Abstract
Bacteria and fungi are ubiquitous in the atmosphere. The diversity and abundance of airborne microbes may be strongly influenced by atmospheric conditions or even influence atmospheric conditions themselves by acting as ice nucleators. However, few comprehensive studies have described the diversity and dynamics of airborne bacteria and fungi based on culture-independent techniques. We document atmospheric microbial abundance, community composition, and ice nucleation at a high-elevation site in northwestern Colorado. We used a standard small-subunit rRNA gene Sanger sequencing approach for total microbial community analysis and a bacteria-specific 16S rRNA bar-coded pyrosequencing approach (4,864 sequences total). During the 2-week collection period, total microbial abundances were relatively constant, ranging from 9.6 × 105 to 6.6 × 106 cells m−3 of air, and the diversity and composition of the airborne microbial communities were also relatively static. Bacteria and fungi were nearly equivalent, and members of the proteobacterial groups Burkholderiales and Moraxellaceae (particularly the genus Psychrobacter) were dominant. These taxa were not always the most abundant in freshly fallen snow samples collected at this site. Although there was minimal variability in microbial abundances and composition within the atmosphere, the number of biological ice nuclei increased significantly during periods of high relative humidity. However, these changes in ice nuclei numbers were not associated with changes in the relative abundances of the most commonly studied ice-nucleating bacteria.
Microbes are abundant in the atmosphere, with both cultivation-dependent and molecular approaches showing that the atmosphere harbors a diverse assemblage of bacteria and fungi, including taxa also commonly found on leaf surfaces (5, 49) and in soil habitats (30). The abundance and composition of airborne microbial communities are variable across time and space (14, 24, 27, 33, 47, 48, 69). However, the atmospheric conditions responsible for driving the observed changes in microbial abundances are unknown. The diversity of airborne microorganisms, and the factors influencing diversity levels, also remains poorly characterized. One reason for these limitations in knowledge is that until recently, culture-based microbiological methods have been the standard, and it is well-recognized that such methods capture only a small portion of the total microbial diversity (59). As demonstrated in a number of recent studies (6, 13, 22, 23, 33, 52, 59, 63, 73), advances in culture-independent techniques allow far more of the microbial diversity present in the atmosphere to be surveyed and the spatiotemporal variability in microbial communities to be examined.
Microbes are often considered passive inhabitants of the atmosphere, dispersing via airborne dust particles. However, recent studies suggest that many atmospheric microbes may be metabolically active (3, 4, 64), even up to altitudes of 20,000 m (34). Some airborne microbes may alter atmospheric conditions directly by acting as cloud condensation nuclei (7, 25, 56) and/or ice nuclei (IN) (19, 41, 56, 57, 61); this hypothesis is supported by the observation that most ice nuclei in snow samples are inactivated by a 95°C heat treatment (16, 17). However, the overall contribution of airborne microbes to atmospheric processes such as ice nucleation remains unclear.
The best-studied ice-nucleating microbes are gram-negative bacteria that have also been isolated from leaf surfaces, including Pseudomonas syringae, Pseudomonas fluorescens, Erwinia herbicola, Xanthomonas campestri, and Sphingomonas spp. (45). These bacteria have been cultured extensively, and their ice-nucleating activity has been traced to a membrane-bound glycoprotein (40, 42, 70). However, their specific influence on atmospheric processes remains, at this point, largely anecdotal. Less is known about the ice-nucleating activities of fungi, but a few studies have shown that fungi can be effective ice nucleators, capable of initiating ice nucleation at temperatures as high as −2°C (41, 61). At this point, all known ice-nucleating microorganisms are amenable to culture-based studies, but given that the vast majority of microorganisms have yet to be cultured, it is likely that other ice-nucleating microbes remain undiscovered.
The work presented here addresses three overarching questions. (i) Are microbial abundances altered by changes in atmospheric conditions? (ii) How is the diversity and composition of airborne microbial communities influenced by changes in atmospheric conditions? (iii) Can we identify known and novel ice-nucleating microbes in the atmosphere by testing for correlations between taxa abundances and the concentrations of biological ice nuclei? To address these questions, we combined epifluorescence microscopy, tagged pyrosequencing, Sanger sequencing, and an ice nucleation assay with atmospheric measurements to characterize the microbial communities at a high-elevation research site.
MATERIALS AND METHODS
Sampling site description.
Air samples were collected from a single location 4 m above ground level on the roof of the Storm Peak Laboratory (SPL). SPL sits at the top of Mt. Werner near Steamboat Springs, CO (40.45°N, 106.73°W) at 3,200 m above sea level. The SPL research station is owned and operated by the Desert Research Institution Division of Atmospheric Sciences, and its unique mountaintop location provides researchers with an opportunity to study the free troposphere and boundary layer atmosphere in a fairly pristine location. Further details on the sampling location and atmospheric conditions at the site can be found in the report of Borys and Wetzel (9). Sampling was conducted at nine time points over a 2-week period in late March and early April 2008 when the ground was still snow covered. The duration of each sampling period and the corresponding weather conditions are presented in Table 1.
TABLE 1.
Description of samples collected and corresponding meteorological conditions
| Sample IDa | Date of collection | Collection time | Observed weather conditions | Temp (°C) | % Relative humidity (range) | Avg wind speed (km/h) | Maximum wind speed (km/h) | Barometric pressure (MPa) | Potental temp (K) |
|---|---|---|---|---|---|---|---|---|---|
| 3-23.CL.N | 3/23/09-3/24/08 | 22:30-06:30 (8.5 h total) | Clear skies, night, free troposphere | −7.0 | 40-57 | 29 | 36 | 6.90 | 300 |
| 3-24.CL.D | 3/24/08 | 10:30-16:00 (5.5 h total) | Clear skies, day, boundary layer | −2.5 | 34-39 | 27 | 37 | 6.89 | 306 |
| 4-1.CD.N | 4/1/08-4/2/08 | 21:00-07:30 (10.5 h total) | Cloudy skies, night, free troposphere, snowed 5 cm | −8.4 | 79-101 | 24 | 32 | 6.85 | 303 |
| 4-2.CD.N | 4/2/08-4/3/08 | 23:00-06:30 (7.5 h total) | Cloudy skies, night, free troposphere, snowed 5 cm | −8.5 | 98-102 | 19 | 23 | 6.82 | 304 |
| 4-3.CD.D | 4/3/08 | 07:00-13:00 (6.0 h total) | In cloud, day, boundary layer, snowed 5 cm | −8.7 | 95-101 | 23 | 31 | 6.84 | 303 |
| 4-3.CL.N | 4/3/08-4/4/08 | 20:15-08:30 (12.25 h total) | Clear skies, night, free troposphere | −12 | 64-99 | 20 | 24 | 6.88 | 297 |
| 4-4.CL.N | 4/4/08-4/5/08 | 20:50-08:50 (12 h total) | Clear skies, night, free troposphere | −5.0 | 39-51 | 27 | 36 | 6.81 | 304 |
| 4-5.CD.D | 4/5/08 | 10:20-17:20 (7 h total) | Cloudy skies, day, boundary layer | −4.8 | 51-102 | 21 | 29 | 6.78 | 309 |
| 4-6.CD.N | 4/6/08-4/7/08 | 21:00-08:00 (11 h total) | In cloud, night, free troposphere | −13 | 92-99 | 25 | 32 | 6.80 | 297 |
| 4-3.SN | 4/3/08 | NAb | Collected after exposed to ∼2 h of sunlight | NA | NA | NA | NA | NA | NA |
| 4-7.SN | 4/7/08 | NA | Collected while still in a cloud | NA | NA | NA | NA | NA | NA |
The sample ID consists of the following information: date the sample was collected, the microbial environment type, clear air (CL), cloudy air (CD), or snow (SN), and the time of day the sample was collected, day (D) or night (N).
NA, not applicable.
Meteorological measurements and particle counts relative to microbiology studies.
SPL's suite of meteorological instruments includes a continuously recording meteorological station that automatically streams data to the Western Regional Climate Center. Meteorological instruments used in this study were a research-grade thermometer, deiced wind vane anemometer, barometer, and relative humidity sensors (Campbell Scientific, Inc.; Met One and Vaisala interfaced to data loggers). An Aerodynamic Particle Sizer (APS, model 3320; TSI, Shoreview, MN) was used to measure total concentration and size distributions of particles with diameters of 530 nm to more than 5 μm.
Airborne microbial sample collection.
Air samples were captured on triplicate sterile 0.22-μm cellulose nitrate filters (Fisher Scientific, Pittsburgh, PA) via vacuum filtration with a flow rate of 7.5 liters min−1. The total volume of air that passed through each filter during each sampling period ranged from 2.5 m3 to 5.4 m3. One filter from each triplicate set was used for each of the following assays: total microbial abundance measurements via epifluorescence microscopy, DNA extraction and the associated microbial community composition analyses, and ice nucleation assays. Additionally, two fresh snow samples were collected following overnight storms at SPL that occurred during the sampling period (Table 1). Snow was melted at 4°C in sterile bags, and 1 liter of snowmelt was filtered through each of three 0.22-μm cellulose nitrate filters. The same three assays described above were also performed on the two collected snow samples.
Total microbial abundance.
Aerosol particles were shaken from the filters at approximately 200 rpm in a small petri dish in 8 ml of high-performance liquid chromatography (HPLC)-grade water for 2 h at 4°C. A subset of the filters was examined under a microscope after this shaking process to assure that most visible particles were removed from the filter. While the efficiency of particle removal was not evaluated for every individual sample, all sample filters were treated identically, and therefore any bias associated with this method should have been held constant across the sample set. The microbial particles suspended in HPLC-grade water were stained with 4′,6′-diamidino-2-phenylindole (DAPI), a DNA binding dye (Kirkegaard and Perry, Gaithersburg, MD) at a final working concentration of 500 μg ml−1 and counted at 1,000× magnification using a Nikon Eclipse E400 epifluorescence microscope following the protocol described by Hernandez et al. (36). Intact cells were counted on 25-mm-diameter black polycarbonate filters with pore sizes of 0.22 μm (GEI-W&PT, Trevose, PA). Microbial abundance is expressed as cells per cubic meter of air, taking into account the dilution, the flow rate, and the number of hours sampled.
Ice nucleation counts.
Particles were removed from the filters using the same protocol described in the total particle count section above. The drop-freeze assay was performed using a modified version of the protocol described by Vali (72). Drop-freeze plates were assembled by coating an aluminum foil-covered copper plate with a solution of 5% (wt/vol) petrolatum in xylene. After the xylene had evaporated, 10-μl drops were arrayed (96 drops per sample) in an 8 by 12 format across the surface of the plate. Extra columns of 10-μl, HPLC-grade water drops (not containing sample) were placed on each side of the array as a plate control. The temperature of the plate was then cooled with a Peltier device at a constant rate of 1°C every 30 s until all of the water drops froze. The drop-freeze plate preparation was considered to be robust if the HPLC-grade water drops did not freeze above −20°C. All sample drops freezing at −10°C and above were recorded as containing ice nuclei and were used to calculate the abundance of ice nuclei in each sample. The total concentration of ice nuclei in each sample was calculated using equation 13 under the cumulative nucleus spectra section of Vali's 1971 method (72). The cumulative nucleus concentration calculation is sensitive to the total number of drops used as well as the volume of each drop in the assay. For example, if the sample is aliquoted into a large number of small-volume drops, more total ice nuclei will be observed than if the same sample were aliquoted into fewer, larger-volume drops. The calculation described for Vali's 1971 method (72) determines the total concentration of ice nuclei per ml of sample volume. The total number of ice nuclei was then calculated per cubic meter of air or per liter of snowmelt for the air and snow samples, respectively. A subset of samples was reanalyzed using replicate filters in order to demonstrate the repeatability of the assay, and the ice nucleation numbers varied by less than 1% between replicate filters.
Total microbial community composition via Sanger sequencing.
We examined the overall microbial community composition of bacterial and fungal communities in air and snow samples using a method similar to that of Lauber et al. (44) by constructing clone libraries of partial rRNA genes followed by Sanger sequencing. DNA from microbial cells trapped on the filters was extracted using the Ultra-Clean plant DNA isolation kit (MoBio Laboratories, Carlsbad, CA). The filters were cut into strips using sterile scissors and forceps under a laminar flow hood and loaded into the bead tube of the DNA extraction kit and heated to 65°C for 10 min, followed by 2 min of vortexing. The remaining steps of the extraction were carried out according to the manufacturer's instructions. Ribosomal sequences from all three domains (Archaea, Bacteria, and Eukarya) were amplified using the universal primer set 515f and 1391r, which amplifies an 850- to 1,100-bp portion of the small-subunit rRNA gene (6). PCRs were performed using the Platinum PCR Supermix (Invitrogen, Carlsbad, CA). Primers were added to a final concentration of 0.2 μM, and 2 μl of DNA template was used in each 25-μl reaction mixture. PCR amplification was performed with an initial denaturation at 94°C for 3 min followed by 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s with a final extension at 72°C for 10 min. Each sample was amplified in triplicate, and the amplicons were pooled by sample and run on a 1.2% agarose gel. The bands were purified with the Qiaquick gel extraction kit (Qiagen, Carlsbad, CA). PCR amplicons were cloned into the TOPO TA cloning vector, transformed into TOP10 chemically competent cells (Invitrogen, Carlsbad, CA), and grown on L-agar supplemented with 50 μg ml−1 ampicillin. Following overnight incubation at 37°C, colonies were picked and M13 colony PCR was performed using identical PCR conditions to those described above. PCR products were sent to Agencourt Bioscience (Beverly, MA) for single-pass sequencing. Negative control filters were extracted and PCR amplified and no-template PCRs were run. These reactions produced no visible PCR product on a SYBR green-stained agarose gel.
Sanger sequence analysis.
Sequences were binned into major taxonomic groups (i.e., bacteria, fungi, and plants) using the BLAST algorithm (2) against the NCBI nonredundant database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequences with expect (E) values greater than 1e−100 were not included in the analysis. Of the 672 Sanger sequences having an average length of 750 bp, 12% were of low quality or chimeric and were therefore removed from the analysis. Approximately 7% of the quality sequences were most closely matched to plants and these sequences were not further analyzed. Bacterial and fungal sequences were analyzed separately, as there is currently no single database that can simultaneously classify both of these groups with confidence. The bacterial sequences were aligned using the NAST aligner (minimum length, 600 bp; minimum identity, 75%), checked for chimeras using Bellerophon (21) against the Greengenes database (20), and classified with the Greengenes Classify tool (http://greengenes.lbl.gov/cgi-bin/nph-classify.cgi) using the Hugenholtz taxonomy. More detailed phylogenetic analysis of specific bacterial groups was performed with the maximum likelihood tree-building program RAxML-7.0.4 (68) using the GTR nucleotide substitution model, and a consensus tree was built from the highest-scoring tree of 100 bootstrapped trees. Fungal sequences were aligned using MUSCLE (26) and classified by performing a local BLAST analysis against the comprehensive 16S/18S Silva database, r96_ssu.fasta file (http://www.arb-silva.de/no_cache/download/archive/release_96_exports/). The number of unique fungal phylotypes was defined as those sequences sharing ≥97% sequence similarity, estimated with the FastGroupII algorithm (74).
Bacterial community composition via tagged pyrosequencing.
We used a newly developed bar-coded pyrosequencing procedure to comprehensively survey the airborne bacterial communities. This protocol is identical to the protocol described by Fierer et al. (32), including both the PCR conditions and primer sequences. Negative controls (both no-template control and template control from unused filters) were included at all steps of the process to check for contamination. PCR amplicons from each sample were pooled at approximately equimolar concentrations into a single tube used for the pyrosequencing reaction. Samples were pyrosequenced at the Environmental Genomics Core Facility at the University of South Carolina on a 454 Life Sciences genome sequence FLX (Roche) machine.
Pyrosequencing phylogenetic analysis.
Sequences were analyzed and processed according to the methods of Hamady et al. (35). Only those sequences that were >200 bp in length with a quality score of >25 with no ambiguous characters were included in the analysis (37). Sequences were assigned to samples by examining the 12-bp bar code (32). Phylotypes were identified using Megablast to identify connected components (nearest neighbor), sets of similar sequences (with the parameters E value of 1e−8, minimum coverage of 99%, and minimum pairwise identity of 97%). A representative sequence was chosen from each phylotype by selecting the most highly connected sequence, i.e., the sequence that had the most hits more significant than the BLAST threshold compared to other sequences in the data set (46). The set of all representative sequences was aligned using NAST (21) (with the parameters of minimum alignment length of 190 and sequence identity of 70%) with a PH lanemask (http://greengenes.lbl.gov/) to screen out hypervariable regions of the sequence. Sequences were checked for chimeras using an in-house BLAST-based chimera checking program, which basically checks to see if the sequence ends hit different phyla. A related neighbor-joining tree was built using Clear-cut (67), employing the Kimura correction. Unweighted UniFrac (50, 51) was run using the resulting tree and the sequences annotated by environment type. The UniFrac algorithm provides an estimate of the overall phylogenetic distance between each pair of communities and therefore avoids some of the pitfalls associated with phylotype-based community analyses (51). Taxonomic identity of the phylotypes was assigned with BLAST against the Greengenes database (20) by using an E value cutoff of 1e−10 and the Hugenholtz taxonomy.
Statistical analysis.
UniFrac was used to quantitatively compare the phylogenetic structures of the individual air and snow samples (51). The UniFrac algorithm was applied to a neighbor-joining tree containing all sequences across all samples together with an environment file used to match sequence identity numbers (IDs) to their corresponding sample ID. Relationships between microbial community phylogenetic distance, as determined using the UniFrac algorithm, and meteorological parameters were investigated using the PRIMER software package (www.primer-e.com) (18). Nonmetric multidimensional scaling was used to visualize the UniFrac phylogenetic distance between the air and snow bacterial communities in each pair of environments. The RELATE function in PRIMER was used to evaluate correlations between community phylogenetic distance and each of the individual weather parameters. The effect of each individual weather parameter (Table 1) on community phylogenetic distance was considered significant if P was <0.05. The variations in total particle abundance, total microbial abundance, and the cumulative ice nuclei between clear or cloudy skies were assessed with Student's t test using Statview v5.01.
Nucleotide sequence accession numbers.
The Sanger sequences have been deposited in the GenBank nr database. The bacterial sequences have accession numbers FJ746985 through FJ747245, and the fungal sequences have accession numbers FJ747246 through FJ747530. The authors are currently working on depositing the pyrosequencing data in the GenBank short-read archive; please contact the corresponding author if the pyrosequencing data for this study are desired.
RESULTS
Meteorological conditions.
Atmospheric conditions were measured across the 2-week sampling period and included both clear and cloudy conditions. This is typical of SPL in winter months, since the facility is above cloud base 25% of the time. Based on numerous field sampling programs that have been conducted at SPL, it has been observed that synoptic-scale storms occur at a frequency of 5 to 7 days, and when storms are present, SPL typically remains in cloud for periods of 24 to 48 h. When SPL is in cloud, supercooled liquid water is typically present (9). Half of the samples were collected during cloudy periods and half during clear periods, with cloudy and clear periods defined as periods of high and low relative humidity, respectively (see Table 1 for a description of samples). Cloudy samples were collected within a cloud or with the cloud immediately overhead; cloudy samples had an average relative humidity of 98% and for clear samples average relative humidity was 63%. There were three snowstorm events of approximately 5 cm each during the nights of April 1 to 2 and April 2 to 3 and during the early morning to early afternoon on April 3 corresponding to samples 4-1.CD.N, 4-2.CD.N, and 4-3.CD.D, respectively (Table 1). Samples 4-5.CD.D and 4-5.CD.N were also cloudy air samples but with no snowfall. The remaining four samples were collected from clear skies (Table 1). Samples collected during the day roughly correspond to samples taken from the boundary layer and samples collected during the night correspond to free troposphere samples (9).
Particle counts in relation to total microbial abundance and total ice nuclei.
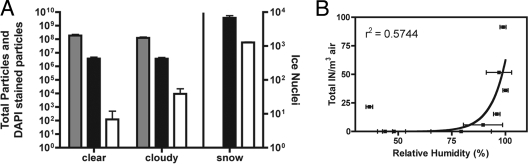
The number of total particles and DAPI-stained particles (i.e., cells) remained relatively constant across all nine sampling periods. Total microbial counts ranged from 9.6 × 105 to 6.6 × 106 cells m−3 of air, and the concentrations of total particles with diameters of 530 nm to more than 5 μm, as determined via APS size fractionation, ranged between 8.2 × 107 and 2.3 × 108 particles m−3 (Fig. 1A). There was no significant difference between clear and cloudy air samples with respect to either cell concentrations (P = 0.5) or total particle counts (P = 0.10). However, the total number of ice nuclei was far more variable across the 2-week sampling period than either the cell counts or total particle counts, ranging from 0 IN/m3 to 91 IN/m3 (Fig. 1). When samples were binned into clear (n = 4) or cloudy (n = 5) air samples, the mean IN concentration was significantly greater in the cloudy air samples (P < 0.05) (Fig. 1A). Similarly, the mean number of IN was positively correlated with relative humidity measurements across all of the sampling events (Fig. 1B).
FIG. 1.
Total APS particle counts, total microbial abundance, and total number of ice nuclei observed in clear and cloudy skies and in replicate snow samples. (A) The left y axis shows APS particle counts (gray bars) (P = 0.09 for clear versus cloudy) and DAPI counts (DNA-containing particles) (black bars) (P = 0.47 for clear versus cloudy) for samples collected in clear air, cloudy air, and from fresh snow. The right y axis shows the number of IN observed per volume of air or melted snow (white bars) (P = 0.05 between cloudy and clear). The air samples are in units of particles or IN m−3 of air, while the snow samples are in units of particles or IN per liter of snow melt. There were no statistics run between the air and snow samples, as the units cannot be compared, and there were no total particle measurements, as the APS instrument was only used for atmospheric aerosols. (B) Relationship between the mean relative humidity during each of the air sampling periods and the total number of ice nuclei m−3 of air.
Characterization of airborne microbial communities via Sanger sequencing.
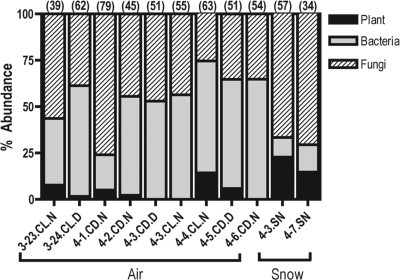
All of the high-quality sequences (590 in total) obtained from the nine air samples and two snow samples could be assigned to one of three major taxonomic groups: plant, bacteria, or fungi (Fig. 2). Plant sequences made up 1.6 to 23% of the sequences from the individual air and snow samples (Fig. 2). The snow possessed the largest number of plant sequences (20% of total Sanger sequence sets), whereas less than 4% of the sequences in the individual air samples belonged to plants (Fig. 2). The plant sequences were predominantly close matches to conifers, particularly Pinus spp., of which most were likely airborne pollen grains. It should also be pointed out that these plant sequences correspond to nuclear 18S rRNA genes and not chloroplast, 16S-like genes.
FIG. 2.
Coarse level taxonomic description of the nine air samples and two snow samples. The numbers in parentheses represent the total number of sequenced clones from each sample (E < 1e−100) that were nonchimeric with an E value of <1e−100. Refer to Table 1 for a complete description of each sample ID listed on the x axis.
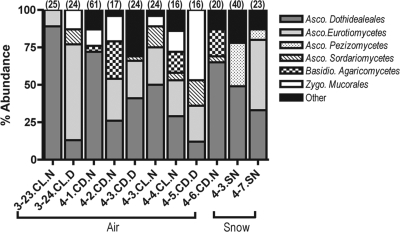
Overall, bacteria/fungi ratios across the nine air samples were fairly consistent, with bacterial and fungal sequences having roughly equivalent representation in the individual libraries (Fig. 2). In contrast, fungal sequences represented 68% of the sequences obtained from the two snow sample libraries (Fig. 2). Since the composition of the bacterial communities was nearly identical across the Sanger sequence data set and the pyrosequencing data set (data not shown), we did not use the Sanger sequences to describe the bacterial communities, as the pyrosequencing data set provided a far more comprehensive survey of the bacterial diversity. The fungal Sanger data set consisted of 289 18S rRNA sequences generated from the 11 samples (9 air and 2 snow). Of the 289 fungal sequences, 126 unique phylotypes were identified at the 97% sequence similarity level. The majority of the sequences were identified as Ascomycota (82% and 92% of the fungal sequences in the air and snow, respectively). Sequences identified as Zygomycota were present in the air samples (14%) but absent from the snow samples, with Basidiomycota sequences making up the remainder of the fungal sequences in the air (4%) and snow (8%) samples. Dothideomycetes was the most abundant fungal class in both the air (53%) and snow (58%) samples, followed by Eurotiomycetes (19%) in the air and both Leotiomycetes (14%) and Lecanoromycetes (11%) in the snow samples. Figure 3 provides further details on the composition of the fungal communities across individual samples. On a per sample basis, Dothideomycetes were the most abundant fungal class in 7 of 11 samples, while Eurotiomycetes dominated three samples (Fig. 3).
FIG. 3.
Composition of the fungal communities as determined using full-length Sanger sequencing. Numbers in parentheses indicate the number of sequences obtained from each sample. Refer to Table 1 for a complete description of each sample listed on the x axis.
Characterization of airborne bacterial communities via pyrosequencing.
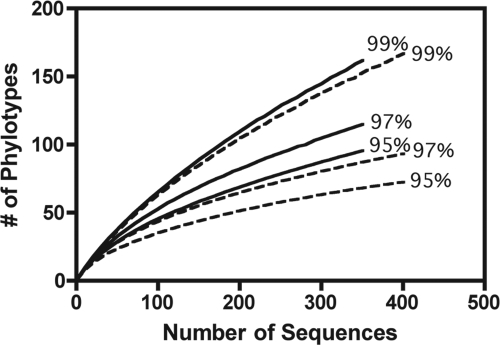
Using the pyrosequencing technique, a total of 4,864 bacterial sequences were generated across all samples. Of the 4,864 sequences, 646 unique phylotypes were observed (with a phylotype defined as those sequences sharing ≥97% identity). Sequences were also grouped at the 95% and 99% sequence similarity levels, which yielded 571 and 1,229 unique phylotypes, respectively. Rarefaction curves were constructed for the most and least diverse samples at each of these sequence similarity levels (Fig. 4). Rarefaction curves at all three similarity levels did not include an asymptote, suggesting that we did not survey the full extent of airborne bacterial diversity, even with an average of 407 sequences per sample (Fig. 4).
FIG. 4.
Rarefaction curves at the 99%, 97%, and 95% sequence similarity levels. Curves shown are from the most diverse (sample ID 4-4.CL.N; solid lines) and least diverse (sample ID 3-24.CL.D; dashed lines) of the nine airborne bacterial samples.
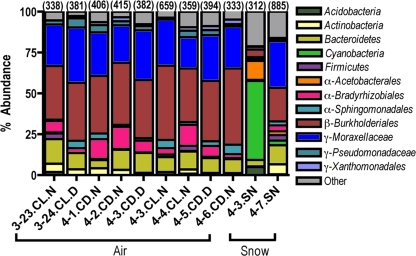
All of the air samples had similar bacterial communities regardless of atmospheric conditions; however, the two snow communities were considerably more variable (Fig. 5). At the phylum level of taxonomic resolution, there were seven major taxa represented throughout the 11 samples. The most abundant phyla present (Fig. 5) in the nine air samples were Proteobacteria (83%) followed by Bacteroidetes (10%), while the snow communities were dominated by Proteobacteria (43%) and Cyanobacteria (26%). The majority of the proteobacterial sequences were similar to the Betaproteobacteria and Gammaproteobacteria subphyla, accounting for 47% and 39% of the air sample proteobacterial sequences and 17% and 14% of the snow proteobacterial sequences, respectively (Fig. 5). The Betaproteobacteria were dominated by Burkholderiales (96% and 91% of the air and snow Betaproteobacteria sequences), and the Gammaproteobacteria were predominantly composed of Moraxellaceae (83% and 92% of the air and snow Gammaproteobacteria sequences) (Fig. 5). Other less common taxa identified from most of the air samples included Enterobacteriaceae, Pseudomonadaceae, and Xanthomonadaceae (each representing <5% of the bacterial sequences in the air and snow samples). Overall, the bacterial communities present in the snow samples differed from those found in the air samples (Fig. 5). In particular, one of the snow samples was dominated by Cyanobacteria sequences (48% of the bacterial sequences).
FIG. 5.
Most abundant bacterial groups identified using bar-coded pyrosequencing. Numbers in parentheses indicate the number of sequences obtained from each sample (E < 1e−100) that were nonchimeric with an E value of <1e−100 from a single tagged pyrosequencing run. Refer to Table 1 for a complete description of each sample ID listed on the x axis. Proteobacterial groups are designated by the Greek symbols α, β, and γ.
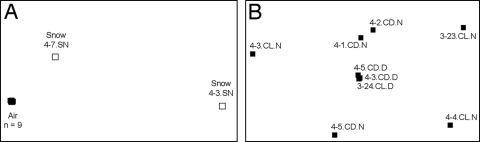
The snow samples harbored phylogenetically distinct bacterial communities compared to those found in the nine air samples (Fig. 6A), and there was minimal variation in the bacterial phylogenetic structure across the nine air samples (Fig. 6B). The minor differences between the air bacterial communities could not be explained by the measured meteorological conditions, as there was no significant correlation between any of the measured meteorological factors (singly or in combination) and the UniFrac distance matrix (P > 0.05 in all cases).
FIG. 6.
Nonmetric multidimensional scaling plot of the UniFrac distance matrix from the tagged pyrosequence bacterial data set. These plots compress the multidimensional UniFrac distance matrix to two dimensions. (A) Air and snow data. Stress value, 0.01. (B) Data for air only; IDs correspond to the IDs listed in Table 1. Stress value, 0.07.
DISCUSSION
Total microbial abundance.
Total airborne microbial abundances, as determined via epifluorescence microscopy, remained relatively stable throughout the sampling period despite the changing atmospheric conditions (Fig. 1A). The total microbial abundances observed at SPL are similar to cell counts reported from other airborne microbial studies (1, 28, 43), suggesting that microbial concentrations are relatively constrained across a range of atmospheric conditions. However, our microbial abundance measurements are significantly higher (approximately an order of magnitude) than those from a similar study reporting bacterial and fungal counts at a high-elevation research site in the Austrian Alps (8). The difference between our study and the work of Bauer et al. (8) is likely methodological, as the Bauer et al. (8) study counted bacteria and fungi separately, did not include pollen in their measurements, and used a different air sampling procedure.
Recently, primary biological particles have been shown to contribute a large fraction to the total atmospheric aerosol abundance (38). Of the total aerosol concentration at SPL, the number of particles with biological characteristics (fluorescing in the presence of the DNA binding dye DAPI) accounted for approximately 5 to 10% of the particles measured by the APS (Fig. 1A). This microbial aerosol fraction is similar to estimates provided in the recent literature. For example, air samples that were collected in Germany (55), the South Atlantic (53), and at a remote location in Siberia (54) contained microbial fractions (percentage of fluorescing particles in relation to the total particle count) ranging from 17% to 30%. While our study did not include measurements spanning the different seasons, the microbial aerosol fraction was relatively constant over the 2-week sampling period (Fig. 1A), which is consistent with the temporal stability observed by the Matthias-Maser (53, 54, 55) and Jaenicke (38) studies mentioned above.
Bacteria/fungi ratios.
The relative abundance of the major taxonomic groups (plant, bacteria, and fungi) varied little over the 2-week sampling period (Fig. 2). The plant sequences were minor constituents of the nine libraries constructed from the air samples, with fungal and bacterial sequences having approximately equal representation in the libraries (Fig. 2). The 1:1 ratio of bacterial and fungal sequences observed in these air samples is consistent with that reported by Wilson et al. (73), who observed a similar ratio in a single air sample collected in California (73). The bacteria/fungi ratios at SPL differed from those of both a recent ribosomal sequencing-based study (33) and an earlier culture-based survey (24), as both studies observed much higher variation in their bacterial to fungal ratios over time. These differences may be attributed to differences in sampling location and/or the time of sampling. The air collected by Fierer et al. (33) was collected at head height on a college campus, and the di Giorgio et al. (24) work was conducted at two cities in France over a year. Both studies were performed at locations that are likely influenced by a number of anthropogenic disturbances that could potentially cause large shifts in microbial abundance and composition over short time periods, whereas the air at SPL is comparatively undisturbed with regard to surface-level microbial sources. While there are approximately equal numbers of bacterial and fungal sequences in our airborne samples, it is important to recognize that these sequence ratios do not necessarily indicate that bacteria and fungi have equivalent biomass concentrations in the atmosphere, as these taxa differ with respect to their cell sizes and the number of small-subunit rRNA copies per cell.
Fungal community composition.
At the level of taxonomic resolution afforded by the 18S rRNA gene, the fungal sequences appear to be more variable than the bacterial communities (Fig. 3), although some of this variability could be attributed to the small number of sequences used to assess the airborne fungal communities. The majority of the samples collected at SPL were dominated by the Dothideales and Eurotiomycetes classes of fungi (Fig. 3), which were also observed to be the dominant fungi colonizing sediment exposed by a receding glacier (39). Culturable fungi that are commonly thought to be abundant in the atmosphere, such as Alternaria, Cladosporium, and Penicillium spp. (15, 66), were not found in the air samples at SPL. These differences may be attributed to culturing bias, although we did not compare cultivation-dependent and cultivation-independent surveys of fungal diversity in this study.
Bacterial diversity.
Just as the relative abundances of airborne bacteria and fungi were moderately constant over time, the types of bacteria present in the atmosphere sampled at SPL also changed very little over the 2-week sampling period (Fig. 5). Although the 4,864 bacterial sequences generated from the bacteria-specific pyrosequencing run represent one of the more comprehensive surveys of airborne bacterial diversity to date (with an average of 407 sequences per sample), we still did not survey the full extent of bacterial diversity within the individual air samples, as evidenced by the lack of asymptotes in Fig. 4. The bacterial diversity levels observed at SPL are considerably lower than those reported in other molecular-based studies of the atmosphere (13) and in other environments such as in soil (31). The differences in diversity levels between the various airborne landscapes may be attributed to sampling location, as Despres et al. (22, 23) observed lower bacterial diversity levels in the air of the high-alpine environment compared to their urban and rural sampling locales (22, 23).
Bacterial community composition.
The bacterial taxa identified from the nine air samples collected at SPL were similar to those found in other studies where air samples were also dominated by Betaproteobacteria and Gammaproteobacteria (22, 23, 31, 52, 63). In particular, we found that most of the Betaproteobacteria sequences were comamonads (members of the order Burkholderiales) (Fig. 5), which are a metabolically and phenotypically diverse group of bacteria that is relatively common in the atmosphere (13, 33). The dominant Gammaproteobacteria taxon, the Moraxellaceae, was highly represented across the nine air samples (Fig. 5), with Psychrobacter spp. being the most abundant genus-level group within this family. Interestingly, members of the genus Psychrobacter have previously been isolated from various low-temperature environments, such as Antarctic soils (10), Antarctic sea ice (11, 12), and Siberian permafrost (60).
Bacteria in the atmosphere are distinct from those found in fresh snow.
Previous studies have used snow samples to predict microbial abundances and the numbers of ice-nucleating particles of biological and nonbiological origin present in the atmosphere (16, 17). This approach would seem legitimate, as precipitation events are known to scavenge and collect particles that eventually get deposited onto the ground (62). However, the bacterial communities found in the air above SPL appear to be distinct when compared to the bacterial communities found in freshly fallen snow (Fig. 5 and 6). The two snow samples harbored distinct bacterial communities, although this variation was likely due to the exposure of sample 4-3.SN to intense sunlight for approximately 2 hours prior to sampling, which may explain the overrepresentation of cyanobacterial sequences in this sample. Snow sample 4-7.SN was much more representative of the bacterial communities found in the air, although there were still a number of bacterial groups found in this snow sample that were not present in any of the air samples (Fig. 5), indicating that freshly fallen snow is not truly representative of the microbial life residing in the atmosphere.
Shifts in total IN in response to shifting weather conditions.
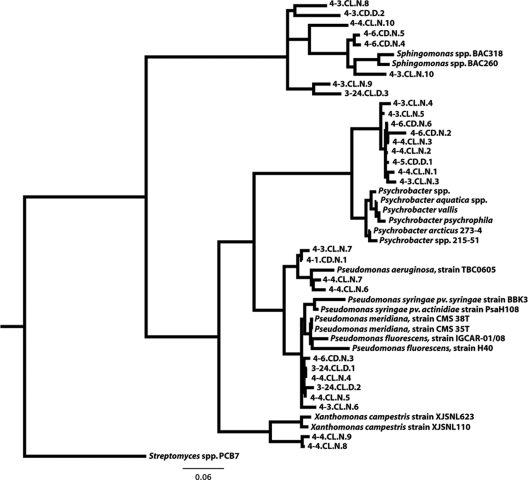
While the types and abundances of microorganisms in the air samples were fairly constant over time and not strongly influenced by changes in atmospheric conditions, the abundance of ice-nucleating particles was far more variable, increasing with increases in relative humidity (Fig. 1). Other studies have also shown that ice nucleation rates in cirrus and in wave clouds respond positively to increases in humidity (71). The ice nuclei counted in this study were counted at temperatures equal to or higher than −10°C, which has recently been used as a temperature cutoff to define ice nucleators of biological origin (16, 17). The most potent nonbiological ice-nucleating particles generally initiate freezing at temperatures below −10°C (65). Based on these studies, we made the assumption that the IN counted via the drop-freeze assay were likely to have been initiated by bacteria, fungi, and/or plant pollen. Throughout our bacterial community analysis, we identified ribosomal DNA gene sequences with homology to the following known ice-nucleating bacteria: P. syringae, P. fluorescens, E. herbicola, and Sphingomonas spp. (Fig. 7). However, these bacterial taxa were not common in either our Sanger sequence or pyrosequence data sets. For instance, the Pseudomonadaceae family, which contains the most extensively studied ice-nucleating species, represented less than 1% of the snow pyrosequences and only 3% of the air pyrosequence data set. However, sequences matching the Psychrobacter genus were relatively common in our data sets, with similar abundance levels across all nine air samples (Fig. 5). Members of this genus have recently been shown to initiate ice nucleation at temperatures as high as −2°C (60), which rivals the ice-nucleating temperature of the well-studied bacterium P. syringae. The phylogeny presented in Fig. 7 illustrates the phylogenetic relatedness between the 16S rRNA sequences generated from this current study to 16S rRNA sequences of many of the verified ice-nucleating bacterial isolates.
FIG. 7.
Phylogenetic distribution of representative full-length Sanger sequences that have similarities to the 16S rRNA gene of known ice-nucleating bacteria. Sequences were aligned using the Greengenes NAST aligner, and the maximum-likelihood tree was made using RAxML-7.0.4. A Streptomyces sequence was used as the outgroup. Each Sanger sequence is represented by its sample ID (see Table 1 for sample descriptions).
Considering that the abundance of IN was significantly higher in the cloudy air parcels than in clear air parcels (Fig. 1) and that the bacterial community composition was well-constrained (Fig. 5), it is likely that the bacteria responsible for this increase in total IN at SPL are either very rare members of the atmospheric microbial community or exhibit variable ice-nucleating capacities. In support of this hypothesis, studies of P. syringae and E. herbicola isolates have shown that these bacterial species have the potential to increase their ice-nucleating activity in response to environmental triggers, including nutrient deprivation and/or temperature reduction (29, 58). Therefore, nontransient bacteria in the atmosphere may adjust their concentrations of IN proteins when conditions become favorable (i.e., high humidity), thus increasing the ice-nucleating potential of these bacterial taxa. Alternatively, fungal spores and/or pollen grains may be responsible for the observed increase in IN abundance during periods of cloud cover, as they have also been shown to possess high-temperature ice-nucleating capabilities (41, 61).
Acknowledgments
We thank Ian McCubbin, Kelly Baustian, and Eszter Horanyi for their assistance at Storm Peak Laboratory. We also thank the Fierer lab for their helpful input on the manuscript and Matt Zelenok for his assistance with the drop-freeze assays.
This study was supported by the CIRES Innovative Research Program. Support was also provided by Steamboat and Resort Corporation. The Desert Research Institute is an equal opportunity service provider and employer and a permittee of the Medicine-Bow Routt National Forests.
Footnotes
Published ahead of print on 5 June 2009.
REFERENCES
- 1.Albrecht, A., R. Witzenberger, U. Bernzen, and U. Jackel. 2007. Detection of airborne microbes in a composting facility by cultivation based and cultivation-independent methods. Ann. Agric. Environ. Med. 14:81-85. [PubMed] [Google Scholar]
- 2.Altschul, S., T. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amato, P., M. Mènager, M. Sancelme, P. Laj, G. Mailhot, and A.-M. Delort. 2005. Microbial population in cloud water at the Puy de Dùme: implications for the chemistry of clouds. Atmos. Environ. 39:4143-4153. [Google Scholar]
- 4.Amato, P., M. Parazols, M. Sancelme, G. Mailhot, P. Laj, and A.-M. Delort. 2007. An important oceanic source of micro-organisms for cloud water at the Puy de Dôme (France). Atmos. Environ. 41:8253-8263. [Google Scholar]
- 5.Andrews, J. H., and R. F. Harris. 2000. The ecology and biogeography of microorganisms on plant surfaces. Annu. Rev. Phytopathol. 38:145-180. [DOI] [PubMed] [Google Scholar]
- 6.Angenent, L. T., S. T. Kelley, A. S. Amand, N. R. Pace, and M. T. Hernandez. 2005. Molecular identification of potential pathogens in water and air of a hospital therapy pool. Proc. Natl. Acad. Sci. USA 102:4860-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer, H., H. Giebl, R. Hitzenberger, A. Kasper-Giebl, G. Reischl, F. Zibuschka, and H. Puxbaum. 2003. Airborne bacteria as cloud condensation nuclei. J. Geophys. Res. 108:4658. [Google Scholar]
- 8.Bauer, H., A. Kasper-Giebl, M. Loflund, H. Giebl, R. Hitzenberger, F. Zibuschka, and H. Puxbaum. 2002. The contribution of bacteria and fungal spores to the organic carbon content of cloud water, precipitation and aerosols. Atmos. Res. 64:109-119. [Google Scholar]
- 9.Borys, R. D., and M. A. Wetzel. 1997. Storm Peak Laboratory: a research, teaching, and service facility for the atmospheric sciences. Bull. Am. Meteorol. Soc. 78:2115-2123. [Google Scholar]
- 10.Bowman, J. P., J. Cavanagh, J. J. Austin, and K. Sanderson. 1996. Novel Psychrobacter species from Antarctic ornithogenic soils. Int. J. Syst. Bacteriol. 46:841-848. [DOI] [PubMed] [Google Scholar]
- 11.Bowman, J. P., D. S. Nichols, and T. A. McMeekin. 1997. Psychrobacter glacincola sp. nov., a halotolerant, psychrophillic bacterium isolated from Antarctic sea ice. Syst. Appl. Microbiol. 20:209-215. [Google Scholar]
- 12.Bozal, N., M. J. Montes, E. Tudela, and J. Guinea. 2003. Characterization of several Psychrobacter strains isolated from Antarctic environments and description of Psychrobacter luti sp. nov. and Psychrobacter fozii sp. nov. Int. J. Syst. Evol. Microbiol. 53:1093-1100. [DOI] [PubMed] [Google Scholar]
- 13.Brodie, E. L., T. Z. DeSantis, J. P. M. Parker, I. X. Zubietta, Y. M. Piceno, and G. L. Andersen. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. USA 104:299-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown, J. K. M., and M. S. Hovmoller. 2002. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297:537-541. [DOI] [PubMed] [Google Scholar]
- 15.Burge, H. A. 2002. An update on pollen and fungal spore aerobiology. J. Allergy Clin. Immunol. 110:544-552. [DOI] [PubMed] [Google Scholar]
- 16.Christner, B. C., R. Cai, C. E. Morris, K. S. McCarter, C. M. Foreman, M. L. Skidmore, S. N. Montross, and D. C. Sands. 2008. Geographic, seasonal, and precipitation chemistry influence on the abundance and activity of biological ice nucleators in rain and snow. Proc. Natl. Acad. Sci. USA 105:18854-18859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christner, B. C., C. E. Morris, C. M. Foreman, R. Cai, and D. C. Sands. 2008. Ubiquity of biological ice nucleators in snowfall. Science 319:1214. [DOI] [PubMed] [Google Scholar]
- 18.Clarke, K. R., and R. M. Warwick. 2001. A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Mar. Ecol. Prog. Ser. 216:265-278. [Google Scholar]
- 19.Constantinidou, H. A., S. S. Hirano, L. S. Baker, and C. D. Upper. 1990. Atmospheric dispersal of ice nucleation-active bacteria: the role of rain. Phytopathology 80:934-937. [Google Scholar]
- 20.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeSantis, T. Z., Jr., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, R. Phan, and G. L. Andersen. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394-W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Despres, V., J. Nowoisky, M. Klose, R. Conrad, M. O. Andreae, and U. Poschl. 2007. Molecular genetics and diversity of primary biogenic aerosol paricles in urban, rural, and high-alpine air. Biogeosciences 4:349-384. [Google Scholar]
- 23.Despres, V. R., J. F. Nowoisky, M. Klose, R. Conrad, M. O. Andreae, and U. Poschl. 2007. Characterization of primary biogenic aerosol particles in urban, rural, and high-alpine air by DNA sequence and restriction fragment analysis of ribosomal RNA genes. Biogeosciences 4:1127-1141. [Google Scholar]
- 24.di Giorgio, C., A. Krempff, H. Guiraud, P. Binder, C. Tiret, and G. Dumenil. 1996. Atmospheric pollution by airborne microorganisms in the city of Marseilles. Atmos. Environ. 30:155-160. [Google Scholar]
- 25.Dingle, A. N. 1966. Pollen as condensation nuclei. J. Rech. Atmos. 2:231-237. [Google Scholar]
- 26.Edgar, R. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elster, J., R. J. Delmas, J.-R. Petit, and K. Reháková. 2007. Composition of microbial communities in aerosol, snow and ice samples from remote glaciated areas (Antarctica, Alps, Andes). Biogeosciences 4:1779-1813. [Google Scholar]
- 28.Fabian, M. P., S. L. Miller, T. Reponen, and M. T. Hernandez. 2005. Ambient bioaerosol indices for indoor air quality assessments of flood reclamation. J. Aerosol Sci. 36:763-783. [Google Scholar]
- 29.Fall, A. L., and R. Fall. 1998. High-level expression of ice nuclei in Erwinia herbicola is induced by phosphate starvation and low temperature. Curr. Microbiol. 36:370-376. [DOI] [PubMed] [Google Scholar]
- 30.Fierer, N., M. A. Bradford, and R. B. Jackson. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354-1364. [DOI] [PubMed] [Google Scholar]
- 31.Fierer, N., M. Breitbart, J. Nulton, P. Salamon, C. Lozupone, R. Jones, M. Robeson, R. A. Edwards, B. Felts, S. Rayhawk, R. Knight, F. Rohwer, and R. B. Jackson. 2007. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl. Environ. Microbiol. 73:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fierer, N., M. Hamady, C. L. Lauber, and R. Knight. 2008. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. USA 105:17994-17999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fierer, N., Z. Liu, M. Rodriguez-Hernandez, R. Knight, M. Henn, and M. T. Hernandez. 2008. Short-term temporal variability in airborne bacterial and fungal populations. Appl. Environ. Microbiol. 74:200-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffin, D. W. 2004. Terrestrial microorganisms at an altitude of 20,000 m in Earth's atmosphere. Aerobiologia 20:135-140. [Google Scholar]
- 35.Hamady, M., J. J. Walker, J. K. Harris, N. J. Gold, and R. Knight. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5:235-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernandez, M., S. L. Miller, D. Landfear, and J. M. Macher. 1999. A combined fluorochrome method for quantitation of metabolocally active and inactive airborne bacteria. Aerosol Sci. Technol. 30:145. [Google Scholar]
- 37.Huse, S. M., J. A. Huber, H. G. Morrison, M. L. Sogin, and M. D. Welch. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaenicke, R. 2005. Abundance of cellular material and proteins in the atmosphere. Science 308:73. [DOI] [PubMed] [Google Scholar]
- 39.Jumpponen, A. 2003. Soil fungal community assembly in a primary successional glacier forefront ecosystem as inferred from rDNA sequence analyses. New Phytol. 158:569-578. [DOI] [PubMed] [Google Scholar]
- 40.Kajava, A. V., and S. E. Lindow. 1993. A model of the three-dimensional structure of ice nucleation proteins. J. Mol. Biol. 232:709-717. [DOI] [PubMed] [Google Scholar]
- 41.Kieft, T., and V. Ahmadjian. 1989. Biological ice nucleation activity in lichen mycobionts and photobionts. Lichenologist 21:355-362. [Google Scholar]
- 42.Kozloff, L. M., M. A. Turner, and F. Arellano. 1991. Formation of bacterial membrane ice-nucleating lipoglycoprotein complexes. J. Bacteriol. 173:6528-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lange, J., P. Thorne, and N. Lynch. 1997. Application of flow cytometry and fluorescent in situ hybridization for assessment of exposures to airborne bacteria. Appl. Environ. Microbiol. 63:1557-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lauber, C. L., M. S. Strickland, M. A. Bradford, and N. Fierer. 2008. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 40:2407-2415. [Google Scholar]
- 45.Lee, R. E. J., G. J. Warren, and L. V. Gusta. 1995. Biological ice nucleation and its applications. APS Press, St. Paul, MN.
- 46.Ley, R. E., M. Hamady, C. Lozupone, P. J. Turnbaugh, R. R. Ramey, J. S. Bircher, M. L. Schlegel, T. A. Tucker, M. D. Schrenzel, R. Knight, and J. I. Gordon. 2008. Evolution of mammals and their gut microbes. Science 320:1647-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lighthart, B. 1997. The ecology of bacteria in the alfresco atmosphere. FEMS Microbiol. Ecol. 23:263-274. [Google Scholar]
- 48.Lighthart, B., and B. Shaffer. 1995. Viable bacterial aerosol particle size distributions in the midsummer atmosphere at an isolated location in the high desert chaparral. Aerobiologia 11:19-25. [Google Scholar]
- 49.Lindow, S. E., and M. T. Brandl. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lozupone, C., M. Hamady, and R. Knight. 2006. UniFrac: an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maron, P. A., D. P. H. Lejon, E. Carvalho, K. Bizet, P. Lemanceau, L. Ranjard, and C. Mougel. 2005. Assessing genetic structure and diversity of airborne bacterial communities by DNA fingerprinting and 16S rDNA clone library. Atmos. Environ. 39:3687-3695. [Google Scholar]
- 53.Matthias-Maser, S., J. Brinkmann, and W. Schneider. 1999. The size distribution of marine atmospheric aerosol with regard to primary biological aerosol particles over the South Atlantic Ocean. Atmos. Environ. 33:3569-3575. [Google Scholar]
- 54.Matthias-Maser, S., and R. Jaenicke. 2000. The size distribution of primary biological aerosol particles in the multiphase atmosphere. Aerobiologia 16:207-221. [Google Scholar]
- 55.Matthias-Maser, S., K. Peters, and R. Jaenicke. 1995. Seasonal variation of primary biological aerosol particles. J. Aerosol Sci. 26:S545-S546. [Google Scholar]
- 56.Mohler, O., P. J. DeMott, G. Vali, and Z. Levin. 2007. Microbiology and atmospheric processes: the role of biological particles in cloud physics. Biogeosciences 4:2559-2591. [Google Scholar]
- 57.Mohler, O., D. G. Georgakopoulos, C. E. Morris, S. Benz, V. Ebert, S. Hunsmann, H. Saathoff, M. Schnaiter, and R. Wagner. 2008. Heterogeneous ice nucleation activity of bacteria: new laboratory experiments at simulated cloud conditions. Biogeosciences 5:1425-1435. [Google Scholar]
- 58.Nemecek-Marshall, M., R. LaDuca, and R. Fall. 1993. High-level expression of ice nuclei in a Pseudomonas syringae strain is induced by nutrient limitation and low temperature. J. Bacteriol. 175:4062-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 60.Ponder, M. A., S. J. Gilmour, P. W. Bergholz, C. A. Mindock, R. Hollingsworth, M. F. Thomashow, and J. M. Tiedje. 2005. Characterization of potential stress responses in ancient Siberian permafrost psychroactive bacteria. FEMS Microbiol. Ecol. 53:103-115. [DOI] [PubMed] [Google Scholar]
- 61.Pouleur, S., C. Richard, J.-G. Martin, and H. Antoun. 1992. Ice nucleation activity in Fusarium acuminatum and Fusarium avenaceum. Appl. Environ. Microbiol. 58:2960-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Radke, L. F., P. V. Hobbs, and M. W. Eltgrowth. 1980. Scavenging of aerosol particles by precipitation. J. Appl. Meteorol. 19:715-722. [Google Scholar]
- 63.Radosevich, J. L., W. J. Wilson, J. H. Shinn, T. Z. DeSantis, and G. L. Andersen. 2002. Development of a high-volume aerosol collection system for the identification of air-borne micro-organisms. Lett. Appl. Microbiol. 34:162-167. [DOI] [PubMed] [Google Scholar]
- 64.Sattler, B., H. Puxbaum, and R. Psenner. 2001. Bacterial growth in supercooled cloud droplets. Geophys. Res. Lett. 28:239-242. [Google Scholar]
- 65.Schaller, R. C., and N. Fukuta. 1979. Ice nucleation by aerosol particles: experimental studies using a wedge-shaped thermal diffusion chamber. J. Atmos. Sci. 36:1788-1802. [Google Scholar]
- 66.Shelton, B. G., K. H. Kirkland, W. D. Flanders, and G. K. Morris. 2002. Profiles of airborne fungi in buildings and outdoor environments in the United States. Appl. Environ. Microbiol. 68:1743-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sheneman, L., J. Evans, and J. A. Foster. 2006. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics 22:2823-2824. [DOI] [PubMed] [Google Scholar]
- 68.Stamatakis, A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688-2690. [DOI] [PubMed] [Google Scholar]
- 69.Tong, Y., and B. Lighthart. 2000. The annual bacterial particle concentration and size distribution in the ambient atmosphere in a rural area of the Willamette Valley, Oregon. Aerosol Sci. Technol. 32:393-403. [Google Scholar]
- 70.Turner, M. A., F. Arellano, and L. M. Kozloff. 1991. Components of ice nucleation structures of bacteria. J. Bacteriol. 173:6515-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vali, G. 1996. Ice nucleation: a review, p. 271-279. In M. Kulmala and P. Wagner (ed.), Nucleation and atmospheric aerosols. Pergamon Press, Oxford, United Kingdom.
- 72.Vali, G. 1971. Quantitative evaluation of experimental results on the heterogeneous freezing nucleation of supercooled liquids. J. Atmos. Sci. 28:402-409. [Google Scholar]
- 73.Wilson, K. H., W. J. Wilson, J. L. Radosevich, T. Z. DeSantis, V. S. Viswanathan, T. A. Kuczmarski, and G. L. Andersen. 2002. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu, Y., M. Breitbart, P. McNairnie, and F. Rohwer. 2006. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinformatics 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]