Abstract
Soils harbor enormously diverse bacterial populations, and soil bacterial communities can vary greatly in composition across space. However, our understanding of the specific changes in soil bacterial community structure that occur across larger spatial scales is limited because most previous work has focused on either surveying a relatively small number of soils in detail or analyzing a larger number of soils with techniques that provide little detail about the phylogenetic structure of the bacterial communities. Here we used a bar-coded pyrosequencing technique to characterize bacterial communities in 88 soils from across North and South America, obtaining an average of 1,501 sequences per soil. We found that overall bacterial community composition, as measured by pairwise UniFrac distances, was significantly correlated with differences in soil pH (r = 0.79), largely driven by changes in the relative abundances of Acidobacteria, Actinobacteria, and Bacteroidetes across the range of soil pHs. In addition, soil pH explains a significant portion of the variability associated with observed changes in the phylogenetic structure within each dominant lineage. The overall phylogenetic diversity of the bacterial communities was also correlated with soil pH (R2 = 0.50), with peak diversity in soils with near-neutral pHs. Together, these results suggest that the structure of soil bacterial communities is predictable, to some degree, across larger spatial scales, and the effect of soil pH on bacterial community composition is evident at even relatively coarse levels of taxonomic resolution.
The biogeographical patterns exhibited by microbial communities have been examined in a wide range of environments, and studies focusing on microbial biogeography continue to be published at a rapid pace. We know that microbial community diversity and composition can vary considerably across space, and this variation is theorized to be linked to changes in a number of biotic or abiotic factors (22, 36, 41). There are numerous overarching reasons for this interest in understanding microbial biogeography. For example, comparing microbial patterns to those commonly observed in plant and animal taxa is of intense theoretical interest (22, 25). From a more practical standpoint, studies of microbial biogeography can often provide key insights into the physiologies, environmental tolerances, and ecological strategies of microbial taxa, particularly those difficult-to-culture taxa that often dominate in natural environments. However, perhaps the most important rationale for studying microbial biogeography is the most basic one: microbes are diverse, ubiquitous, and abundant, yet their biogeographical patterns and the factors driving these spatial patterns often remain poorly understood.
No single biogeographical pattern is shared by all microorganisms, just as there is no single biogeographical pattern followed by all “macrobial” (i.e., plant and animal) communities (31). The specific biogeographical patterns exhibited by microorganisms are variable and highly dependent on a number of factors, including the taxonomic group in question (29), the degree of phylogenetic resolution at which the communities are examined (e.g., Pseudomonas) (7), and the spatial scale of the study (40). However, some common patterns emerge if we specifically examine the biogeography of soil microorganisms. In particular, the structure and diversity of soil bacterial communities have been found to be closely related to soil environmental characteristics (5, 37, 47), and soil pH is often correlated with the observed biogeographical patterns (19, 24). However, due to the paucity of detailed and comprehensive studies of soil bacterial biogeography, particularly across larger spatial scales, our understanding of soil microbial biogeography remains incomplete.
Previous studies of soil bacterial biogeography have focused on either surveying a few soils in detail or surveying a larger number of soils by techniques that offer less detailed phylogenetic information. For example, a few recent studies used pyrosequencing or Sanger sequencing-based techniques to deeply survey the diversity and composition of the bacterial communities within a single soil or a few soils (1, 14, 20, 39, 42). Such studies are valuable in that they provide our best assessments of overall bacterial diversity and community structure and the relative abundances of specific bacterial taxa within soils. However, because such studies often examine only a limited number of soils, they do not allow for robust assessment of biogeographical patterns and the factors that may drive these patterns. Other studies have examined bacterial communities across a larger number of soils, using more limited techniques, such as fingerprinting methods that offer little specific phylogenetic information on bacterial community structure or techniques that describe communities at very coarse levels of taxonomic resolution (18, 19). A comprehensive assessment of the biogeographical patterns exhibited by soil bacterial communities requires both depth (individual communities surveyed at a reasonable level of phylogenetic detail) and breadth (examining a sufficiently large number of samples to assess spatial patterns). With the recent development of the bar-coded pyrosequencing technique (23), we need not sacrifice depth for breadth, or vice versa. This was demonstrated in several recent studies (2, 12, 17, 28) that used bar-coded pyrosequencing to simultaneously analyze relatively large numbers of individual samples, surveying the bacterial community in each sample to an extent that would be difficult (or prohibitively expensive) using standard cloning and Sanger sequencing techniques.
Here we apply the bar-coded pyrosequencing technique to examine the structure and diversity of bacterial communities in 88 soils collected from across North and South America. This work expands on a previous fingerprinting-based survey of bacterial communities across a similar set of soils (19), using the pyrosequencing technique to extend the analyses and to answer the following questions. Which taxa are most abundant in soil? How does the phylogenetic structure of bacterial communities vary across the continental scale? Which environmental factors best predict bacterial community structure and diversity? Are some soil bacterial phyla more diverse than others?
MATERIALS AND METHODS
Sample collection, DNA extraction, and soil characterization.
Samples were collected from 88 sites representing a wide range of ecosystem types as described by Fierer and Jackson (19). Briefly, all soils were collected near the height of the plant growing season from nonagricultural soils that were minimally disturbed and unsaturated for the majority of the year. At each site, soil from the top 5 cm of mineral soil was collected from 5 to 10 randomly selected locations within an area of ≈100 m2. Soil samples were composited, stored, and shipped at 4°C for 1 to 3 days before the samples were sieved through 4-mm mesh to thoroughly homogenize and remove roots and plant detritus from the samples. Soils were archived at −80°C until DNA extraction. The following soil and site characteristics were determined for each sample and used in the subsequent statistical analyses: mean annual temperature (MAT), potential evapotranspiration rate, soil moisture deficit (an index of average annual soil moisture), gravimetric soil moisture content (at the time of sampling), soil texture (% silt plus clay), organic carbon content (% OC), soil carbon/nitrogen ratio (C:N), extractable NH4+ and NO3− levels, pH, and potential carbon mineralization rate (μg C-CO2 g soil−1 day−1). Details on these soil and site characteristics, the methods used to determine these characteristics, and each of the samples included in this study have been described previously (19).
For this study, we focused on intersite variability, not the variability in bacterial communities within individual plots, and thus DNAs were extracted and amplified from a single, composited sample per site. Approximately 10 g of soil from each site was homogenized in a mortar and pestle with liquid N2, and DNAs were extracted from a 0.5-g subsample by use of a MoBio PowerSoil DNA extraction kit following the manufacturer's instructions, with an additional incubation step at 65°C for 10 min followed by 2 min of bead beating to limit DNA shearing. Eluted DNAs were stored at −20°C.
Bar-coded pyrosequencing.
Amplification, pooling, and pyrosequencing were performed as described by Fierer et al. (17). Briefly, a portion of the 16S small-subunit ribosomal gene (positions 27 to 338 [V1 and V2]; Escherichia coli numbering) was amplified using a 27F primer with a Roche 454 A pyrosequencing adapter, while the 338R primer contained a 12-bp bar-code sequence, a TC linker, and a Roche 454 B sequencing adapter. The targeted gene region has been shown to be the most appropriate for the accurate taxonomic classification of bacterial sequences, as other regions of the 16S rRNA gene can lead to significant misclassification of sequences (30). The bar code for each sample was unique and error correcting to facilitate sorting of sequences from a single pyrosequencing run (23). PCRs were conducted with 30 μM of each forward and reverse primer, 1.5 μl template DNA, and 22.5 μl Platinum PCR SuperMix (Invitrogen, Carlsbad, CA). Amplification was performed as described by Fierer et al. (17). Each sample was amplified in triplicate, pooled, and cleaned using a MoBio 96 htp PCR cleanup kit. Equal amounts of PCR product for each sample were combined in a single tube and sent to the Environmental Genomics Core Facility at the University of South Carolina to be run on a Roche FLX 454 pyrosequencing machine.
Processing of pyrosequencing data.
Data were processed by following the procedure described by Hamady et al. (23) and Fierer et al. (17). Briefly, low-quality sequences were removed (those sequences of <200 bp in length with an average quality score of >25), and the 12-bp bar code was examined in order to assign sequences to samples. Phylotypes were identified using Megablast, with a phylotype defined at the 97% sequence similarity level. A representative sequence from each phylotype was aligned using NAST (11), with a relaxed neighbor-joining tree built using Clear-Cut (46). The difference in overall community composition between each pair of samples was determined from the neighbor-joining tree by use of the unweighted UniFrac algorithm (32, 33). UniFrac calculates the fraction of branch length unique to a sample or environment across a phylogenetic tree constructed from each pair of samples or environments. Since UniFrac provides a phylogenetic metric of community distance, it avoids some of the pitfalls associated with comparing communities at only a single level of taxonomic resolution and provides a more robust index of community distances. The taxonomic identity of each phylotype was determined using RDPII taxonomy (9). All sequences have been deposited in the GenBank short-read archive.
We used two indices to compare community-level bacterial diversity across all 88 soils. First we compared the number of phylotypes (richness), with the number of phylotypes defined at the 97% sequence similarity level as described above. This index of diversity is limited in that it characterizes diversity at only a single level of taxonomic resolution. For this reason, we also estimated diversity using Faith's index of phylogenetic diversity (Faith's PD) (15), which provides an integrated index of the phylogenetic breadth contained within each community. In both cases, we calculated the diversity metrics for a randomly selected subset of 1,200 sequences per soil, as diversity is unavoidably correlated with the number of sequences collected. By using a set number of sequences, we could compare general diversity patterns even though it is highly unlikely that we surveyed the full extent of diversity in each community (45).
The UniFrac and diversity metrics described above were also applied to specific lineages of bacteria (Acidobacteria, Actinobacteria, Alphaproteobacteria, Beta/Gammaproteobacteria, and Bacteroidetes). These lineage-specific analyses were distinct from those described above in that we compared the diversity and phylogenetic composition of these individual taxa across the collected soils, not just the overall patterns evident from examining all taxa together. These five taxa were the most abundant groups of bacteria in the total sequence data set, and for reasons of clarity, we refer to these five taxonomic groups as phyla, recognizing that we are using the term “phyla” in a general manner. The beta- and gammaproteobacterial groups were not analyzed separately, as these groups are often combined in certain taxonomic schemes. For the lineage-specific UniFrac analyses, we limited the number of sequences to 250, 200, 100, 100, and 100 randomly selected sequences per soil for Acidobacteria, Alphaproteobacteria, Bacteroidetes, Beta/Gammaproteobacteria, and Actinobacteria, respectively. Normalizing the number of sequences per soil allows us to control for the effects of survey effort (number of sequences per phylum per soil) in comparing the lineage-specific UniFrac distances across the sample set. Because some soils did not have the required number of sequences per phylum, these lineage-specific analyses were conducted on only 57 to 69 of the 88 samples (see Table 2), excluding those soils where the individual phyla were relatively rare.
TABLE 2.
Correlations between unweighted UniFrac distances and environmental parameters (Spearman rank correlations estimated using Mantel tests)a
| Phylum (no. of soils) | Correlation
|
||||||
|---|---|---|---|---|---|---|---|
| Soil pH | Soil moisture deficit | C/N ratio | % OC | MAT (°C) | Potential C mineralization rate (μg C-CO2 g soil−1 day−1) | % Silt plus clay | |
| Full community (88) | 0.79 | 0.50 | 0.26 | 0.23 | 0.16 | 0.10 | 0.08 |
| Acidobacteria (69) | 0.72 | 0.30 | 0.05 | 0.05 | 0.11 | 0.06 | 0.20 |
| Actinobacteria (66) | 0.63 | 0.40 | 0.18 | 0.16 | 0.15 | 0.13 | 0.07 |
| Alphaproteobacteria (69) | 0.70 | 0.37 | 0.10 | 0.17 | 0.12 | 0.03 | 0.11 |
| Beta/Gammaproteobacteria (69) | 0.53 | 0.34 | 0.06 | 0.19 | 0.21 | 0.11 | 0.09 |
| Bacteroidetes (57) | 0.37 | 0.45 | 0.29 | 0.12 | 0.24 | 0.06 | 0.11 |
Significant correlations are indicated in bold (P < 0.01). Other soil and site characteristics that were tested but were not included in this table because there were no significant correlations (P > 0.15 in all cases) include mean annual precipitation, extractable NH4+ and NO3− levels, total microbial biomass, potential evapotranspiration, and latitude. Multivariate correlations did not increase the correlation coefficients.
Statistical analyses.
Pairwise UniFrac distances calculated for both the lineage-specific and total community analyses were visualized using nonmetric multidimensional scaling plots as implemented in PRIMER v6 (8). Statistical analyses were performed in a similar manner to those described by Lauber et al. (29) and Fierer and Jackson (19). Correlations between the diversity estimates and soil characteristics were tested for significance by use of SYSTAT 11.0. Best-fit modeling of PD and individual phyla were performed in SigmaPlot, using linear, polynomial (quadratic), and power law functions. ANOSIM analyses were conducted using PRIMER v6 (8), as were Mantel-type tests to find correlations between UniFrac distances and soil/site characteristics. Rarefaction curves were produced using EstimateS (version 8.0) (http://purl.oclc.org/estimates).
RESULTS
Distribution of taxa and phylotypes across 88 soils.
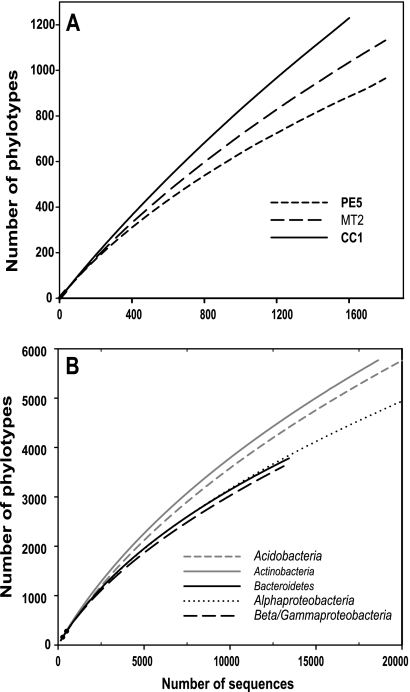
Across all 88 samples, we obtained 152,359 quality sequences, with an average read length of 232 bp and a range of 207 to 277 bp. Of these sequences, 132,090 (87%) were able to be classified. On average, each individual sample was represented by 1,501 classifiable sequences, with a range of 1,047 to 2,167 sequences per sample. When grouped at the 97% similarity level, there were 49,944 phylotypes in the complete data set, with an average of 1,017 phylotypes per sample. The majority of phylotypes (65%) were represented by a single sequence, and 75% of the phylotypes were found in only a single soil. The most abundant single phylotype across the entire sample (classified as a member of the Alphaproteobacteria [Rhizobiales]) set was represented by only 531 sequences (approximately 0.34% of the total number of classifiable sequences). Likewise, within an individual soil, the most abundant phylotype accounted for a maximum of 11% of the sequences from that soil (a member of the Betaproteobacteria of the Gallionella genus in sample TL1). Even at this depth of sequencing, we did not survey the full extent of taxonomic diversity within individual soils at the 97% similarity level of taxonomic resolution. This is evidenced by the lack of asymptotes in the rarefaction curves for the representative low-, intermediate-, and high-diversity (as measured by phylotype richness) soils shown in Fig. 1A.
FIG. 1.
(A) Rarefaction results for soils with low (PE5), average (MT2), and high (CC1) levels of diversity. The same three soils were also used for Fig. 3. (B) Rarefaction results for the five dominant bacterial phyla across all soils combined. To make the patterns clear, we have shown rarefaction curves for only the first 20,000 sequences per group. PE5, Manu National Park, Peru, tropical forest soil, pH 3.6; MT2, Missoula, MT, temperate coniferous forest, pH 6.7; CC1, Cedar Creek LTER, United States, temperate grassland, pH 6.0.
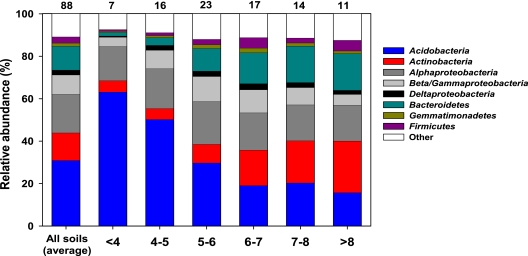
Of the classifiable sequences, 25 phyla were identified across the sample set (listed in Table S1A in the supplemental material). The dominant phyla were Acidobacteria, Alphaproteobacteria, Actinobacteria, Bacteroidetes, and Beta/Gammaproteobacteria, representing approximately 31%, 18%, 13%, 11%, and 9.1% of the sequences that could be classified below the domain level, respectively (Fig. 2; see Table S1A in the supplemental material). As described in Materials and Methods, these five phyla were selected for more detailed lineage-specific analyses because of their relatively high abundances in nearly all of the soils examined. Phyla that were less abundant but still were found in most of the soils examined included the Firmicutes (3%), Deltaproteobacteria (2%), Gemmatimonadetes (1.5%), TM7 (1.0%), Verrucomicrobia (0.9%), and Cyanobacteria (0.7%) (see Table S1A in the supplemental material). We identified sequences from an additional 11 rare phyla, which we define here as those phyla represented by <0.5% of the sequences, and details on the occurrence of these rare phyla are provided in Table S1A in the supplemental material.
FIG. 2.
Relative abundances of dominant bacterial taxa in all soils combined and in soils with different pH levels. The numbers above the columns indicate the number of soils included in each category. Relative abundances were estimated from the proportional abundances of classifiable sequences, excluding those sequences that could not be classified below the domain level. For additional taxonomic descriptions of the soil bacterial communities, see Tables S1A and S1B in the supplemental material.
Across all soils, acidobacterial sequences were most abundant. This phylum was dominated by members of groups 1 to 4 (see Table S1B in the supplemental material). Most of the actinobacterial sequences were classified as belonging to the Actinobacteridae and Rubrobacteridae taxa, with the Sphingobacteria taxon dominating the Bacteroidetes phylum. Rhizobiales and Burkholderiales were the most abundant groups within the Alphaproteobacteria and Beta/Gammaproteobacteria phyla, respectively. More complete information on the relative abundances of the 25 phyla and the dominant subphyla is provided in Tables S1A and S1B in the supplemental material.
Variability in bacterial community diversity.
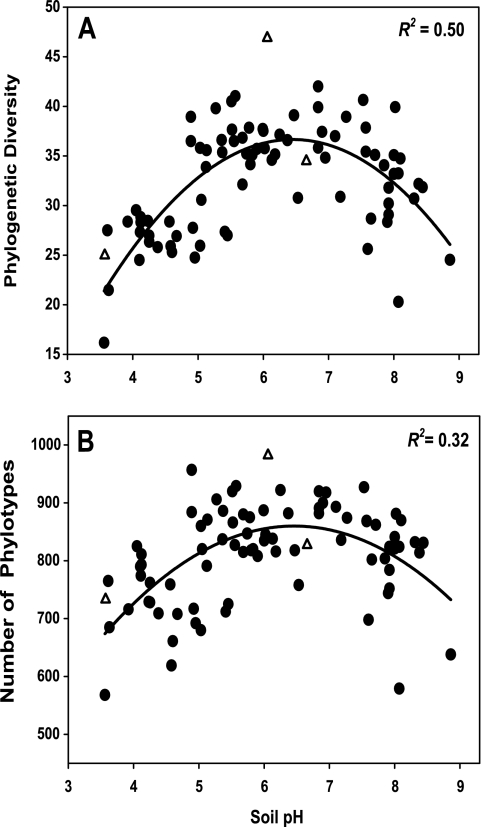
When we compared soil bacterial communities at the same level of surveying effort (1,200 sequences per soil), we found that community-level diversity was highly variable with respect to both overall phylogenetic diversity (Fig. 3A) and phylotype richness (Fig. 3B). Of those soil and site characteristics considered, only soil pH was significantly correlated (P < 0.05) with either phylotype richness (R2 = 0.32) or Faith's PD (R2 = 0.50) (Fig. 3; Table 1), and multivariate models did not lead to a significant improvement in correlation values over those for pH alone. With both metrics, diversity was highest in soils with near-neutral pHs (Fig. 3). Soils with the lowest levels of diversity were found in either deserts with soil pHs of >8 or temperate and tropical forest soils with pHs of <4.5. The soil with the highest level of diversity, regardless of the metric used, was a soil from the Cedar Creek LTER, a grassland site in Minnesota, that had a pH of 6.1 (Fig. 3). Although sample collection was not precisely standardized across sites, these data do indicate that pH is a reasonably good predictor of overall bacterial diversity across many soils, although the trend across fewer samples may be less evident.
FIG. 3.
Relationship between soil pH and soil bacterial diversity, measuring using Faith's PD (A) and the number of phylotypes (B), with phylotypes defined at the 97% sequence similarity level. Lines represent the best-fit quadratic model to the data. Unfilled triangles represent the three soils shown in Fig. 1A. Diversity indices were calculated using 1,200 sequences per soil sample.
TABLE 1.
Relationships between phylogenetic diversity (Faith's PD) and measured soil characteristics for the full sequence set and the five most abundant phylaa
| Phylum | Correlation
|
||||||
|---|---|---|---|---|---|---|---|
| Soil pH | Soil moisture deficit | C/N ratio | % OC | MAT (°C) | C mineralization rate (μg C-CO2 g soil−1 day−1) | % Silt plus clay | |
| Full community | 0.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acidobacteria | 0.23 | 0.04 | 0 | 0 | 0.05 | 0.01 | 0 |
| Actinobacteria | 0.31 | 0.14 | 0.01 | 0.06 | 0.01 | 0.03 | 0.03 |
| Alphaproteobacteria | 0.27 | 0.08 | 0.08 | 0.04 | 0.03 | 0 | 0 |
| Beta/Gammaproteobacteria | 0.25 | 0.03 | 0.04 | 0.01 | 0.01 | 0.02 | 0.03 |
| Bacteroidetes | 0.06 | 0.08 | 0.02 | 0.06 | 0.02 | 0.03 | 0 |
Those relationships that are significant are indicated in bold. We tested three models to describe the relationships (linear, quadratic, and power), and the quadratic function provided the best fit in all cases. The relationships between Faith's PD and soil pH for the full community and the individual phyla are shown in Fig. 3A and Fig. S1 in the supplemental material.
Soil pH was also the best predictor of variability in diversity levels within each of the five dominant phyla. The estimates of Faith's PD for the individual phyla had significant regression coefficients with soil pH for four of the five phyla (for Acidobacteria, R2 = 0.23; for Actinobacteria, R2 = 0.31; for Alphaproteobacteria, R2 = 0.27; and for Beta/Gammaproteobacteria, R2 = 0.25), but the relationships were weaker than those for overall bacterial community diversity (Table 1; see Fig. S1 in the supplemental material). If we compare the number of phylotypes across all soils included in the lineage-specific analyses, we find that the Acidobacteria and Actinobacteria exhibited higher levels of diversity at the 97% similarity level than the other three dominant phyla did. However, if we compare the average numbers of phylotypes per soil or the average levels of phylogenetic diversity per soil (with both metrics calculated from 100 randomly selected sequences per soil per phylum), we see no apparent differences between the five phyla (see Table S2 in the supplemental material). In other words, some phyla are more diverse than others if we consider all 88 soils together, but the levels of diversity for any given phyla seem to be relatively similar within individual soils.
Variability in overall bacterial community composition.
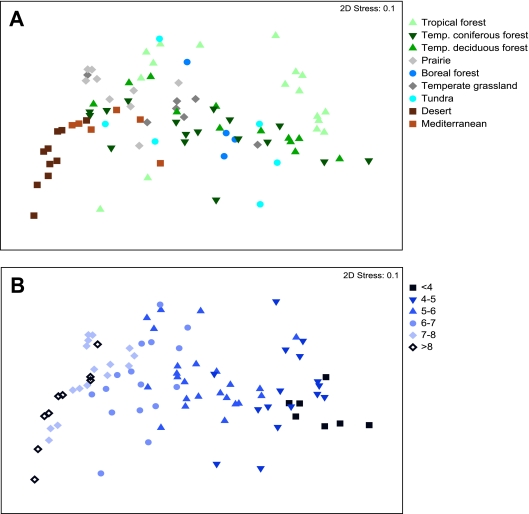
The composition of bacterial communities was highly variable across the soils represented in this study (Fig. 2 and 4). On average, each pair of soils shared only 0.9% of their phylotypes (at the 97% similarity level), although this degree of community overlap is likely to be an underestimate given that we did not identify all phylotypes present in a given sample (Fig. 1A). Visualization of the pairwise UniFrac distances on nonmetric multidimensional scaling plots indicates significant variability within and across the biomes. Except for the desert soils and perhaps the soils from Mediterranean-type biomes, soils from similar biomes do not necessarily harbor similar bacterial communities, as the variability between biomes exceeded the variability within a given biome. This pattern is evident in Fig. 4 and was confirmed by a nonsignificant ANOSIM P value (P > 0.05) for biome effects on UniFrac distances. Similarly, there was no significant correlation between UniFrac distances and the pairwise geographic distances between sampling locations (r = 0.05; P = 0.4), indicating that soils collected from distant locations did not necessarily harbor more distinct communities than those collected in close proximity.
FIG. 4.
Nonmetric multidimensional scaling plots derived from pairwise Unifrac distances between soils, with symbols coded by general ecosystem type (A) and pH category (B).
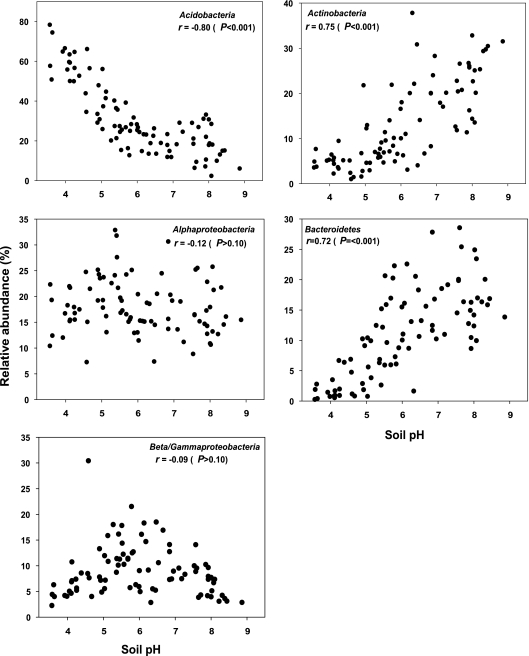
Of the edaphic soil characteristics measured, pH was most strongly correlated with the overall UniFrac distances between soils (Table 2). UniFrac distances show minimal overlap among communities that differ by more than 2 pH units when samples are viewed by pH category (Fig. 4B). With the exception of soil moisture deficit, which is strongly correlated with soil pH (r = 0.65), Mantel tests of the remaining edaphic characteristics indicated no significant relationship with the overall UniFrac distances between communities (Table 2). Multivariate models did not lead to a significant increase in correlation values over models that considered pH alone. If we examine the phylum-level relative abundances, the strong influence of pH on overall bacterial community composition is clearly evident (Fig. 2 and 5). Relative abundances of Acidobacteria, Actinobacteria, and Bacteroidetes were significantly correlated with soil pH (P < 0.05 in all cases), with acidobacterial abundances decreasing with pH and the abundances of Actinobacteria and Bacteroidetes positively correlated with soil pH (Fig. 2 and 5). In contrast, Alpha- and Beta/Gammaproteobacteria abundances showed no significant relationship to soil pH (P > 0.1 in both cases) (Fig. 5).
FIG. 5.
Correlations between relative abundances of the five dominant bacterial phyla and soil pH. Pearson correlation coefficients (r) are shown for each taxon, with the associated Bonferroni-corrected P values.
Variability in the composition of the dominant taxa.
As with overall bacterial community composition, if we examined the five dominant phyla individually, we also found that the phylogenetic structure within these groups was most strongly correlated with soil pH (Table 2). Correlations between UniFrac distances and soil pHs were significant (P < 0.05 in all cases) within all five phyla but were stronger for the Acidobacteria (r = 0.72), Alphaproteobacteria (r = 0.70), and Actinobacteria (r = 0.63) than for the Beta/Gammaproteobacteria (r = 0.53) and the Bacteroidetes (r = 0.37). Only soil moisture deficit and pH were significantly correlated with pairwise UniFrac distances within each of these phyla (Table 2). In other words, the data demonstrate that not only was the overall community composition structured by soil pH, a pattern that is evident if we examine the relative abundances of the different phyla across the pH gradient (Fig. 5), but the differences in the phylogenetic structures of the individual phyla were also significantly correlated with soil pH. These patterns are evident by comparing the relative abundances of subphyla (percentage of sequences within a given phyla represented by a given subphylum) in soils from different pH categories (see Table S1B in the supplemental material). Acidobacteria subgroups 1, 2, and 3 decreased in relative abundance as soil pH increased, while groups 4 and 6 showed the opposite pattern (see Table S1B in the supplemental material). The relative abundance of the Actinobacteridae decreased with pH, while the Rubrobacteridae were the most prevalent actinobacterial subphylum in higher-pH soils. Within the alphaproteobacterial phylum, the relative abundance of Sphingomonadales increased with pH, while various other alphaproteobacterial groups showed the opposite pattern (see Table S1B in the supplemental material).
DISCUSSION
General characteristics of soil bacterial communities.
Although we collected an average of 1,501 sequences per soil, we still have not surveyed the full extent of bacterial diversity within individual soils, so we can only conclude that the typical soil harbors more than 1,000 phylotypes (if we define phylotypes at the 97% sequence similarity level). This result is to be expected; a number of other studies have used modeling approaches (21) or more extensive surveys than those described here (48) to demonstrate that soil bacterial communities harbor an enormous number of unique taxa. Since most soil bacterial taxa can be considered rare (according to Elshahed et al. [14]), it is not feasible to document the full extent of bacterial diversity in a given soil, even if a full pyrosequencing run is devoted to a single soil (20, 42). Not only do individual soils harbor a large amount of phylogenetic diversity, but at our survey depths, soil communities shared a small percentage of their phylotypes, and most phylotypes were found in only a single soil. Again, this is not surprising, as other studies have also shown a high degree of endemism at finer levels of taxonomic resolution (20). However, it is important to recognize that the degree of overlap between pairs of soils would likely increase if the individual soils were surveyed more comprehensively.
All of the communities were dominated by five major groups (Acidobacteria, Actinobacteria, Proteobacteria, Bacteroidetes, and Firmicutes), with these five groups accounting for more than 90% of the sequences in each of the soils examined. The relative abundances of the dominant taxa (Fig. 2; see Table S1A in the supplemental material) roughly correspond with those reported in a meta-analysis conducted by Janssen (27). Although we focus here on the variability in bacterial communities across a range of soil habitats, it is important to recognize that soils are more similar to one another than to other microbial habitats (34), as all of the soils were largely composed of the same five bacterial groups and the rarer phyla never had abundances of >10% (see Table S1A in the supplemental material). It is also important to note that the primer 338R has some biases, especially toward the Verrucomicrobia, Planctomycetes, and Chlamydiae, which are routinely found using universal primers but were found in low abundance in this study. Although many other studies have made a similar observation, it is still noteworthy given that all bacterial phyla are phylogenetically diverse yet there also must be broad metabolic differences between these phyla that allow some phyla to dominate in soil while others nearly always remain quite rare.
Although each of the five dominant phyla had similar average levels of diversity within individual soils (see Table S2 in the supplemental material), the Acidobacteria and Actinobacteria were more diverse across the entire sample set than the proteobacterial and Bacteroidetes phyla (Fig. 1). These differences may be due to the strong pH influence on acidobacterial and actinobacterial community composition, whereby the relative abundances of certain taxa within these groups were strongly influenced by changes in soil pH (Fig. 2; see Table S1B in the supplemental material), with soils of distinct pH levels exhibiting minimal taxon co-occurrence.
Utility of bar-coded pyrosequencing methodology.
The bar-coded pyrosequencing technique used here yields results that are qualitatively similar to those of a fingerprint-based analysis of soil bacterial communities (19), but the technique provides a far more robust description of the changes in bacterial community structure across the sample set. A pyrosequencing run is not cheap, and the large number of custom primers required only adds to this cost, but the technique does allow hundreds of samples to be analyzed simultaneously, with each community analyzed in considerable detail. Although the phylogenetic structure and composition of the surveyed communities can be determined with a high degree of accuracy (20, 26, 42), the method does not allow us to identify bacterial taxa at the finest levels of taxonomic resolution. However, with ever-increasing read lengths, this constraint will gradually become less relevant. Of course, the bar-coded pyrosequencing technique itself is of limited utility unless it is accompanied by appropriate tools for sequence analysis (44). The phylogenetic approaches described here (e.g., UniFrac distances and Faith's PD) are more powerful than standard phylotype-based approaches where community structure and diversity are compared at a single level of sequence similarity because they take into account different levels of similarity between different pairs of taxa (33). In particular, comparing communities by grouping sequences into phylotypes defined at the 97% similarity level has limitations in that such surveys will be far from comprehensive (Fig. 1A), and overarching patterns evident by comparing overall phylogenetic structure (Fig. 4) may be more difficult to discern and quantify.
Soil pH as a predictor of bacterial community structure.
Although there was a high degree of variability in bacterial community composition across the range of soils examined here, overall bacterial community composition and (to a lesser extent) diversity were surprisingly predictable at this scale of inquiry by considering only a single parameter, soil pH. This influence of soil pH on overall community composition was evident even at a very coarse level of taxonomic resolution, where we saw the relative abundances of certain bacterial phyla (e.g., Actinobacteria, Bacteroidetes, and Acidobacteria) changing in a consistent manner across the soil pH gradient. Even if we used lineage-specific analyses to examine shifts in community composition and diversity within individual phyla, we still found that pH was often significantly correlated with the structure of these phyla across the range of soils examined (Table 2; Fig. 5). Although soil pH was the best predictor of bacterial community composition and diversity compared to the other soil and site characteristics that were measured, a large amount of the variability in bacterial community structure remains unexplained. Although we measured a standard suite of soil characteristics and included all of these characteristics in our statistical analyses, it is entirely possible that the consideration of other soil or site variables not measured here would improve our ability to predict changes in community structure across these spatial scales. For instance, salinity has been shown to be a significant driver of global distributions of bacteria, but it is not routinely measured in most soil studies (34). Likewise, cation exchange capacity is often overlooked as an important determinant of microbial community diversity and composition. However, we are not suggesting that soil pH can always be considered a universal predictor of bacterial community structure. Depending on the types of soils included in an individual survey, other factors may be more useful for predicting community patterns, particularly in examining temporal or spatial changes in bacterial communities across soils that share similar pH levels.
The apparent influence of soil pH on soil bacterial community structure has also been documented in a number of other studies using various methods to characterize microbial communities (3, 4, 10). In particular, a recent study by Hartman et al. (24) found that pH was the best predictor of changes in soil bacterial communities, and they observed similar changes in phylum-level abundances across the pH gradient to those observed here for Acidobacteria and Actinobacteria. Other studies have also shown changes in the relative abundances of Actinobacteria, Acidobacteria, and Bacteroidetes across soils of different pHs that parallel those shown in Fig. 2 (13, 38, 43, 49). However, the results reported do differ somewhat from those of Fierer et al. (16), where the relative abundances of various taxa were determined by quantitative PCR for a subset of the soils included in this survey. Specifically, they found that the abundances of Bacteroidetes, Betaproteobacteria, and Acidobacteria were most strongly related to estimated carbon availability, not soil pH. This disparity between the studies could be related to the larger number of soils included in this study, but it is most likely due to differences in methodologies. Although the pyrosequencing technique does have certain limitations, it is better suited for assessing changes in the relative abundances of major taxa, because phylum-specific quantitative PCR assays have been shown to exclude many taxa within these phyla (28).
The correlations between soil pH and bacterial community structure and diversity observed here and in a number of other studies are striking. However, we cannot identify the specific mechanism or mechanisms responsible for generating the observed patterns. There are at least two general explanations that may explain, singly or in combination, why soil pH was the best predictor of community composition and diversity across the range of samples included here. First, soil pH may not directly alter bacterial community structure but may instead function as an integrating variable that provides an integrated index of soil conditions. There are a number of soil characteristics (e.g., nutrient availability, cationic metal solubility, organic C characteristics, soil moisture regimen, and salinity) that are often directly or indirectly related to soil pH (6), and these factors may drive the observed changes in community composition as the hydrogen ion concentration varies by many orders of magnitude across the soils in this study. Although it would be nearly impossible to alter soil pH without simultaneously altering a wide range of other soil edaphic factors, more controlled experiments could be used to test the validity of this scenario and to determine whether changes in soil pH alone can lead to the patterns observed here. A second hypothesis is that pH directly imposes a physiological constraint on soil bacteria, altering competitive outcomes or reducing the net growth of individual taxa unable to survive if the soil pH falls outside a certain range. Many bacteria have intracellular pH levels close to neutral (35), and therefore extreme pHs may impose a significant stress that certain taxa may tolerate better than others. If soil pH does lead directly to taxon sorting, it should be noted that the effects of pH appear to be evident at even coarse levels of taxonomic resolution (Fig. 2 and 5). Although individual phyla harbor considerable phylogenetic diversity, the members of certain phyla appear to share a common set of attributes that largely constrain those taxa to a certain pH range (Fig. 5). In contrast, the relative abundances of other taxa (e.g., the proteobacterial taxa) (Fig. 5; see Table S1A in the supplemental material) are not well correlated with pH, suggesting that these taxa do not share similar pH responses or that the abundances of these groups are predominantly influenced by factors other than pH. Thus, although pH appears to be a driver of many patterns in soil microbial diversity, the influence of other factors will need to be understood in order to develop a model that accurately predicts soil microbial community structure across larger spatial scales.
Supplementary Material
Acknowledgments
We thank members of the Fierer and Knight labs for their technical and intellectual assistance with this study. We also thank all of the individuals who donated their time and energy to help us collect the soil samples included in this study. In particular, we acknowledge Rob Jackson, Ben Colman, and Josh Schimel for their assistance with sample collection and associated soil analyses.
This work was supported by funding provided to N.F. from the National Science Foundation (MCB0610970 and DBI0301773) and the Andrew W. Mellon Foundation.
Footnotes
Published ahead of print on 5 June 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acosta-Martínez, V., S. Dowd, Y. Sun, and V. Allen. 2008. Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem. 40:2762-2770. [Google Scholar]
- 2.Andersson, A. F., M. Lindberg, H. Jakobsson, F. Bäckhed, P. l. Nyrén, and L. Engstrand. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE 3:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bääth, E., and T. H. Anderson. 2003. Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol. Biochem. 35:955-963. [Google Scholar]
- 4.Blagodatskaya, E. V., and T. H. Anderson. 1998. Interactive effects of pH and substrate quality on the fungal-to-bacterial ratio and qCO2 of microbial communities in forest soils. Soil Biol. Biochem. 30:1269-1274. [Google Scholar]
- 5.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brady, N. C., and R. R. Weil. 2007. The nature and properties of soil, 13th ed. Prentice Hall, Upper Saddle River, NJ.
- 7.Cho, J. C., and J. M. Tiedje. 2000. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke, K. R., and R. M. Warwick. 2001. A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Mar. Ecol. Prog. Ser. 216:265-278. [Google Scholar]
- 9.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cookson, W. R., M. Osman, P. Marschner, D. A. Abaye, I. Clark, D. V. Murphy, E. A. Stockdale, and C. A. Watson. 2007. Controls on soil nitrogen cycling and microbial community composition across land use and incubation temperature. Soil Biol. Biochem. 39:744-756. [Google Scholar]
- 11.DeSantis, T. Z., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, R. Phan, and G. L. Andersen. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394-W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dethlefsen, L., S. Huse, M. L. Sogin, and D. A. Relman. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6:2383-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichorst, S. A., J. A. Breznak, and T. M. Schmidt. 2007. Isolation and characterization of soil bacteria that define Teniglobus gen. nov., in the phylum Acidobacteria. Appl. Environ. Microbiol. 73:2708-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elshahed, M. S., N. H. Youssef, A. M. Spain, C. Sheik, F. Z. Najar, L. O. Sukharnikov, B. A. Roe, J. P. Davis, P. D. Schloss, V. L. Bailey, and L. R. Krumholz. 2008. Novelty and uniqueness patterns of rare members of the soil biosphere. Appl. Environ. Microbiol. 74:5422-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faith, D. P. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61:1-10. [Google Scholar]
- 16.Fierer, N., M. A. Bradford, and R. B. Jackson. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354-1364. [DOI] [PubMed] [Google Scholar]
- 17.Fierer, N., M. Hamady, C. L. Lauber, and R. Knight. 2008. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. USA 105:17994-17999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fierer, N., J. A. Jackson, R. Vilgalys, and R. B. Jackson. 2005. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71:4117-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fierer, N., and R. B. Jackson. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 103:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulthorpe, R. R., L. F. W. Roesch, A. Riva, and E. W. Triplett. 2008. Distantly sampled soils carry few species in common. ISME J. 2:901-910. [DOI] [PubMed] [Google Scholar]
- 21.Gans, J., M. Wolinsky, and J. Dunbar. 2005. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387-1390. [DOI] [PubMed] [Google Scholar]
- 22.Green, J. L., B. J. M. Bohannan, and R. J. Whitaker. 2008. Microbial biogeography: from taxonomy to traits. Science 320:1039-1043. [DOI] [PubMed] [Google Scholar]
- 23.Hamady, M., J. J. Walker, J. K. Harris, N. J. Gold, and R. Knight. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5:235-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartman, W. H., C. J. Richardson, R. Vilgalys, and G. L. Bruland. 2008. Environmental and anthropogenic controls over bacterial communities in wetland soils. Proc. Natl. Acad. Sci. USA 105:17842-17847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horner-Devine, M. C., M. Lage, J. B. Hughes, and B. J. M. Bohannan. 2004. A taxa-area relationship for bacteria. Nature 432:750-753. [DOI] [PubMed] [Google Scholar]
- 26.Huse, S. M., L. Dethlefsen, J. A. Huber, D. M. Welch, D. A. Relman, and M. L. Sogin. 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4:e1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, R. T., M. S. Robeson, C. L. Lauber, M. Hamady, R. Knight, and N. Fierer. 2009. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 4:442-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauber, C. L., M. S. Strickland, M. A. Bradford, and N. Fierer. 2008. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 40:2407-2415. [Google Scholar]
- 30.Liu, Z. Z., C. Lozupone, M. Hamady, F. D. Bushman, and R. Knight. 2007. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 35:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lomolino, M., B. Riddle, and J. Brown. 2006. Biogeography, 3rd ed. Sinauer Associates, Sunderland, MA.
- 32.Lozupone, C., M. Hamady, and R. Knight. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform. 7:731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozupone, C. A., and R. Knight. 2007. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. USA 104:11436-11440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madigan, M. T., and J. M. Martinko. 2006. Brock biology of microorganisms, 11th ed. Pearson Benjamin Cummings, San Francisco, CA.
- 36.Martiny, J. B. H., B. J. M. Bohannan, J. H. Brown, R. K. Colwell, J. A. Fuhrman, J. L. Green, M. C. Horner-Devine, M. Kane, J. A. Krumins, C. R. Kuske, P. J. Morin, S. Naeem, L. Ovreas, A. L. Reysenbach, V. H. Smith, and J. T. Staley. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102-112. [DOI] [PubMed] [Google Scholar]
- 37.McCaig, A. E., L. A. Glover, and J. I. Prosser. 2001. Numerical analysis of grassland bacterial community structure under different land management regimens by using 16S ribosomal DNA sequence data and denaturing gradient gel electrophoresis banding patterns. Appl. Environ. Microbiol. 67:4554-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendez, M. O., J. W. Neilson, and R. M. Maier. 2008. Characterization of a bacterial community in an abandoned semiarid lead-zinc mine tailing site. Appl. Environ. Microbiol. 74:3899-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morales, S. E., T. F. Cosart, J. V. Johnson, and W. E. Holben. 2009. Extensive phylogenetic analysis of a soil bacterial community illustrates extreme taxon evenness and the effects of amplicon length, degree of coverage, and DNA fractionation on classification and ecological parameters. Appl. Environ. Microbiol. 75:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noguez, A. M., H. T. Arita, A. E. Escalante, L. J. Forney, F. Garcia-Oliva, and V. Souza. 2005. Microbial macroecology: highly structured prokaryotic soil assemblages in a tropical deciduous forest. Global Ecol. Biogeogr. 14:241-248. [Google Scholar]
- 41.Ramette, A., and J. M. Tiedje. 2007. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb. Ecol. 53:197-207. [DOI] [PubMed] [Google Scholar]
- 42.Roesch, L. F. W., R. R. Fulthorpe, A. Riva, G. Casella, A. K. M. Hadwin, A. D. Kent, S. H. Daroub, F. A. O. Camargo, W. G. Farmerie, and E. W. Triplett. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sait, M., K. E. R. Davis, and P. H. Janssen. 2006. Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl. Environ. Microbiol. 72:1852-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schloss, P. D. 2008. Evaluating different approaches that test whether microbial communities have the same structure. ISME J. 2:265-275. [DOI] [PubMed] [Google Scholar]
- 45.Shaw, A. K., A. L. Halpern, K. Beeson, B. Tran, J. C. Venter, and J. B. H. Martiny. 2008. It's all relative: ranking the diversity of aquatic bacterial communities. Environ. Microbiol. 10:2200-2210. [DOI] [PubMed] [Google Scholar]
- 46.Sheneman, L., J. Evans, and J. A. Foster. 2006. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics 22:2823-2824. [DOI] [PubMed] [Google Scholar]
- 47.Singh, B. K., S. Munro, E. Reid, B. Ord, J. M. Potts, E. Paterson, and P. Millard. 2006. Investigating microbial community structure in soils by physiological, biochemical and molecular fingerprinting methods. Eur. J. Soil Sci. 57:72-82. [Google Scholar]
- 48.Tringe, S. G., C. von Mering, A. Kobayashi, A. A. Salamov, K. Chen, H. W. Chang, M. Podar, J. M. Short, E. J. Mathur, J. C. Detter, P. Bork, P. Hugenholtz, and E. M. Rubin. 2005. Comparative metagenomics of microbial communities. Science 308:554-557. [DOI] [PubMed] [Google Scholar]
- 49.Upchurch, R., C. Y. Chi, K. Everett, G. Dyszynski, D. C. Coleman, and W. B. Whitman. 2008. Differences in the composition and diversity of bacterial communities from agricultural and forest soils. Soil Biol. Biochem. 40:1294-1305. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.