according to standard protocols, and a PCR product of the appropriate length (about 2.25 kbp) was amplified, purified, and sequenced by using an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA). Upstream and downstream sequences were identified using inverse PCR (IPCR). Templates for IPCR were obtained from about 1 μg of total DNA digested with enzymes that did not cut inside of the sequence previously obtained. The digested DNA preparations (500 ng) were ligated overnight at 14°C in a volume of 50 μl with 3 U of T4 DNA ligase (Roche). DNA flanking this sequenced region was amplified by PCR with specific reverse and forward primers for 5′ and 3′ sequences, respectively. The PCR products obtained were purified and sequenced. Database searches and sequence analyses were performed using the BLAST program (1).
Cloning of arsenite oxidation genes in strain 5bvl1.
The aoxAB and aoxABcytC genes were amplified by PCR using specific forward and reverse primers containing additional KpnI and BamHI recognition sites, respectively. The PCR products were purified after digestion with KpnI and BamHI and ligated into the corresponding sites of a pBBR1MCS-5 vector (18). Then, each construct was transformed into competent E. coli S17-1 cells. These plasmids were mobilized into strain 5bvl1 by biparental conjugation according to standard protocols (9). All strain-plasmid combinations were analyzed for their As(III) oxidation abilities.
As(III) oxidation analysis.
O. tritici strains were checked for oxidation of arsenite to arsenate by the AgNO3 method. Agar plates were flooded with a solution of 0.1 M AgNO3. A brownish color revealed the presence of arsenate in the medium, while the presence of arsenite was detected by a yellow color.
The conversion of As(III) to As(V) was also measured in liquid medium using a colorimetric method. As(V), in contrast to As(III) but similarly to phosphate, can react with the molybdate, producing a complex of blue color that has an absorbance peak at 820 nm (2). Then, this complex is measured using a UV/Vis spectrophotometer. Controls employing the culture medium were performed to exclude any phosphate interference in arsenate determination. A standard curve correlating absorbance and concentrations of As(V) was prepared to convert the A820 into As(V) concentrations.
Preparation of total protein extracts and enzyme assays.
For preparation of total protein extracts, O. tritici SCII24 was grown in the presence or absence of As(III) to late log phase and cells were harvested by centrifugation and resuspended in cold 50 mM morpholineethanesulfonic acid (pH 6.0). Cells were disrupted by double passage through a French press. Unbroken cells were removed by centrifugation at 30,000 × g (4°C). The arsenite oxidase activity was measured as described previously by Anderson and coworkers (3). The reduction of the artificial electron acceptor 2,4-dichlorophenolindophenol (60 μM) was monitored at 600 nm in the presence of 200 μM As(III). The Bradford reagent was used to determine protein concentrations.
Mutagenesis.
The aoxB gene was cloned into the suicide plasmid pK18mob and was disrupted by insertion of a tetracycline resistance gene into its HindIII restriction site. This plasmid, pΔaoxB, was transformed into E. coli S17-1 and transferred into O. tritici SCII24 by conjugation. Transconjugants selected from plates with tetracycline (15 μg/ml) and ampicillin (100 μg/ml) were tested for their ability to oxidize arsenite. Positive mutants (O. tritici ΔaoxB) were confirmed by PCR.
Redox activity assay.
The reduction abilities of c-type cytochromes upon addition of As(III) were measured as previously described (4, 8). Cell extracts of the O. tritici type strain and O. tritici ΔaoxB were obtained by disruption through a French press (Thermo Electron Corporation) at 12,000 lb/in2 in Tris-HCl (100 mM; pH 7.2), followed by centrifugation at 19,000 × g for 10 min at 4°C to remove unbroken cells and debris. The extracts (26.3 to 32.4 mg protein/ml) were flushed with nitrogen and 1 ml was transferred to a cuvette. The extracts were completely oxidized by adding 40 μl potassium ferrocyanide (100 mM) together with 90 μl potassium cyanide (0.5 M). Arsenite (10 mM) was added to the extracts to test for reduction of the c-type cytochromes. Sodium dithionite was added to completely reduce cytochromes at the end of the assays. Optical absorption spectra were measured between 400 and 650 nm (6405 UV/Vis spectrophotometer; Jenway).
RT-PCR experiments.
RNA was prepared from O. tritici SCII24 cells grown in MDM to exponential phase. Total RNA was extracted with RNeasy total RNA (Qiagen) according to the manufacturer's instructions and was treated with DNase I (Roche). Reverse transcription-PCRs (RT-PCRs) were carried out using the AccessQuick RT-PCR system (Promega) according to the manufacturer's instructions. The primers used in these RT-PCRs were the following: aoxRf, 5′-ACTCGGCATTTCACGCACAAC-3′; aoxAf, 5′-TCCGTTGAGCTATTCGGCGGA-3′; aox1Bf, 5′-ATCGTTTGGCAATCTGCCTTTC-3′; aox2Bf, 5′-TCGACAAATCGACCTTGCGCC-3′; cytCf, 5′-ACACTATCGTTATGCGTGCTTATG-3′; aoxAr, 5′-ACGGTTGACAGGATACTCGAG-3′; aox1Br, 5′-AGCTTGTCGGCTGCATCTGGCC-3′; aox2Br, 5′-TCCGTATAGAGACGCTGGGTG-3′; cytCr, 5′-TCATGGTGTCTGGAATGTTTTAA-3′. RT-PCR products were examined by agarose gel electrophoresis, and the absence of DNA contamination of the RNA was confirmed by PCR using Taq polymerase without reverse transcriptase.
Nucleotide sequence accession number.
The nucleotide sequence determined in this study for O. tritici SCII24 has been deposited in GenBank under accession number FJ465505.
RESULTS
Growth of strain O. tritici SCII24 in medium with arsenite.
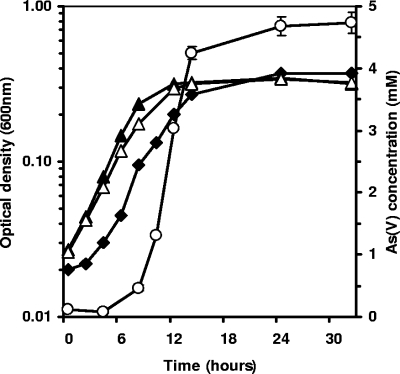
Strain O. tritici SCII24 was not able to grow in minimal medium without or with arsenite as electron donor (see Fig. S1A in the supplemental material). Inoculation of this strain into minimal medium supplemented with organic matter (0.05% yeast extract) and in the presence or absence of As(III) resulted in bacterial growth (see Fig. S1B in the supplemental material). In addition, O. tritici in medium with a high concentration of arsenite (5 mM) was able to oxidize arsenite to arsenate. As shown in Fig. 1, this strain revealed a remarkable arsenite-oxidizing rate, being able to oxidize almost 5 mM As(III) within 14 h of bacterial growth. However, this As(III) oxidizer was only able to grow in the presence of an extra source of energy and carbon (yeast extract). This result indicates that this organism cannot gain energy from As(III) oxidation.
FIG. 1.
Growth of O. tritici SCII24 and O. tritici 5bvl1 in MDM supplemented with yeast extract (0.05%). Results shown include the optical density at 600 nm of O. tritici SCII24 in the absence (Δ) and presence (⧫) of 5 mM As(III), the optical density of O. tritici 5bvl1 in medium without arsenite (▴), and the concentration of arsenate resulting from O. tritici SCII24 growth (○).
Sequencing of the arsenite oxidase-encoding genes.
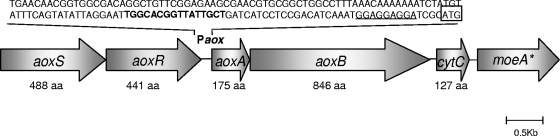
Using degenerate oligonucleotide primers for the large subunit of the arsenite oxidases followed by IPCR and primer walking techniques, different open reading frames (ORFs) related to the genes involved in arsenite oxidation were found in the DNA of O. tritici SCII24. Based on the BLAST analysis, the cluster is comprised by several ORFs which encode proteins that belong to families previously reported to be involved in arsenite oxidation, such as AoxS, AoxR, AoxA, AoxB, CytC, and MoeA (Fig. 2). Upstream of the aoxA gene, we identified a putative promoter that comprised a consensus sequence, TGGCACX5TTGCW, that was also found in strain NT-26 (29).
FIG. 2.
Genetic organization of the aox gene cluster in O. tritici SCII24. Upstream of aoxA is a putative promoter sequence (Paox). The consensus sequence is indicated in bold, the ribosome-binding site is underlined, and the start codon is in a box.
The aoxB gene encodes the large subunit of the arsenite oxidase protein (AoxB), with 846 amino acids, and shows clear homologies to AoxB proteins from different organisms. Alignment with the different proteins showed the highest identity (92%) with the larger subunit of the arsenite oxidase from Agrobacterium tumefaciens 5A (AoxB) (17) and Rhizobium sp. strain NT-26 (AroA) (27). The [3Fe-4S] cluster-binding conserved motif, C-X2-C-X3-C-X70-S, of Alcaligenes faecalis arsenite oxidase, in which most of the residues interact with the molybdenum center (11), are conserved in AoxB of O. tritici. Moreover, the four amino acids (H, E, R, and H) that have been predicted to bind As(III) (11) also appear in O. tritici AoxB (His199, Glu207, Arg488, and His452).
Immediately upstream from aoxB is the aoxA gene, which encodes the arsenite oxidase small subunit. The predicted protein AoxA (175 amino acids) displayed 93% identity and 91% identity with the AroB and AoxA from strains NT-26 and A. tumefaciens, respectively. The SignalP prediction software (12) predicted a cleavage site located between Ala40 and Ala41. This 40-amino-acid-long, predicted TAT signal sequence contains the usual conserved motifs: a positively charged region harboring the conserved motif RRXFL, a region rich in hydrophobic amino acids, and a motif in which a cleavage site is located. The residues coordinating the [2Fe-2S] Rieske cluster (C-X-H-X15-C-X2-H) detected in other small subunits of arsenite oxidases are also conserved in AoxA of O. tritici. Downstream from aoxB, there is an ORF which, based on a BLASTP search, encodes a cytochrome c (127 amino acids) that displays high sequence identity to the strain NT-26 (75%) and A. tumefaciens (74%) c-type cytochrome 552. The N-terminal sequence of the cytochrome contains a conserved motif, C-X2-C-H, which is characteristic of heme-binding sites. Based on analysis with the SignalP program (12), the c552 has a predicted signal peptide cleavage site between Ala20 and Glu21. Since it does not contain any other transmembrane domains, it is assumed to be a periplasmic protein. Further downstream, an additional ORF (partial) was detected which encodes a protein homologous to the members of MoeA family, which usually are involved in biosynthesis of the molybdenum cofactor, an important cofactor of a diverse group of enzymes, including AoxAB.
Upstream from aoxA and the putative promoter was found an ORF (aoxR) that encodes a 441-amino-acid protein homologous to a putative transcriptional regulator from A. tumefaciens (89% identity). This product (AoxR) has the amino acid structural features common to regulators of two-component signal transduction systems. This protein also contains the same residues as A. tumefaciens, Asp13, Asp14, Asp58, and Lys107, that are considered to be important in the process of the phosphorylation signal essential for regulatory function (17). Immediately upstream of aoxR was an ORF (aoxS) coding for a 488-amino-acid protein that displays high identity to a putative sensor histidine kinase of A. tumefaciens (82%).
Arsenite oxidase activity of O. tritici SCII24.
Arsenite oxidase activity was determined in crude extracts of cells grown in minimal medium containing yeast extract (0.05%) and in the presence or absence of As(III) (5 mM). It was found that the optimum pH for arsenite oxidase activity (average of triplicate measurements) was at pH 6.0. Arsenite oxidase activity was found exclusively associated with the soluble fraction, with 0.065 and 0.0054 μmol As(III) oxidized min−1 mg of protein−1 in cells grown in the presence or absence of As(III), respectively, while no activity was detected in the membrane fraction. The arsenite oxidase enzyme is located most probably in the periplasm in O. tritici SCII24, as has been observed in Hydrogenophaga sp. strain NT-14 (35) and strain NT-26 (28), while it is located in the membrane fraction in A. faecalis (3) and Herminiimonas arsenicoxydans strain ULPAs1 (21).
Ability of aox gene products to confer arsenite oxidation.
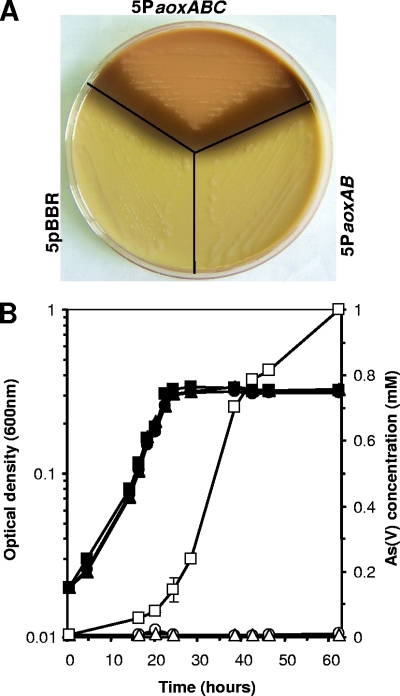
Our previous studies showed that the wild-type O. tritici SCII24 is arsenate resistant and contains two operons coding for two different arsenate reductases (6). Nevertheless, the strain exhibits an As(III)-oxidizing phenotype in the presence of arsenite. Different constructs of the aox genes were used to evaluate the role of these genes in the arsenite oxidation process by O. tritici 5bvl1, a non-As(III) oxidizer. Silver nitrate staining of the strain 5bvl1 carrying several constructs showed that only 5PaoxABC could convert As(III) to As(V) (Fig. 3A). In contrast, an arsenite-oxidizing phenotype was not observed in cells of strain 5bvl1 carrying the empty vector (5pBBR) or carrying the construct PaoxAB (5PaoxAB). The importance of the genes aoxA-B-cytC on the arsenite oxidation mechanism was also confirmed when strain 5bvl1 carrying this group of genes, in contrast to other constructs, was able to convert As(III) to As(V) (Fig. 3B). However, the onset of arsenate production seems to occur later in growth in strain 5PaoxABC compared to strain SCII24 (Fig. 1). We also sought to determine whether these strain-plasmid combinations were more resistant to As(III) than the wild-type strain 5bvl1. In liquid-grown cultures amended with 1 mM As(III), there was no difference in growth phenotype between the native strain without any arsenite oxidase gene and the strain 5bvl1 harboring the PaoxAB or PaoxABC constructs (Fig. 3B).
FIG. 3.
Heterologous expression of aox genes in O. tritici 5bvl1. (A) Comparison of the As(III) oxidation phenotypes of strain 5bvl1 carrying the different clones (empty vector, PaoxAB, and PaoxABcytC). The presence of As(V) is indicated by the dark color, whereas the presence of As(III) is indicated by yellow. (B) Growth of several constructs in MDM with yeast extract (0.05%) amended with 1 mM As(III). Culture growth is shown with filled symbols and As(V) concentration is shown with open symbols: 5pBBR (circles), 5PaoxAB (triangles), and 5PaoxABC (squares). Error bars represent standard errors of the means calculated from triplicate experiments.
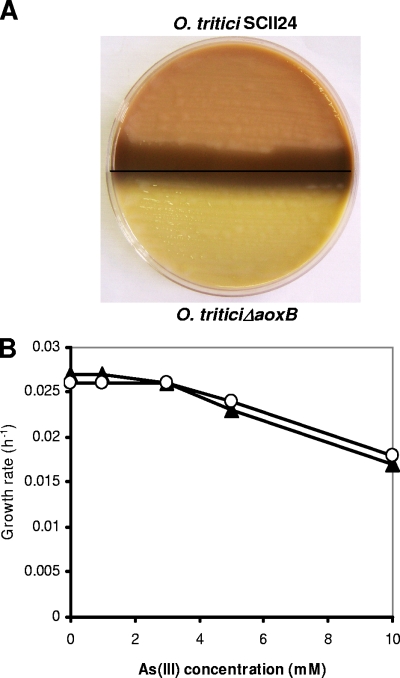
The aox mutant (O. tritici ΔaoxB) failed to oxidize As(III) to As(V), as was revealed by the yellow color associated with agar inoculated with the mutant (Fig. 4A). Even though that mutant did not show arsenite oxidase activity, the mutation did not seem to affect the arsenite resistance, given that the mutant was able to grow in the presence of high arsenite concentrations (Fig. 4B).
FIG. 4.
(A) Effect of aoxB mutation on the arsenite oxidation phenotype of O. tritici SCII24. The phenotype of mutant O. tritici ΔaoxB was compared with wild type. (B) Growth rates for the wild type (○) and aox mutant (▴) in response to increasing arsenite concentrations. Cultures were grown overnight in MDM supplemented with yeast extract (0.05%).
Redox activities.
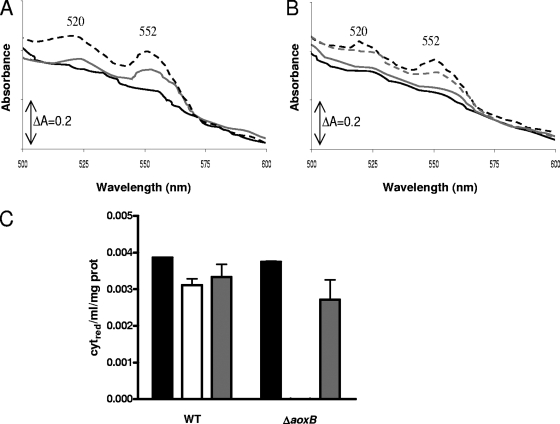
The ability of c-type cytochromes to serve as electron acceptors during As(III) oxidation by the arsenite oxidase was investigated by spectroscopy. The source of both arsenite oxidase and cytochrome c was crude cell homogenate, which also included membrane components. The spectrum obtained from the samples showed three peaks, as expected, at 417, 523, and 552 nm, characteristic of ferrous c-type cytochromes. In Fig. 5A, the cytochromes of the O. tritici type strain cell extract were totally oxidized by potassium ferrocyanide, and after addition of 10 mM arsenite showed reduction within 1 min. The response to 10 mM arsenite was maximal, as higher concentrations of arsenite did not increase the peak intensity. The same was observed after the addition of dithionite. The mutant O. tritici ΔaoxB, under the same experimental conditions, did not exhibit the same spectra, failing to show cytochrome c reduction upon addition of arsenite (Fig. 5B). However, the following addition of dithionite caused reduction of the c-type cytochromes present in the cell extract, resulting in cytochrome c reduction to an extent similar to the one observed with the wild-type strain (Fig. 5C).
FIG. 5.
Effects of arsenite on cell extracts of O. tritici type strain (A) and mutant O. tritici ΔaoxB (B) containing c-type cytochromes and relative cytochrome c reduction extension per milligram of protein by arsenite in O. tritici wild-type (WT) and O. tritici ΔaoxB mutant cells (C). The black broken line represents the extract at the beginning of the assay, prior to oxidation with ferrocyanide. The black solid line shows the totally oxidized extract, after addition of ferrocyanide and potassium cyanide. The gray line shows the spectrum obtained after addition of 10 mM arsenite. The gray broken line represents the spectrum obtained after full reduction by dithionite and is not shown in panel A, as it superposes with the gray solid line. The relative cytochrome c reduction was determined from the difference between the change in absorbance at 552 and 570 nm [ΔA(552-570)] in the assay and the ΔA(552-570) when totally oxidized by ferrocyanide. The black bar represents cytochrome c reduction at the beginning of the assay; the white bar represents cytochrome c reduction after complete oxidation with ferrocyanide followed by addition of 10 mM As(III); the gray bar represents cytochrome c reduction after addition of sodium dithionite. Results are averages of three independent assays.
Transcriptional analysis of the aox operon by RT-PCR.
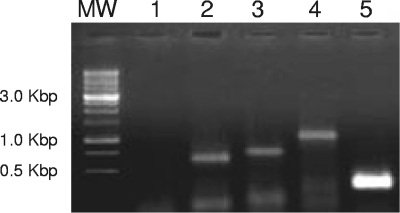
The genetic organization of the aox operon suggests that the genes aoxS-R-A-B-cytC might be transcribed together. To analyze this possibility, O. tritici SCII24 was grown and RNA was isolated and used in RT-PCR experiments. As can be observed in Fig. 6, no fragment was detected using primers directed to the intergenic region between aoxR and aoxA. However, a fragment corresponding to the intergenic region between aoxB and cytC was observed. These results suggest the existence of coexpression of aoxA-B-cytC independent of the remaining genes, aoxS and aoxR.
FIG. 6.
RT-PCR analysis of the arsenite oxidase gene cluster. Lanes show the RT-PCR products from total RNA extracted from O. tritici SCII24. Lanes: 1, no amplification using primers (aoxRf and aoxAr) for intergenic region aoxR-aoxA; 2, successful amplification using primers (aoxAf and aox1Br) for intergenic region aoxA-aoxB; 3, successful amplification using primers (aoxB1f and cytCr) for intergenic region aoxB-cytC; 4, amplification of an inner region from the aoxB gene using the primers aox2Bf and aox2Br; 5, amplification of the cytC gene using the primers cytCf and cytCr. MW, molecular weight standards.
DISCUSSION
This study describes the contribution of the heterotrophic alphaproteobacterium O. tritici SCII24 to the arsenic cycle. This organism was previously reported to be able to resist up to 50 mM As(III), 10 mM Sb(III), and more than 200 mM As(V), which makes it one of the most resistant microorganisms described to date (6). This resistance was attributed to the presence of two functional ars operons (ars1 and ars2). The ars1 operon confers resistance to arsenite and antimonite, and ars2 is responsible for resistance to arsenite and arsenate. At the time, it was not overruled that other mechanisms in the cell could participate in arsenic resistance. Subsequent studies reported here led to the identification in this strain of an operon coding for an arsenite oxidase and a cytochrome c and demonstrated that these genes are essential for arsenite oxidation in O. tritici strain SCII24.
The physiological importance of arsenite oxidation in O. tritici strain SCII24 is not completely clear, since this process does not seem to be directly implicated in the energetic metabolism but mainly seems to be a detoxification process for the cell. Interestingly, an insertion mutation in the arsenite oxidase gene did not prevent the organism from growing in the presence of arsenite, suggesting that ars1 and ars2 (6) are the most important operons for the O. tritici SCII24 arsenic resistance phenotype. Both reactions, As(III) oxidation and As(V) reduction, may occur simultaneously in the wild-type strain, but the results indicate that the rate of As(III) oxidation exceeded As(V) reduction, resulting in an arsenite-oxidizing phenotype.
The genetic organization of aoxAB, with the small Rieske subunit gene upstream from the catalytic molybdopterin subunit, is conserved in all the arsenite-oxidizing prokaryotes analyzed to date, while no obvious conservation was observed in the ORFs flanking these two genes. However, in A. tumefaciens (17), Thiomonas sp. strain 3As (10), strain NT-26 (27), and O. tritici SCII24 (this work), the aoxAB genes have been shown to be cotranscribed with the downstream gene, a c-type cytochrome. Redox studies with O. tritici SCII24 and a mutant demonstrated that arsenite oxidation requires the presence of cytochrome c and a functional arsenite oxidase. Therefore, cytochrome c is essential in As(III) oxidation, and most probably the cytochrome c and the arsenite oxidase (the molybdenum and Rieske Fe-S subunits) constitute an electron transport system in O. tritici SCII24. This result was unexpected, since in strain NT-26, cytC expression is not considered essential for arsenite oxidation (29). In spite of the detection of cytC in the genome of some As(III)-oxidizing bacteria, little work has been performed to clarify the exact role of cytochrome c in arsenite oxidation. Consequently, it is not possible to know whether this essential role of CytC in the oxidation of arsenite is a particular characteristic of O. tritici strain SCII24 or can be extended to other organisms.
The genetic organization of the arsenite oxidase gene clusters of O. tritici strain SCII24 and A. tumefaciens are similar, although the location of the putative promoter upstream from aoxAB and the absence of cotranscription of aoxR-aoxA indicate that the aox operon of O. tritici could be regulated differently from previously described operons. In the case of A. tumefaciens, a large operon, aoxSRABcytC, was required to confer arsenite oxidase ability (17), and in H. arsenicoxydans the inactivation of aoxRS led to a complete loss of arsenite oxidase activity (20). The constructions in O. tritici strain 5bvl1 showed that aoxABcytC genes were essential for efficient arsenite oxidation, and the expression of aoxS or aoxR genes was not necessary. This finding also indicates that other potential downstream operon components may not be required for As(III) oxidation in this organism. Possible interference of native AoxSR proteins from strain 5bvl1 was not considered, since Southern hybridization analysis of total DNA from strain 5bvl1 to detect aoxSR homologues gave a negative result (see Fig. S2 in the supplemental material). The As(III) oxidation ability of strain 5PaoxABC was observed only in the late exponential growth phase, suggesting that transcription of the aox operon in O. tritici could involve other factors, such as those related to quorum sensing. In fact, a quorum-sensing-based response was shown to be a second regulatory circuit for aox transcription in A. tumefaciens (17).
Bacteria metabolizing toxic elements represent an attractive tool to remediate contaminated sites. In this case, the extremely important ability of O. tritici strain SCII24 to oxidize As(III) to As(V), a less toxic and more easily immobilized form, can be considered a useful step within arsenic bioremediation strategy. This capacity, associated with high arsenic resistance conferred by ars genes, emphasizes the contribution of this strain to restore livable conditions in environments contaminated with arsenic.
Supplementary Material
Acknowledgments
We thank A. Schäfer for providing the plasmid pK18mob.
This research was founded by Fundação para a Ciência e Tecnologia, Portugal, under project PPCDT/AMB/60909/2004. Rita Branco and Romeu Francisco were supported by Ph.D. grants from Fundação para a Ciência e Tecnologia.
Footnotes
Published ahead of print on 12 June 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames, B. N. 1966. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8:115-118. [Google Scholar]
- 3.Anderson, G. L., J. Williams, and R. Hille. 1992. The purification and characterization of arsenite oxidase from Alcaligenes faecalis, a molybdenum-containing hydroxylase. J. Biol. Chem. 267:23674-23682. [PubMed] [Google Scholar]
- 4.Bäcklund, A. S., J. Bohlin, N. Gustavsson, and T. Nilsson. 2009. Periplasmic c cytochromes and chlorate reduction in Ideonella dechloratans. Appl. Environ. Microbiol. 75:2439-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharjee, H., and B. P. Rosen. 2007. Arsenic metabolism in prokaryotic and eukaryotic microbes, p. 371-406. In D. H. Nies and S. Silver (ed.), Molecular microbiology of heavy metals. Springer-Verlag, Berlin, Germany.
- 6.Branco, R., A. P. Chung, and P. V. Morais. 2008. Sequencing and expression of two arsenic resistance operons with different functions in the highly arsenic-resistant strain Ochrobactrum tritici SCII24T. BMC Microbiol. 8:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, L., G. Liu, C. Rensing, and G. Wang. 2009. Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol. 9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Commack, R. 1995. Redox enzymes. Splitting molecular hydrogen. Nature 373:556-557. [DOI] [PubMed] [Google Scholar]
- 9.de Lourenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 10.Duquesne, K., A. Lieutaud, J. Ratouchniak, D. Muller, M.-C. Lett., and V. Bonnefoy. 2008. Arsenite oxidation by a chemoautotrophic moderately acidophilic Thiomonas sp.: from the strain isolation to the gene study. Environ. Microbiol. 10:228-237. [DOI] [PubMed] [Google Scholar]
- 11.Ellis, P. J., T. Conrads, R. Hille, and P. Kuhn. 2001. Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 Å and 2.03 Å. Structure 9:125-132. [DOI] [PubMed] [Google Scholar]
- 12.Emanuelsson, O., S. Brunak, G. von Heijne, and H. Nielsen. 2007. Locating proteins in the cell using TargetP, SignalP, and related tools. Nat. Protoc. 2:953-971. [DOI] [PubMed] [Google Scholar]
- 13.Gihring, T. M., and J. F. Banfield. 2001. Arsenite oxidation and arsenate respiration by a new Thermus isolate. FEMS Microbiol. Lett. 204:335-340. [DOI] [PubMed] [Google Scholar]
- 14.Gihring, T. M., G. K. Druschel, R. B. McCleskey, R. J. Hamers, and J. F. Banfield. 2001. Rapid oxidation by Thermus aquaticus and Thermus thermophilus: field and laboratory investigations. Environ. Sci. Technol. 35:3857-3862. [DOI] [PubMed] [Google Scholar]
- 15.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 16.Inskeep, W. P., R. E. Macur, N. Hamamura, T. P. Warelow, S. A. Ward, and J. M. Santini. 2007. Detection, diversity and expression of aerobic bacterial arsenite oxidase genes. Environ. Microbiol. 9:934-943. [DOI] [PubMed] [Google Scholar]
- 17.Kashyap, D. R., L. M. Botero, W. L. Franck, D. J. Hassett, and T. R. McDermott. 2006. Complex regulation of arsenite oxidation in Agrobacterium tumefaciens. J. Bacteriol. 188:1081-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, I. I., and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 19.Mukhopadhyay, R., B. P. Rosen, L. Phung, and S. Silver. 2002. Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol. Rev. 26:311-325. [DOI] [PubMed] [Google Scholar]
- 20.Muller, D., C. Médigue, S. Koechler, V. Barbe, M. Barakat, E. Talla, V. Bonnefoy, E. Krin, F. Arsène-Ploetze, C. Carapito, M. Chandler, B. Cournoyer, S. Cruveiller, C. Dossat, S. Duval, M. Heymann, E. Leize, A. Lieutaud, D. Lièvremont, Y. Makita, S. Mangenot, W. Nitschke, P. Ortet, N. Perdial, B. Schoepp, P. Siguier, D. D. Simeonova, Z. Rouy, B. Segurens, E. Turlin, D. Vaellenet, A. V. Dorsselaer, S. Weiss, J. Weissenbach, M.-C. Lett, A. Danchin, and P. N. Bertin. 2007. A tale of two oxidation states: bacterial colonization of arsenic-rich environments. PLoS Genet. 3:0518-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller, D., D. Lièvremont, D. D. Simeonova, J.-C. Hubert, and M.-C. Lett. 2003. Arsenite oxidase aox genes from a metal-resistant α-proteobacterium. J. Bacteriol. 185:135-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oremland, R. S., and J. F. Stolz. 2003. The ecology of arsenic. Science 300:939-944. [DOI] [PubMed] [Google Scholar]
- 23.Oremland, R. S., and J. F. Stolz. 2005. Arsenic, microbes and contaminated aquifers. Trends Microbiol. 13:45-49. [DOI] [PubMed] [Google Scholar]
- 24.Oremland, R. S., S. E. Hoeft, J. M. Santini, N. Bano, R. A. Hollibaugh, and J. T. Hollibaugh. 2002. Anaerobic oxidation of arsenite in Mono Lake water and by a facultative, arsenite-oxidizing chemoautotroph strain MLHE-1. Appl. Environ. Microbiol. 68:4795-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quéméneur, M., A. Heinrich-Salmeron, D. Muller, D. Lièvremont, M. Jauzein, P. N. Bertin, F. Garrido, and C. Joulian. 2008. Diversity surveys and evolutionary relationships of aoxB genes in aerobic arsenite-oxidizing bacteria. Appl. Environ. Microbiol. 74:4567-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhine, E. D., S. M. Ní Chadhain, G. J. Zylstra, and L. Y. Young. 2007. The arsenite oxidase genes (aroAB) in novel chemoautotrophic arsenite oxidizers. Biochem. Biophys. Res. Commun. 354:662-667. [DOI] [PubMed] [Google Scholar]
- 27.Santini, J. M., and R. N. vanden Hoven. 2004. Molybdenum-containing arsenite oxidase of the chemolithoautotrophic arsenite oxidizer NT-26. J. Bacteriol. 186:1614-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santini, J. M., L. I. Sly, R. D. Schnagl, and J. M. Macy. 2000. A new chemolithoautotrophic arsenite-oxidizing bacterium isolated from a gold mine: phylogenetic, physiological and preliminary biochemical studies. Appl. Environ. Microbiol. 66:92-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santini, J. M., U. Kappler, S. A. Ward, M. J. Honeychurch, R. N. vanden Hoven, and P. V. Bernhardt. 2007. The NT-26 cytochrome c552 and its role in arsenite oxidation. Biochem. Biophys. Acta 1767:189-196. [DOI] [PubMed] [Google Scholar]
- 30.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 31.Sharp, R. J., and R. A. Williams. 1988. Properties of Thermus ruber strains isolated from Icelandic hot springs and DNA:DNA homology of Thermus ruber and Thermus aquaticus. Appl. Environ. Microbiol. 54:2049-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silver, S., and L. T. Phung. 2005. Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 71:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, A. H., P. A. Lopipero, M. N. Bates, and C. M. Steinmaus. 2002. Public health. Arsenic epidemiology and drinking water standards. Science 296:2145-2146. [DOI] [PubMed] [Google Scholar]
- 34.Stolz, J. F., P. Basu, J. M. Santini, and R. S. Oremland. 2006. Arsenic and selenium in microbial metabolism. Annu. Rev. Microbiol. 60:107-130. [DOI] [PubMed] [Google Scholar]
- 35.vanden Hoven, R. N., and J. M. Santini. 2004. Arsenite oxidation by the heterotroph Hydrogenophaga sp. str. NT-14: the arsenite oxidase and its physiological electron acceptor. Biochem. Biophys. Acta 1656:148-155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.