Abstract
The whole-genome sequence of the endosymbiotic bacterium Azorhizobium caulinodans ORS571, which forms nitrogen-fixing nodules on the stems and roots of Sesbania rostrata, was recently determined. The sizes of the genome and symbiosis island are 5.4 Mb and 86.7 kb, respectively, and these sizes are the smallest among the sequenced rhizobia. In the present study, a whole-genome microarray of A. caulinodans was constructed, and transcriptomic analyses were performed on free-living cells grown in rich and minimal media and in bacteroids isolated from stem nodules. Transcriptional profiling showed that the genes involved in sulfur uptake and metabolism, acetone metabolism, and the biosynthesis of exopolysaccharide were highly expressed in bacteroids compared to the expression levels in free-living cells. Some mutants having Tn5 transposons within these genes with increased expression were obtained as nodule-deficient mutants in our previous study. A transcriptomic analysis was also performed on free-living cells grown in minimal medium supplemented with a flavonoid, naringenin, which is one of the most efficient inducers of A. caulinodans nod genes. Only 18 genes exhibited increased expression by the addition of naringenin, suggesting that the regulatory mechanism responding to the flavonoid could be simple in A. caulinodans. The combination of our genome-wide transcriptional profiling and our previous genome-wide mutagenesis study has revealed new aspects of nodule formation and maintenance.
The symbiosis between rhizobia and legumes results in the formation of nitrogen-fixing nodules. The symbiotic interaction begins with the induction of bacterial nod genes by flavonoids secreted from the plant roots (8). The nod genes encode proteins that synthesize the nodulation (Nod) factor, which initiates many developmental changes, such as root hair curling and root cell division required for the formation of the nodule primordium in the host plant early during the nodulation process (8, 24, 50). Bacteria are entrapped in the curled root hair and subsequently infect the root hair through infection threads made of the plant cell wall. Upon release from the infection threads, bacteria invade the plant cell cytoplasm, where they differentiate into bacteroids and provide ammonium to the host plant by reducing atmospheric dinitrogen in exchange for carbon and amino acid compounds (16, 49, 53). It has been deduced that multiple stages exist in the establishment of nitrogen-fixing symbiosis. To identify novel genes involved in various stages of symbiosis, transcriptomic studies based on complete genome sequences were performed using Sinorhizobium (1, 2, 5, 9), Mesorhizobium (71), and Bradyrhizobium (10, 42, 54).
Azorhizobium caulinodans ORS571 is a microsymbiont of Sesbania rostrata (18-20). Nitrogen-fixing nodules are formed by A. caulinodans on the stems as well as on the roots of S. rostrata. Stem nodules occur at the site of adventitious root primordia located on the stems after crack-entry invasion by A. caulinodans (70). During crack-entry invasion, bacteria proliferate in the epidermal fissures at the adventitious root primordia on the stems (27). Cortical infection pockets are formed by Nod factor-dependent induction of local cell death and subsequent colonization by bacteria (13). From the infection pockets, infection threads guide bacteria toward the cells in the nodule primordia for symbiotic uptake (14).
A. caulinodans is taxonomically different from other rhizobia and belongs to the family Xanthobacteraceae, including Xanthobacter autotrophicus, although it is relatively close to Bradyrhizobium (44). Recently, the whole-genome sequence of A. caulinodans was determined (43), and it was revealed to consist of a single circular chromosome of 5.37 Mb, which is the smallest among the sequenced genomes of rhizobia. The size of the symbiosis island is only 87.6 kb and contains nod genes but not nif and fix genes. With the analysis of the genome sequence, we concurrently performed a large-scale screening of rhizobial factors involved in nodule development using 10,080 mutants of A. caulinodans created by random Tn5 mutagenesis. We analyzed a total of 108 Tn5 mutants and identified 86 genes involved in symbiosis (68). The list of mutants, which showed aberrant nodule development at various stages, shows the complexity of nodule development and maturation. Next, we investigated which genes in A. caulinodans were important for establishing symbiosis with host plants. To this end, we constructed a whole-genome microarray of A. caulinodans and analyzed the gene expression in free-living cells versus that in bacteroids isolated from stem nodules. In addition, we examined the effect of a flavonoid, naringenin, which is one of the most efficient inducers of A. caulinodans nod genes (26), on gene expression in free-living cells. In the present report, we discuss the symbiotic mechanism of A. caulinodans based on the results of the transcriptional profiling experiments described herein and our previous transposon mutagenesis study.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A. caulinodans ORS571 (18) was grown at 37°C in MMO medium (5 g/liter sodium lactate, 5 g/liter disodium succinate 6H2O, 1.67 g/liter K2HPO4, 0.87 g/liter KH2PO4, 0.1 g/liter MgSO4·7H2O, 0.1 g/liter NaCl, 1 g/liter (NH4)2SO4, 0.04 g/liter CaCl2·2H2O, 2 mg/liter biotin, 4 mg/liter pantothenic acid, and 4 mg/liter nicotinic acid [pH 6.8]) (26) as a minimal medium or in TY medium (5 g/liter tryptone, 3 g/liter yeast extract, and 0.83 g/liter CaCl2·2H2O [pH 6.8]) (6) as a rich medium. To isolate RNA from free-living cells, A. caulinodans was grown in 100 ml of medium up to an optical density at 600 nm of 0.5 to 0.7. To analyze the effect of flavonoids, naringenin (20 μM) was added to the MMO medium 6 h before harvesting cells. Cultured cells were harvested by centrifugation at 8,000 × g at 4°C for 5 min. The pellets were frozen in liquid nitrogen and stored at −80°C until RNA was isolated.
Plant growth, inoculation, and bacteroid isolation.
S. rostrata seeds were treated with concentrated sulfuric acid for 1 h, rinsed with sterile water, and soaked in sterile water on trays. The trays were stored for 3 days at 37°C under dark conditions. After germination, S. rostrata were grown for 2 weeks before bacterial inoculation at 35°C under a 24-h light regime at an intensity of 50,000 lx, as described in our previous report (68). Bacterial cultures grown overnight were inoculated onto the stems. At 7 days postinoculation, the stem nodules on the second stem internode of plants were harvested, immediately frozen in liquid nitrogen, and stored at −80°C. To isolate bacteroids, the stem nodules were vigorously homogenized with MMS buffer (40 mM MOPS [morpholinepropanesulfonic acid], 20 mM KOH, 2 mM MgSO4, and 0.3 M sucrose [pH 7.0]) (55) containing 4% polyvinylpyrrolidone. The resulting homogenate was passed through three layers of Miracloth (Calbiochem), and the filtrate was centrifuged at 500 × g for 5 min to remove plant cell fractions and other debris. Then, the supernatant was centrifuged at 5,000 × g for 10 min, and the pellet containing bacteroids was collected. To remove the green layer containing the contaminated chloroplasts from the surface of the white pellet, MMS buffer containing 25% polyvinylpyrrolidone was added onto the pellet, and the surface of the pellet was washed by vigorous pipetting; the buffer was then removed. After this washing procedure, the pellet was resuspended in MMS buffer without polyvinylpyrrolidone and centrifuged at 5,000 × g for 2 min, and the supernatant was removed. This procedure was performed twice. The white pellet was used as a bacteroid sample for RNA isolation.
A. caulinodans ORS571 DNA microarray.
The AzcNXa520550F whole-genome array (designed by Affymetrix) contains 7,096 probe pair sets (45 for target preparation controls, 4,725 for ORFs, and 2,326 for intergenic regions tiled in both directions) corresponding to 4,703 ORFs and 1,163 intergenic regions of A. caulinodans ORS571, based on the genome sequence (43). The nucleotide sequence of the entire genome of A. caulinodans ORS571 is available in the DDBJ/EMBL/GenBank databases under the accession number AP009384. The probe length was 25 mer, and for all ORFs, 16 probe pairs were chosen. Every probe pair set contained 16 perfect-match probes and mismatch probes. For the intergenic sequences, probes were selected at a density of 16 to 24 bp to fill the remaining space on the chip. As target preparation controls, the poly(A) controls (dap, lys, phe, thr, and trp from Bacillus subtilis) were included in the gene chip. Furthermore, the hybridization controls (bioB, bioC, and bioD from Escherichia coli and cre from the P1 bacteriophage) were also included.
RNA isolation, cDNA synthesis, and labeling.
Total RNA was isolated from cultured and bacteroid cells using Isogen (Nippon Gene, Japan) according to the manufacturer's instructions. The isolated RNA was treated with DNase I (Invitrogen) and purified by a phenol-chloroform-isoamyl alcohol (25:24:1) extraction and ethanol precipitation. The purified RNA was analyzed by agarose gel electrophoresis, and distinct bands of 16S and 23S rRNAs were detected. In the case of RNA samples from bacteroids, bands of 18S and 28S rRNAs of plants were not detected. To synthesize cDNA, 10 μg of total RNA was mixed with 2 μl of the 260-fold diluted poly(A) target preparation control containing dap, lys, phe, and thr mRNA (Affymetrix) and was reverse transcribed by SuperScript III transcriptase (Invitrogen). After the remaining RNA was digested with RNase H for 20 min at 37°C, cDNA was collected with the MinElute kit (Qiagen, Germany). Fragmentation was performed with DNase I with 1× One-Phor-All buffer (GE Healthcare, United Kingdom). Fragmented cDNA was end labeled with terminal deoxynucleotidyl transferase (Promega) and GeneChip DNA labeling reagent (Affymetrix).
Hybridization, scanning, and microarray data analyses.
Hybridization, washing, staining, and scanning were performed using the GeneChip hybridization, wash, and stain kit (Affymetrix), the GeneChip Hybridization Oven 640 (Affymetrix), the GeneChip Fluidics Station 450 (Affymetrix), and the GeneChip Scanner 3000 (Affymetrix) according to the Affymetrix manual. For hybridization, 1.0 to 1.5 μg of labeled cDNA was mixed with Control Oligo B2 (3 nM; Affymetrix) in a volume of 150 μl hybridization solution. Hybridization was performed overnight at 50°C for 16 h. Primary signal extraction and comparative analyses were performed using GeneChip Operating Software (GCOS) version 1.4 (Affymetrix). To normalize signals between arrays, global scaling was performed by Microarray Suite version 5.0 (Affymetrix) using all probe sets. The target intensity value was set to 500, and the scaling factors were between 1.5 and 4.9. Default statistical parameters of GCOS (Alpha1 = 0.04; Alpha2 = 0.06; Tau = 0.015; Gamma1L = 0.001111; Gamma1H = 0.001111; Gamma2L = 0.001333; Gamma2H = 0.001333; perturbation = 1.1) were applied. A probe pair set was called “present” (i.e., expressed) when the detection P value was lower than the Alpha1 value, i.e., when the intensity of the perfect-match probe cell was significantly greater than that of the corresponding mismatch probe cell. When the detection P value was greater than or equal to the Alpha2 value, i.e., when the intensity of the perfect-match probe cell was significantly less than that of the corresponding mismatch probe cell, the probe pair set was called “absent” (i.e., not expressed). When the detection P value was between Alpha1 and Alpha2, the probe pair set was called “marginal” (i.e., unable to call the transcript present or absent). Data were then processed using Microsoft Excel. For statistical comparison, a Student's t test was applied to the log2 signal with a P value threshold of ≤0.05. To evaluate the differentially expressed genes, the weighted average difference (WAD) method (36, 37) was also applied to the log2 signal. Genes were considered differentially expressed if |WAD| was ≥0.5.
Computational sequence analyses.
Homology searches based on amino acid sequences were performed using the BlastP programs on the National Center for Biotechnology Information (NCBI) server (www.ncbi.nlm.nih.gov/BLAST/). Searches for protein signatures were performed using the InterProScan program on the European Bioinformatics Institute (EBI) server (www.ebi.ac.uk/InterProScan/).
Microarray data accession number.
The entire set of microarray data was deposited in the Center for Information Biology Gene Expression Database (CIBEX) (http://cibex.nig.ac.jp/index.jsp) under the accession number CBX88.
RESULTS AND DISCUSSION
Overview.
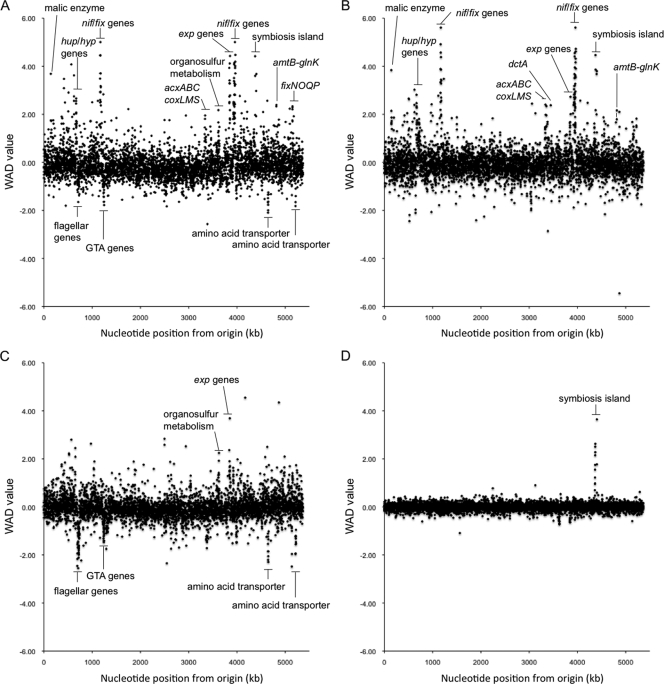
Transcriptional analysis of A. caulinodans was performed under four conditions, and three biological replicates were applied in all conditions. In all replicates, the positive-control probes (dap, lys, phe, and thr from B. subtilis), which were hybridized with the poly(A) target preparation control, were called present with the appropriate ratio of signal intensity; furthermore, the negative-control probes (trp from B. subtilis; bioB, bioC, and bioD from E. coli; and cre from the P1 bacteriophage) were called absent in all arrays (data not shown). This result indicates that the experiments were correctly performed. Table 1 shows the average correlation of replicate microarray hybridizations under each experimental condition, and all replicates showed good correlation. All transcriptional profiling data, such as the log2-transformed signal intensity of each gene, together with the P values (Student's t test) and WAD values, are listed in the supplemental material (see Table S1). We evaluated the differentially expressed genes by using the WAD method (36, 37). In microarray studies, several methods, such as the fold-change method, were employed to identify differentially expressed genes under different experimental conditions. However, evaluations based on the fold-change method have a disadvantage because genes showing lower expression levels tend to be falsely detected as differentially expressed genes. Alternatively, the WAD method that is an improved fold-change-based method can be used to overcome this shortcoming of the fold-change method. Evaluation with the WAD method employs relative-average signal intensity (i.e., the importance of an expressed gene under tested conditions) and average difference. Thus, this method enables us to select differentially expressed genes more accurately. Table 2 shows the number of differentially expressed genes (P ≤ 0.05; |WAD| ≥ 0.5) in each functional category defined in our previous genome sequence study (43). Figure 1 shows the WAD values for each gene plotted against position within the genome for each pairwise comparison.
TABLE 1.
Average correlation of replicate microarray hybridizations for each experimental condition
| Experimental condition | Correlation coefficient (mean ± SD) (n = 3) | No. of genes called present in all replicates |
|---|---|---|
| Free-living cells in TY medium | 0.90 ± 0.07 | 2,375 |
| Free-living cells in MMO medium | 0.95 ± 0.03 | 2,342 |
| Free-living cells in MMO medium supplied with 20 μM naringenin | 0.94 ± 0.04 | 2,394 |
| Bacteroids in stem nodules | 0.99 ± 0.01 | 3,011 |
TABLE 2.
Overview of differential expression patterns based on gene functional categories
| Gene category (no. of genes) | No. of genes differentially expresseda (increased/decreased)
|
|||
|---|---|---|---|---|
| Bacteroid vs TY | Bacteroid vs MMO | MMO vs TY | Naringenin addition vs no addition | |
| Amino acid biosynthesis (132) | 15/19 | 15/20 | 14/6 | 1/0 |
| Cofactor, prosthetic group, and carrier biosynthesis (159) | 22/34 | 18/22 | 21/14 | 0/2 |
| Cell envelope (174) | 17/54 | 15/48 | 29/9 | 0/4 |
| Cellular process (200) | 17/45 | 45/23 | 13/58 | 0/0 |
| Central intermediary metabolism (161) | 71/18 | 62/16 | 25/16 | 1/0 |
| Energy metabolism (303) | 49/45 | 53/28 | 24/51 | 0/0 |
| Fatty acid, phospholipid, and sterol metabolism (136) | 14/35 | 15/20 | 16/13 | 0/0 |
| Purine, pyrimidine, nucleoside, and nucleotide (66) | 2/13 | 3/11 | 7/8 | 0/0 |
| Regulatory function (384) | 31/70 | 26/61 | 29/21 | 0/1 |
| DNA replication, recombination, and repair (79) | 7/1 | 4/17 | 5/2 | 0/0 |
| Transcription (49) | 8/5 | 6/4 | 5/2 | 1/0 |
| Signal transduction (39) | 1/9 | 2/7 | 2/4 | 0/0 |
| Translation (227) | 22/60 | 24/29 | 20/27 | 0/0 |
| Transport and binding protein (714) | 49/186 | 66/114 | 59/82 | 1/4 |
| Other categories (765) | 87/143 | 75/90 | 59/75 | 13/2 |
| Hypothetical protein (954) | 145/144 | 122/153 | 115/79 | 1/1 |
| Unknown protein (175) | 25/28 | 22/30 | 23/10 | 0/0 |
| Total (4,717) | 582/919 | 573/693 | 466/477 | 18/14 |
A differentially expressed gene has a log2 signal with a P value of ≤0.05 (Student's t test) and a |WAD| of ≥0.5.
FIG. 1.
WAD values for each gene plotted against position within the genome, for each pairwise comparison. (A) Expressional difference between bacteroid and free-living cells grown in TY medium. (B) Expressional difference between bacteroid and free-living cells grown in MMO medium. (C) Expressional difference between free-living cells grown in MMO medium with 20 μM naringenin and MMO medium. (D) Expressional difference between free-living cells grown in MMO medium and TY medium.
The most striking differences were observed in the comparison between bacteroids and free-living cells grown in rich medium (Table 2 and Fig. 1A). Thirty-two percent of the genes were differentially expressed; the expression levels of 582 genes were higher in bacteroids than in free-living cells, and the expression levels of 919 genes were lower in bacteroids than in free-living cells. Among the highly expressed genes in bacteroids, 71 genes, including nif, fix, hup, and hyp genes, belonged to the functional category “central intermediary metabolism.” Of the genes belonging to the functional category “transport and binding proteins,” which consists of solute-binding proteins, permeases, and ATP-binding cassette (ABC) transporters, 26%, including two kinds of ABC transporter genes for branched-chain amino acids (AZC_4072 to AZC_4080 and AZC_4583 to AZC_4589), were expressed to a lesser degree in bacteroids than in free-living cells. In contrast, about 7% of the transport and binding protein genes, including sitABC (AZC_0562 to AZC_0564), which encodes an Fe-Mn ABC transporter, and dctA (AZC_3014), which encodes a C4-dicarboxylate transporter, was expressed to a greater extent in bacteroids than in free-living cells.
The nutritional environment also drastically affected the gene expression of A. caulinodans (Table 2 and Fig. 1C). It was noted that 20% of the genes were differentially expressed in free-living cells grown in minimal and rich media; 466 genes were highly expressed, while 477 genes were expressed to a lesser extent in minimal medium than in rich medium. In minimal medium, the genes of the gene clusters involved in exopolysaccharide (EPS) production (AZC_3319 to AZC_3332) and organosulfur metabolism (AZC_3157 to AZC_3180) were expressed to a greater extent than in free-living cells grown in rich medium. The expression levels of these genes were also higher in bacteroids than in free-living cells grown in rich medium (Fig. 1A). On the other hand, a large gene cluster of flagellar biosynthesis genes (AZC_0619 to AZC_0666), a gene cluster of the gene transfer agent (GTA) genes (AZC_1104 to AZC_1127), and two clusters containing branched-chain amino acid transporter genes (AZC_4072 to AZC_4080 and AZC_4583 to AZC_4589) were expressed to a lesser extent in minimal medium (Fig. 1D). The expression levels of these gene clusters were also lower in bacteroids than in free-living cells grown in rich medium (Fig. 1A).
The addition of naringenin influenced the expression of 32 genes; 18 genes were highly expressed and 14 genes were expressed to a lesser extent in the cells supplied with naringenin (Table 2 and Fig. 1D). Of the 18 genes with increased expression, 13 were in the symbiosis island. Of the 14 genes with decreased expression, 7 were in the gene cluster related to organosulfur acquisition, and 5 were in the gene cluster related to EPS synthesis. Of the two remaining genes, one was a gene putatively encoding MarR-like transcriptional factor (AZC_1832) and the other was a gene putatively encoding molybdopterin oxidoreductase (AZC_1874). In the case of Bradyrhizobium japonicum USDA110, about 100 genes were induced in response to genistein, a flavonoid secreted by soybeans (42). Thus, it can be assumed that the regulatory mechanism responding to a flavonoid might be simpler in A. caulinodans than in B. japonicum.
The previous screening of Tn5 mutants showed that 86 genes, including nif, fix, and nod genes, were required for A. caulinodans to establish symbiosis (68). The expression profiling of these genes is listed in the supplemental material (see Table S2). Among these genes, 17 genes, such as nod and noe genes (AZC_3809 and AZC_3811), nif and fix genes (AZC_1038, AZC_1040, AZC_1049, AZC_3414, AZC_3446, AZC_3448, and AZC_3449), a putative transcriptional factor gene (phrR; AZC_0013), a trehalose-6-phosphate phosphatase gene (otsA; AZC_0483), a Lon protease gene (AZC_1610), a C4 dicarboxylate transporter gene (dctA; AZC_3014), EPS synthesis genes (AZC_3319 and AZC_3322), a cysteine desulfurase gene (sufS; AZC_3616), and a fatty acid coenzyme A (CoA) ligase gene (AZC_4536), showed higher expression in bacteroids than in free-living cells grown in both types of media. The expressions of 42 genes, including genes involved in lipopolysaccharide biosynthesis (rfaF [AZC_2251] and rfaD [AZC_2253]), were not significantly different. Five genes (AZC_1714, AZC_1894, AZC_2209, AZC_2370, and AZC_3038) showed lower expression in bacteroids than in free-living cells. Five genes (AZC_0859, AZC_1796, AZC_2131, AZC_2759, and AZC_3904) were called absent or marginal in all replicates applied in this study (data not shown), suggesting that these genes showed little or no expression under all experimental conditions. The nodule-defective phenotypes of the mutants of these 10 genes might have developed due to the polar effect of the Tn5 transposon or unlinked second mutations.
Hereafter, we discuss important features of the genome and the expression profiling of A. caulinodans. WAD values for the genes mentioned below are included in Table S3 in the supplemental material.
Symbiosis island.
As previously described (43), the symbiosis island of A. caulinodans is flanked by tRNA-Gly and interspersed by multiple transposases and integrases, which separate the symbiosis island into several clusters (see Fig. S1 in the supplemental material). Three of these clusters are nod loci (nodD, AZC_3792; the nodABCSUIJZ-noeCHO operon, AZC_3818 to AZC_3808; and nolK, AZC_3850). Two of the clusters (AZC_3826 to AZC_3844 and AZC_3856 to AZC_3877) contain conjugation-related genes. The cluster AZC_3856 to AZC_3877 also contains genes related to amino acid transport. Moreover, a cluster (AZC_3794 to AZC_3800) consists of genes related to nitrile hydratase (NHase).
The expression levels of the nodABCSUIJZ-noeCHO and nolK genes in free-living cells supplied with naringenin were much higher than in those without naringenin (WAD, 0.70 to 3.64) (Fig. 1D). Furthermore, the expression levels of the nodABCSZ-noeHO and nolK genes were higher in bacteroids than in free-living cells grown in both rich and minimal medium (WAD values were 0.73 to 1.25 in a comparison of expression levels in bacteroids and free-living cells grown in minimal medium). The expression of the genes related to conjugation and amino acid transport located in the symbiosis island did not show significant change under any experimental conditions in this study.
A. caulinodans harbors 45 transposase genes on the genome, 19 of which are located in the symbiosis island. Nine of the 45 transposase genes were highly expressed in bacteroids, compared to the levels in free-living cells grown in minimal medium, and 7 of these 9 genes were located in the symbiosis island (AZC_3806, AZC_3821, AZC_3824, AZC_3825, AZC_3846, AZC_3848, and AZC_3851; WAD, 0.63 to 3.86). These results suggest that the symbiosis island is the region where gene transfer has most frequently occurred. Although the high expression of transposase genes in bacteroids has been reported in the transcriptomic analysis of other rhizobia, the biological significance is unknown (9, 71). Interestingly, a transposase gene (AZC_3851) located in the symbiosis island was highly expressed in free-living cells supplied with naringenin. This result suggests that the transposase encoded by AZC_3851 could be activated in the rhizosphere. Such transcriptional behavior of transposase genes has not been reported in other transcriptomic research of rhizobia.
The gene cluster AZC_3794 to AZC_3800 contains the genes AZC_3797 and AZC_3798 encoding proteins with 53% and 30% identity, respectively, to the NHase α and β subunits of Agrobacterium tumefaciens. The expression levels of the genes within this cluster were higher in bacteroids than in free-living cells grown in both rich and minimal media (WAD values were 0.67 to 1.37 in a comparison of expression levels in bacteroids and free-living cells grown in rich medium, and WAD values were 0.56 to 1.62 in a comparison of expression levels in bacteroids and free-living cells grown in minimal medium). In A. tumefaciens and some rhizobia, it is known that NHases are able to convert indole-3-acetonitrile to indole-3-acetamide, and indole-3-acetamide is converted to indole-3-acetic acid by amidase (40). Accordingly, there is a possibility that auxin production may be enhanced in bacteroids.
nif, fix, and suf gene clusters.
Four gene clusters, including nif and fix genes, were found in the genome of A. caulinodans (see Fig. S2 in the supplemental material). The first cluster contained nif genes, along with sufE and sufBCDS genes (AZC_1042 and AZC_1044 to AZC_1047), which are involved in iron-sulfur (Fe-S) cluster assembly, and phn genes, which are involved in phosphonate metabolism. The second cluster contained nif and fix genes, along with phn genes. The first and second gene clusters were conserved among A. caulinodans, X. autotrophicus, and photosynthetic bradyrhizobia, although the syntenies were different (see Fig. S3 in the supplemental material). Most of the nif and fix genes within the first, second, and third clusters were strongly expressed in bacteroids compared to the expression levels in free-living cells (WAD, 0.79 to 5.01 in rich medium and 1.99 to 5.61 in minimal medium) (Fig. 1A and B).
The sufBCDS and sufE genes in the first cluster were strongly expressed in bacteroids (WAD, 1.48 to 3.70 in rich medium and 1.93 to 3.86 in minimal medium). In addition to these suf genes, A. caulinodans has other suf genes outside this cluster, namely, sufSDBCS (AZC_3612 to AZC_3616). The expression levels of these suf genes were also higher in bacteroids than in free-living cells (WAD, 0.90 to 1.82 in rich medium and 0.53 to 1.37 in minimal medium). SufS is cysteine desulfurase, similar to NifS, and supplies sulfur for Fe-S centers (47, 52). The SufBCD complex activates the cysteine desulfurase activity of SufS in conjunction with the SufE that is a sulfur acceptor protein (51). The high expression of these suf genes suggests that bacteroids require a large amount of Fe-S clusters and that Fe-S clusters provided by SufS proteins may be used to sustain nitrogen fixation as well as the NifS protein. In the previous screening, a Tn5 mutant (Ao15-F04) with disruption in the sufS gene (AZC_3616) was obtained, and this mutant formed small stem nodules lacking nitrogen-fixing ability (68). This result also supports the importance of suf genes in stem nodule formation.
Ammonium assimilation.
Unexpectedly, the expression levels of a putative glnK-amtB operon (AZC_4227 to AZC_4226) encoding a nitrogen regulatory protein PII and an ammonium transporter, respectively, were higher in bacteroids than in free-living cells (WAD values were 2.39 and 2.33 in a comparison of expression levels in bacteroids and free-living cells in rich medium, and WAD values were 2.16 and 1.79 in a comparison of expression levels in bacteroids and free-living cells grown in minimal medium) (Fig. 1A and B). In the free-living state, the fixed nitrogen is assimilated by glutathione synthase (17), whereas during symbiosis, the fixed nitrogen is exported from the bacteroids to the plant cells by passive diffusion and assimilated by the host. It has been reported that the expression levels of genes encoding the PII-like protein of Rhizobium etli and Sinorhizobium meliloti were lower in bacteroids than in free-living cells (1, 69). However, our result contradicted these findings. Furthermore, transcriptomic analysis of B. japonicum showed that the expression level of the glnK-amtB operon was high in nodules (54). Although the biochemical function of glnK-amtB in nodules is unknown, it is possible that the transcriptional regulation of glnK-amtB in A. caulinodans and B. japonicum is different from that of R. etli and S. meliloti.
Carbon uptake and metabolism.
A. caulinodans, as well as other rhizobia, uses C4-dicarboxylates as primary carbon sources and has multiple C4-dicarboxylate transport systems. Prokaryotes have three types of C4-dicarboxylate transport systems, that is, a major facilitator superfamily (MFS)-type transporter, an ABC-type transporter, and a tripartite ATP-independent periplasmic transporter. A. caulinodans has four genes (AZC_1325, AZC_2673, AZC_3014, and AZC_3353) putatively encoding an MFS-type transporter (DctA). Searches performed using BlastP revealed that the protein encoded by AZC_3014 was 81% identical to S. meliloti DctA and that the proteins encoded by the other three genes were about 50% identical to S. meliloti DctA. On the genome of S. meliloti, there is a single dctA gene, and a dctA mutant of S. meliloti forms ineffective nodules lacking nitrogen-fixing ability (23). A. caulinodans has 21 genes putatively encoding any of the DctP, -Q, and -M components of tripartite ATP-independent periplasmic C4-dicarboxylate transporters. Of the multiple C4-dicarboxylate transporter genes of A. caulinodans, only AZC_3014 was highly expressed in bacteroids compared to the expression level in free-living cells (WAD, 1.49 in rich medium and 2.38 in minimal medium). We have isolated a Tn5 mutant (Ao62-E02) with disruption in AZC_3014, and this mutant was observed to form inefficient stem nodules lacking nitrogen-fixing ability (68). These results suggest that the MFS transporter encoded by AZC_3014 is the most important C4-dicarboxylate transporter in bacteroids. Thus, C4-dicarboxylate uptake by bacteroids may depend mainly on this transporter.
In addition to C4-dicarboxylates, the array data suggest the possibility that bacteroids of A. caulinodans are supplied with other carbon compounds. A. caulinodans contains AZC_2923 to AZC_2921 genes putatively encoding acetone carboxylase subunits (AcxA, -B, and -C), which are 89, 88, and 79% identical to AcxA, -B, and -C of X. autotrophicus, respectively. It is known that X. autotrophicus has the ability to use acetone as a carbon source. X. autotrophicus produces acetoacetate by the carboxylation of acetone in an ATP-dependent manner (62, 74). The high similarities of amino acid sequences suggest that A. caulinodans may have the ability to use acetone as a carbon source. The gene AZC_2924 adjacent to the acxABC genes encodes a protein with 55% identity to the AcxR protein of X. autotrophicus, which is a transcriptional activator for acxABC genes (63). In the previous screening, a Tn5 mutant (Ao22-A06) with disruption in AZC_2924 was isolated, and this mutant was deficient in nodule formation. It is interesting that the expression levels of acetone carboxylase genes were higher in bacteroids than in free-living cells (WAD, 1.01 to 1.95 in rich medium and 1.48 to 2.40 in minimal medium) (Fig. 1A and B). Furthermore, it was reported that acetone was produced in soybean nodules (67). Although it is uncertain whether acetone is also produced in the nodules of S. rostrata, if acetone production is a general event in the nodules of legumes, A. caulinodans bacteroids may utilize acetone as a carbon source in addition to C4-dicarboxylates.
A. caulinodans contains genes (AZC_2937 to AZC_2939) putatively encoding carbon monoxide dehydrogenase (CODH) subunits (CoxM, -L, and -S) with 34, 38, and 61% identities, respectively, to the CoxM, -L, and -S of Oligotropha carboxidovorans, which is the most intensively studied representative of the carboxydotrophic bacteria. CODH is an O2-stable molybdenum-iron-sulfur-flavin hydroxylase that catalyzes the oxidation of CO to CO2. Carboxydotrophic bacteria constitute a small but diverse group of aerobes (48), and putative coxLMS genes have been identified in the genomes of bacteria that are not known to be CO oxidizers. Among the rhizobia, the B. japonicum strain 110 spc4 is known to have the ability to grow using CO as a sole carbon and energy source under aerobic conditions (45). The expression levels of putative coxLMS genes of A. caulinodans were higher in bacteroids than in free-living cells (WAD, 0.84 to 1.37 in rich medium and 1.50 to 2.34 in minimal medium) (Fig. 1A and B). If these genes really encode functional CODH, there is a possibility that bacteroids of A. caulinodans may oxidize CO in nodules to use CO as a carbon source.
Malic enzyme (ME) catalyzes the reversible oxidative decarboxylation of malate to pyruvate and CO2. The generation of acetyl-CoA via ME and pyruvate dehydrogenase is important for nitrogen-fixing bacteroids in order to metabolize C4-dicarboxylates via the trichloroacetic acid cycle. S. meliloti contains NAD(P)+- and NADP+-dependent ME encoded by dme and tme, respectively (21, 22). A dme mutant of S. meliloti forms nodules showing no nitrogen-fixing ability, while a tme mutant forms wild-type nodules (21, 22). On the genome of A. caulinodans, two genes (AZC_0119 and AZC_3656) putatively encoding ME were identified by BlastP analyses. The protein encoded by AZC_0119 was similar to plant or animal NADP+-dependent ME (44% identical to NADP-ME 1 of Arabidopsis thaliana). The protein encoded by AZC_3656 was 46% identical to TME of S. meliloti. Two Tn5 mutants (Ao30-B02 and Ao58-F11) with disruption in AZC_3656 formed nodules lacking nitrogen-fixing ability (68), suggesting that this putative ME similar to S. meliloti TME is essential for nodule formation by A. caulinodans, although TME is not essential for nodule formation by S. meliloti. The expression levels of AZC_3656 were not different between bacteroids and free-living cells. In contrast, the expression levels of AZC_0119 were much higher in bacteroids than in free-living cells (WAD, 3.68 in rich medium and 3.84 in minimal medium) (Fig. 1A and B). It is important to determine whether AZC_0119 is essential for nitrogen fixation or not. Therefore, the functional assignment of each ME encoded by AZC_0119 and AZC_3656 should be determined.
Sulfur uptake.
Sulfur is a primary component of sulfur-containing amino acids (cysteine and methionine) and plays an essential role as a substituent in certain enzyme cofactors. In rhizobia-legume symbiosis, sulfur has important roles. In S. meliloti, sulfation of the Nod factor is essential to elicit the nodule formation of host plants (56), and sulfated Nod factors are more resistant to degradation than nonsulfated ones (64). Furthermore, nitrogen fixation requires Fe-S clusters. Bacteria typically utilize inorganic sulfate or cysteine as their primary sulfur source, and they also utilize sulfonates or sulfate esters under sulfur-limited conditions (39). Sulfate uptake mechanisms have been well analyzed in E. coli (32, 59, 60). In E. coli, the ABC-type sulfate/thiosulfate transporters are encoded by the cysPTWA genes, with the unlinked sbp gene located separately on the genome. CysT and CysW are permease components, and CysA is an ATP-binding protein. CysP and Sbp are substrate-binding proteins with high affinity for thiosulfate and sulfate, respectively. However, CysP and Sbp possess an overlapping substrate range and share the CysTWA system. The genes related to sulfonate utilization by bacteria have been characterized in B. subtilis (72), E. coli (73), and Pseudomonas putida (38). E. coli contains two types of sulfonate transport and desulfonation gene clusters, ssuABCDE and tauABCD. SsuABC and TauABC are ABC transporters that import alkanesulfonates and taurine, respectively. Alkanesulfonates and taurine are desulfurized by a two-component reduced flavin mononucleotide-dependent monooxygenase (SsuDE) and an α-ketoglutarate-dependent dioxygenase (TauD), respectively. Sulfate ester transporter is characterized as an ABC-type transporter encoded by atsRBC genes in Pseudomonas aeruginosa (33).
BlastP analyses revealed that A. caulinodans contained two genes (AZC_0302 and AZC_0922) encoding proteins that were 50 to 72% identical to E. coli CysP/Sbp. The genes (AZC_0921 to AZC_0919) downstream of AZC_0922 encoded proteins that were 48%, 52%, and 57% identical to E. coli CysT, -W, and -A, respectively. It is noteworthy that a large gene cluster (AZC_3157 to AZC_3180) containing putative ssu genes (AZC_3157 and AZC_3160 to AZC_3164) and ats genes (AZC_3168 to AZC_3171, AZC_3173, and AZC_3177) was identified on the genome. Each protein encoded by these genes was 31 to 61% identical to the corresponding proteins of E. coli or P. aeruginosa (see Table S4 in the supplemental material). AZC_3159 located next to the putative ssu genes (AZC_3160 to AZC_3164) was previously annotated as actinorhodin polyketide dimerase. However, an InterProScan search revealed that the protein encoded by AZC_3159 contained a flavin mononucleotide-binding domain (data not shown), and a BlastP search revealed that this protein was 29% identical to E. coli RutF, a putative flavin, i.e., NADH component of oxidoreductase (see Table S4 in the supplemental material). From these results, it appears that the protein encoded by AZC_3159 might act as an NAD(P)H-flavin oxidoreductase, like SsuE, in cooperation with a putative SsuD encoded by AZC_3157.
The expression levels of putative cysP and sbp genes (AZC_0302 and AZC_0922) were higher in bacteroids (WAD, 0.54 and 1.09, respectively) and free-living cells grown in minimal medium (WAD, 1.04 and 1.61, respectively) than in free-living cells grown in rich medium. The expression levels of 12 genes (AZC_3157 to AZC_3161, AZC_3163, AZC_3165, AZC_3172 to AZC_3174, and AZC_3179 to AZC_3180) within the AZC_3157 to AZC_3180 cluster, including the ssu and ats genes, were also higher in bacteroids than in free-living cells grown in rich medium (WAD, 0.55 to 2.17). The expression levels of 14 genes (AZC_3157 to AZC_3161, AZC_3163, AZC_3167, AZC_3169 to AZC_3174, and AZC_3177) in free-living cells were higher in minimal medium than in rich medium (WAD, 0.57 to 2.25) (Fig. 1A and C). Furthermore, a Tn5 mutant (Ao61-B09) with disruption in AZC_3159 formed stem nodules lacking nitrogen-fixing ability (68). It is unclear whether these genes are related to sulfur acquisition; however, if they are, these data suggest that bacteroids as well as free-living cells grown in minimal medium might actively acquire organosulfur compounds in addition to inorganic sulfate/thiosulfate. In addition, organosulfur uptake may have advantages in obtaining carbon sources.
Vitamins.
Biotin is an essential cofactor in carboxylation, decarboxylation, and transcarboxylation reactions. Generally, biotin is synthesized via the conserved four enzymes encoded by bioF, bioA, bioD, and bioB starting from pimeloyl-CoA (see Fig. S4 in the supplemental material), although pimeloyl-CoA is synthesized in various ways (34, 57, 65). Bacteria have the ability not only to synthesize biotin but also to acquire biotin from the environment. It was proposed that bioYMN genes consisting of an operon are involved in biotin uptake in R. etli (30), and recently, it was found that BioY is a biotin transporter and that BioN and BioM are dedicated to being energy-coupling modules (31). The bioY genes have been identified on a variety of bacterial genomes, and bioY genes are not always adjacent to bioMN genes. A. caulinodans is biotin auxotrophic, and BlastP analyses revealed that A. caulinodans had a gene (AZC_0361) encoding a protein 57% identical to E. coli BioB, but no bioF, bioA, or bioD genes were identified by BlastP analyses. It was also revealed that A. caulinodans contained three genes (AZC_0362, AZC_2177, and AZC_3757) encoding proteins 34%, 31%, and 52% identical to R. etli BioY, respectively (see Fig. S4 in the supplemental material). AZC_0362 is located next to the putative bioB gene (AZC_0361), AZC_2177 is an orphan, and AZC_3757 is located next to bioMN genes (AZC_3755 and AZC_3756). The expression levels of AZC_0361 to AZC_0362 (bioBY cluster) were higher in bacteroids than in free-living cells (WAD values were 3.08 and 3.47 in a comparison of expression levels in bacteroids and free-living cells grown in rich medium, and WAD values were 1.70 and 1.96 in a comparison of expression levels in bacteroids and free-living cells grown in minimal medium), but those of AZC_2177 (orphan bioY) and AZC_3755 to AZC_3757 (bioMNY cluster) were not. This result suggests that the biotin uptake of bacteroids depends mainly on the biotin transporter encoded by AZC_0361, if this protein is really a functional biotin transporter. Why was the putative bioB gene highly expressed in bacteroids, even though A. caulinodans has only this gene among biotin synthesis genes? It is speculated that the BioY-like protein encoded by AZC_0362 may act as a dethiobiotin transporter, and dethiobiotin obtained by AZC_0362 may be converted to biotin by BioB encoded by AZC_0361.
Thiamine is an essential cofactor for several important enzymes of carbohydrate metabolism, including the trichloroacetic acid cycle. Thiamine synthesis and the related genes of bacteria are well studied in E. coli and B. subtilis, as reviewed in references 35 and 58. BlastP analyses revealed that A. caulinodans contained genes homologous to the thiamine-related genes of E. coli and B. subtilis (31 to 65% similar, based on the amino acid sequences [see Fig. S5 in the supplemental material]). The expression of putative thiCGSO genes (AZC_2607 and AZC_3552 to AZC_3554) was higher in bacteroids (WAD, 0.63 to 1.22) and in cells grown in minimal medium (WAD, 1.01 to 2.52) than in those grown in rich medium. We have isolated three Tn5 mutants (Ao5-E01, Ao55-D02, and Ao73-E10) with disruption in the putative thiC gene, and these mutants form stem nodules showing lower nitrogen-fixing activity than the wild-type strain (68). However, the nitrogen-fixing activities of these mutants grown in minimal medium were not significantly different from that of the wild-type strain (68). From these results, it can be deduced that highly activated de novo synthesis of thiamine is essential for bacteroids but not for free-living cells grown in minimal medium, although thiCGSO genes were highly expressed under both conditions.
EPS synthesis.
Rhizobial EPS is one of the important factors in the establishment of symbiosis. Rhizobial EPSs are linear or branched heteropolysaccharides that consist of common sugars (61). A. caulinodans produces a linear homopolysaccharide of α-1,3-linked 4,6-O-(1-carboxyethylidene)-d-galactopyranoside residues (15). S. meliloti produces two structurally distinct EPSs: EPS I (succinoglycan) and EPS II (galactoglycan). These EPSs are essential for the establishment of nitrogen-fixing symbiosis (3). S. meliloti contains two gene clusters (exo-exs and exp gene clusters), directing the biosynthesis of EPS I and EPS II, respectively (3).
BlastP searches suggested that no gene cluster corresponding to the S. meliloti exo-exs cluster existed on the genome of A. caulinodans (data not shown). On the contrary, two gene clusters putatively corresponding to the S. meliloti exp gene cluster, which is composed of five putative operons designated as expA (nine genes), expC (one gene), expD (two genes), expE (eight genes), and expG (one gene) (4, 25), were identified on the genome of A. caulinodans (see Table S5 in the supplemental material). One is composed of AZC_1831 to AZC_1834 encoding proteins that are 45 to 61% identical to ExpA7, -10, -9, and -8 of S. meliloti, respectively. The other is AZC_3319 to AZC_3332. AZC_3319 to AZC_3331 encodes proteins that are 21 to 64% identical to ExpA5, ExpE1 to -7, ExpD1 and -2, ExpG, ExpC, and ExpA4 of S. meliloti, respectively. AZC_3332 encodes a hypothetical protein. The former and latter clusters are designated as exp cluster I and II, respectively. Genes homologous to expA1, expA23, and expE8 of S. meliloti are not contained in these clusters.
There were no significant changes in the expression levels of the exp cluster I genes under any experimental conditions in this study. The expression levels of the exp cluster II genes were higher in free-living cells grown in minimal medium than in those grown in rich medium (WAD, 0.81 to 3.69) (Fig. 1C). In S. meliloti, the expression of the exo gene cluster increased in minimal medium, although the expression of the exp gene cluster did not (2), suggesting that the regulatory system of the exp genes of A. caulinodans is different from that of S. meliloti. Furthermore, the expression levels of the exp cluster II genes of A. caulinodans were higher in bacteroids than in free-living cells (WAD, 1.63 to 4.43 in rich medium and 0.58 to 2.73 in minimal medium) (Fig. 1A and B). In contrast, the expression of most exo genes of S. meliloti did not increase in bacteroids, and the expression of exoX, a posttranscriptional inhibitor of EPS synthesis, increased in bacteroids (2). These results suggest that EPS might be actively synthesized in bacteroids of A. caulinodans but not in bacteroids of S. meliloti. We have isolated two Tn5 mutants (Ao3-G08 and Ao11-A12) with disruption in the putative expA5 (AZC_3319) and expE5 (AZC_3322) genes, respectively (68). Ao3-G08 formed very small nodules called bumps, and Ao11-A12 formed small nodules showing nitrogen-fixing activity. Future studies on the EPS synthesis of these mutants will help to clarify the function of EPS in nodulation formation by A. caulinodans.
GTA genes.
It is noteworthy that a gene cluster related to an unusual class of a virus-like genetic exchange element, the GTA, which was discovered in Rhodobacter capsulatus (46), was identified on the genome of A. caulinodans. BlastP analyses revealed that 15 genes in the gene cluster of AZC_1104 to AZC_1127 encoded proteins 28 to 55% identical to the GTA proteins of R. capsulatus (see Fig. S6 in the supplemental material). The structure of GTA is similar to a small, tailed phage (77), but GTA seems to function only in the transfer of small, random pieces of genomic DNA between cells (41). Recent genome sequencing projects have revealed that GTA gene clusters are present exclusively in alphaproteobacteria (7, 41). More than half of the sequenced alphaproteobacteria genomes contain a GTA-like gene cluster, including Rhizobiales. However, among the symbiotic rhizobia, only A. caulinodans contains the GTA-like gene cluster.
The expression levels of the putative GTA genes of A. caulinodans were lower in bacteroids (WAD, −0.56 to −1.78) and free-living cells grown in minimal medium (WAD, −0.55 to −1.47) than in those grown in rich medium (Fig. 1A and C). As mentioned above, among the rhizobia, GTA genes were found only in A. caulinodans, and the expression of these genes was suppressed in bacteroids. From these results, GTA may have adverse effects on the establishment of symbiosis—or GTA may simply be unnecessary for symbiosis. To investigate which hypotheses are true, it will be necessary to introduce GTA genes into other rhizobia or to enhance artificially the expression of GTA genes in A. caulinodans.
Concluding remarks.
So far, the genome sequencing of A. caulinodans and the screening of genetic factors required for stem nodule formation have been carried out (43, 68). In the present study, we conducted the transcriptional profiling of A. caulinodans in free-living and symbiotic states. The data obtained from these studies enabled us to ascertain some aspects of the mechanism of stem nodule formation by A. caulinodans. Many of the genes we have mentioned in the present report, such as nod, nif, fix, hup, and hyp, were previously reported to be involved in nodule formation; however, transcriptional profiling also revealed that A. caulinodans adapts to the endosymbiotic state by uptaking a variety of compounds. The expression patterns of ssu genes and acxABC genes, especially, indicate that A. caulinodans uses a wide variety of compounds for the acquisition of sulfur and carbon sources. The active acquisition and metabolism of sulfur and carbon sources should reflect the higher ability of nitrogen fixation of stem nodules formed by A. caulinodans. Furthermore, the expression patterns of genes involved in EPS biosynthesis indicate that EPS has an extra function in stem nodules. In addition, A. caulinodans has some distinctive features among the rhizobia: the ability to induce stem nodules after crack-entry infection; the ability to fix nitrogen both in the symbiotic and free-living states even at relatively high concentrations of dissolved oxygen (up to 12 μM) (12); and the ability to colonize the xylem of wheat, rice, tomato, and Arabidopsis plants (11, 28, 29, 66, 75, 76). These traits of A. caulinodans make this bacterium one of the most qualified candidates for the application of rhizobia to form nitrogen-fixing nodules on plants other than legumes. The findings of the present study and further studies will accelerate not only the elucidation of the symbiotic mechanism of legumes and rhizobia but also their applications in agriculture in the future.
Supplementary Material
Acknowledgments
This work was supported by the Bio-Oriented Technology Research Advancement Institution (BRAIN) of Japan (T.A. and H.O.) and the Japan Society for the Promotion of Science (no. 20780231) (T.A.).
Footnotes
Published ahead of print on 19 June 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ampe, F., E. Kiss, F. Sabourdy, and J. Batut. 2003. Transcriptome analysis of Sinorhizobium meliloti during symbiosis. Genome Biol. 4:R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, M. J., C. J. Toman, R. F. Fisher, and S. R. Long. 2004. A dual-genome Symbiosis Chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. USA 101:16636-16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, A., S. Rüberg, B. Baumgarth, P. A. Bertram-Drogatz, I. Quester, and A. Pühler. 2002. Regulation of succinoglycan and galactoglucan biosynthesis in Sinorhizobium meliloti. J. Mol. Microbiol. Biotechnol. 4:187-190. [PubMed] [Google Scholar]
- 4.Becker, A., S. Rüberg, H. Küster, A. A. Roxlau, M. Keller, T. Ivashina, H. P. Cheng, G. C. Walker, and A. Pühler. 1997. The 32-kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J. Bacteriol. 179:1375-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergès, H., E. Lauber, C. Liebe, J. Batut, D. Kahn, F. J. de Bruijn, and F. Ampe. 2003. Development of Sinorhizobium meliloti pilot macroarrays for transcriptome analysis. Appl. Environ. Microbiol. 69:1214-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 7.Biers, E. J., K. Wang, C. Pennington, R. Belas, F. Chen, and M. A. Moran. 2008. Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl. Environ. Microbiol. 74:2933-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broughton, W. J., S. Jabbouri, and X. Perret. 2000. Keys to symbiotic harmony. J. Bacteriol. 182:5641-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capela, D., C. Filipe, C. Bobik, J. Batut, and C. Bruand. 2006. Sinorhizobium meliloti differentiation during symbiosis with alfalfa: a transcriptomic dissection. Mol. Plant-Microbe Interact. 19:363-372. [DOI] [PubMed] [Google Scholar]
- 10.Chang, W. S., W. L. Franck, E. Cytryn, S. Jeong, T. Joshi, D. W. Emerich, M. J. Sadowsky, D. Xu, and G. Stacey. 2007. An oligonucleotide microarray resource for transcriptional profiling of Bradyrhizobium japonicum. Mol. Plant-Microbe Interact. 20:1298-1307. [DOI] [PubMed] [Google Scholar]
- 11.Cocking, E. C. 2001. Xylem colonization of tomato by Azorhizobium caulinodans ORS571. Acta Biol. Hung. 52:189-194. [DOI] [PubMed] [Google Scholar]
- 12.de Bruijn, F. J. 1989. The unusual symbiosis between the diazotrophic stem-nodulating bacterium Azorhizobium caulinodans ORS571 and its host, the tropical legume Sesbania rostrata, p. 457-504. In T. Kosuge and E. W. Nester (ed.), Plant-microbe interactions, molecular and genetic perspectives, vol. 3. McGraw-Hill Publishing Co., New York, NY. [Google Scholar]
- 13.D'Haeze, W., R. De Rycke, R. Mathis, S. Goormachtig, S. Pagnotta, C. Verplancke, W. Capoen, and M. Holsters. 2003. Reactive oxygen species and ethylene play a positive role in lateral root base nodulation of a semiaquatic legume. Proc. Natl. Acad. Sci. USA 100:11789-11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Haeze, W., M. Gao, R. De Rycke, M. Van Montagu, G. Engler, and M. Holsters. 1998. Roles for Azorhizobial Nod factors and surface polysaccharides in intercellular invasion and nodule penetration, respectively. Mol. Plant-Microbe Interact. 11:999-1008. [Google Scholar]
- 15.D'Haeze, W., J. Glushka, R. De Rycke, M. Holsters, and R. W. Carlson. 2004. Structural characterization of extracellular polysaccharides of Azorhizobium caulinodans and importance for nodule initiation on Sesbania rostrata. Mol. Microbiol. 52:485-500. [DOI] [PubMed] [Google Scholar]
- 16.D'Haeze, W., and M. Holsters. 2002. Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 12:79R-105R. [DOI] [PubMed] [Google Scholar]
- 17.Donald, R. G., and R. A. Ludwig. 1984. Rhizobium sp. strain ORS571 ammonium assimilation and nitrogen fixation. J. Bacteriol. 158:1144-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dreyfus, B., J. L. Garcia, and M. Gillis. 1988. Characterization of Azorhizobium cauhodans gen. nov., sp. nov., a stem-nodulating nitrogen-fixing bacterium isolated from Sesbania rostrata. Int. J. Syst. Bacteriol. 38:1988. [Google Scholar]
- 19.Dreyfus, B. L., and Y. R. Dommergues. 1981. Nitrogen-fixing nodules induced by Rhizobium on the stem of the tropical legume Sesbania rostrata. FEMS Microbiol. Lett. 10:313-317. [Google Scholar]
- 20.Dreyfus, B. L., C. Elmerich, and Y. R. Dommergues. 1983. Free-living Rhizobium strain able to grow on N2 as the sole nitrogen source. Appl. Environ. Microbiol. 45:711-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Driscoll, B. T., and T. M. Finan. 1993. NAD+-dependent malic enzyme of Rhizobium meliloti is required for symbiotic nitrogen fixation. Mol. Microbiol. 7:865-873. [DOI] [PubMed] [Google Scholar]
- 22.Driscoll, B. T., and T. M. Finan. 1996. NADP+-dependent malic enzyme of Rhizobium meliloti. J. Bacteriol. 178:2224-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelke, T., D. Jording, D. Kapp, and A. Pühler. 1989. Identification and sequence analysis of the Rhizobium meliloti dctA gene encoding the C4-dicarboxylate carrier. J. Bacteriol. 171:5551-5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gage, D. J. 2004. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68:280-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glazebrook, J., and G. C. Walker. 1989. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661-672. [DOI] [PubMed] [Google Scholar]
- 26.Goethals, K., M. Gao, K. Tomekpe, M. Van Montagu, and M. Holsters. 1989. Common nodABC genes in Nod locus 1 of Azorhizobium caulinodans: nucleotide sequence and plant-inducible expression. Mol. Gen. Genet. 219:289-298. [DOI] [PubMed] [Google Scholar]
- 27.Goormachtig, S., W. Capoen, E. K. James, and M. Holsters. 2004. Switch from intracellular to intercellular invasion during water stress-tolerant legume nodulation. Proc. Natl. Acad. Sci. USA 101:6303-6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gopalaswamy, G., S. Kannaiyan, K. J. O'Callaghan, M. R. Davey, and E. C. Cocking. 2000. The xylem of rice (Oryza sativa) is colonized by Azorhizobium caulinodans. Proc. Biol. Sci. 267:103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gough, C., C. Galera, J. Vasse, G. Webster, E. C. Cocking, and J. Dénarié. 1997. Specific flavonoids promote intercellular root colonization of Arabidopsis thaliana by Azorhizobium caulinodans ORS571. Mol. Plant-Microbe Interact. 10:560-570. [DOI] [PubMed] [Google Scholar]
- 30.Guillén-Navarro, K., G. Araíza, A. García-de los Santos, Y. Mora, and M. F. Dunn. 2005. The Rhizobium etli bioMNY operon is involved in biotin transport. FEMS Microbiol. Lett. 250:209-219. [DOI] [PubMed] [Google Scholar]
- 31.Hebbeln, P., D. A. Rodionov, A. Alfandega, and T. Eitinger. 2007. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc. Natl. Acad. Sci. USA 104:2909-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hryniewicz, M., A. Sirko, A. Pałucha, A. Böck, and D. Hulanicka. 1990. Sulfate and thiosulfate transport in Escherichia coli K-12: identification of a gene encoding a novel protein involved in thiosulfate binding. J. Bacteriol. 172:3358-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hummerjohann, J., S. Laudenbach, J. Rétey, T. Leisinger, and M. A. Kertesz. 2000. The sulfur-regulated arylsulfatase gene cluster of Pseudomonas aeruginosa, a new member of the cys regulon. J. Bacteriol. 182:2055-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ifuku, O., H. Miyaoka, N. Koga, J. Kishimoto, S. Haze, Y. Wachi, and M. Kajiwara. 1994. Origin of carbon atoms of biotin. 13C-NMR studies on biotin biosynthesis in Escherichia coli. Eur. J. Biochem. 220:585-591. [DOI] [PubMed] [Google Scholar]
- 35.Jenkins, A. H., G. Schyns, S. Potot, G. Sun, and T. P. Begley. 2007. A new thiamin salvage pathway. Nat. Chem. Biol. 3:492-497. [DOI] [PubMed] [Google Scholar]
- 36.Kadota, K., Y. Nakai, and K. Shimizu. 2008. A weighted average difference method for detecting differentially expressed genes from microarray data. Algorithms Mol. Biol. 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadota, K., Y. Nakai, and K. Shimizu. 2009. Ranking differentially expressed genes from Affymetrix gene expression data: methods with reproducibility, sensitivity, and specificity. Algorithms Mol. Biol. 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kahnert, A., P. Vermeij, C. Wietek, P. James, T. Leisinger, and M. A. Kertesz. 2000. The ssu locus plays a key role in organosulfur metabolism in Pseudomonas putida S-313. J. Bacteriol. 182:2869-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kertesz, M. A. 2000. Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol. Rev. 24:135-175. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi, M., T. Suzuki, T. Fujita, M. Masuda, and S. Shimizu. 1995. Occurrence of enzymes involved in biosynthesis of indole-3-acetic acid from indole-3-acetonitrile in plant-associated bacteria, Agrobacterium and Rhizobium. Proc. Natl. Acad. Sci. USA 92:714-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang, A. S., and J. T. Beatty. 2007. Importance of widespread gene transfer agent genes in α-proteobacteria. Trends Microbiol. 15:54-62. [DOI] [PubMed] [Google Scholar]
- 42.Lang, K., A. Lindemann, F. Hauser, and M. Göttfert. 2008. The genistein stimulon of Bradyrhizobium japonicum. Mol. Genet. Genomics 279:203-211. [DOI] [PubMed] [Google Scholar]
- 43.Lee, K. B., P. De Backer, T. Aono, C. T. Liu, S. Suzuki, T. Suzuki, T. Kaneko, M. Yamada, S. Tabata, D. M. Kupfer, F. Z. Najar, G. B. Wiley, B. Roe, T. T. Binnewies, D. W. Ussery, W. D'Haeze, J. D. Herder, D. Gevers, D. Vereecke, M. Holsters, and H. Oyaizu. 2008. The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genomics 9:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, K. B., C. T. Liu, Y. Anzai, H. Kim, T. Aono, and H. Oyaizu. 2005. The hierarchical system of the “Alphaproteobacteria”: description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 55:1907-1919. [DOI] [PubMed] [Google Scholar]
- 45.Lorite, M. J., J. Tachil, J. Sanjuán, O. Meyer, and E. J. Bedmar. 2000. Carbon monoxide dehydrogenase activity in Bradyrhizobium japonicum. Appl. Environ. Microbiol. 66:1871-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marrs, B. 1974. Genetic recombination in Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. USA 71:971-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mihara, H., M. Maeda, T. Fujii, T. Kurihara, Y. Hata, and N. Esaki. 1999. A nifS-like gene, csdB, encodes an Escherichia coli counterpart of mammalian selenocysteine lyase. Gene cloning, purification, characterization and preliminary x-ray crystallographic studies. J. Biol. Chem. 274:14768-14772. [DOI] [PubMed] [Google Scholar]
- 48.Mörsdorf, G., K. Frunzke, D. Gadkari, and O. Meyer. 1992. Microbial growth on carbon monoxide. Biodegradation 3:61-82. [Google Scholar]
- 49.Oke, V., and S. R. Long. 1999. Bacteroid formation in the Rhizobium-legume symbiosis. Curr. Opin. Microbiol. 2:641-646. [DOI] [PubMed] [Google Scholar]
- 50.Oldroyd, G. E. D., and J. A. Downie. 2004. Calcium, kinases and nodulation signalling in legumes. Nat. Rev. Mol. Cell Biol. 5:566-576. [DOI] [PubMed] [Google Scholar]
- 51.Outten, F. W., M. J. Wood, F. M. Munoz, and G. Storz. 2003. The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J. Biol. Chem. 278:45713-45719. [DOI] [PubMed] [Google Scholar]
- 52.Patzer, S. I., and K. Hantke. 1999. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. J. Bacteriol. 181:3307-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perret, X., C. Staehelin, and W. J. Broughton. 2000. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64:180-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pessi, G., C. H. Ahrens, H. Rehrauer, A. Lindemann, F. Hauser, H. M. Fischer, and H. Hennecke. 2007. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol. Plant-Microbe Interact. 20:1353-1363. [DOI] [PubMed] [Google Scholar]
- 55.Planque, K., I. R. Kennedy, G. E. DeVries, A. Quispel, and A. A. N. Van Brussel. 1977. Location of nitrogenase and ammonia assimilation enzymes in bacteroids of Rhizobium leguminosarum and Rhizobium lupini. J. Gen. Microbiol. 102:95-104. [Google Scholar]
- 56.Roche, P., F. Debellé, F. Maillet, P. Lerouge, C. Faucher, G. Truchet, J. Dénarié, and J. C. Promé. 1991. Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipo-oligosaccharide signals. Cell 67:1131-1143. [DOI] [PubMed] [Google Scholar]
- 57.Rodionov, D. A., A. A. Mironov, and M. S. Gelfand. 2002. Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res. 12:1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2002. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J. Biol. Chem. 277:48949-48959. [DOI] [PubMed] [Google Scholar]
- 59.Sirko, A., M. Hryniewicz, D. Hulanicka, and A. Böck. 1990. Sulfate and thiosulfate transport in Escherichia coli K-12: nucleotide sequence and expression of the cysTWAM gene cluster. J. Bacteriol. 172:3351-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sirko, A., M. Zatyka, E. Sadowy, and D. Hulanicka. 1995. Sulfate and thiosulfate transport in Escherichia coli K-12: evidence for a functional overlapping of sulfate- and thiosulfate-binding proteins. J. Bacteriol. 177:4134-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skorupska, A., M. Janczarek, M. Marczak, A. Mazur, and J. Król. 2006. Rhizobial exopolysaccharides: genetic control and symbiotic functions. Microb. Cell Factories 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sluis, M. K., and S. A. Ensign. 1997. Purification and characterization of acetone carboxylase from Xanthobacter strain Py2. Proc. Natl. Acad. Sci. USA 94:8456-8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sluis, M. K., R. A. Larsen, J. G. Krum, R. Anderson, W. W. Metcalf, and S. A. Ensign. 2002. Biochemical, molecular, and genetic analyses of the acetone carboxylases from Xanthobacter autotrophicus strain Py2 and Rhodobacter capsulatus strain B10. J. Bacteriol. 184:2969-2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Staehelin, C., L. S. Forsberg, W. D'Haeze, M. Y. Gao, R. W. Carlson, Z. P. Xie, B. J. Pellock, K. M. Jones, G. C. Walker, W. R. Streit, and W. J. Broughton. 2006. Exo-oligosaccharides of Rhizobium sp. strain NGR234 are required for symbiosis with various legumes. J. Bacteriol. 188:6168-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stok, J. E., and J. De Voss. 2000. Expression, purification, and characterization of BioI: a carbon-carbon bond cleaving cytochrome P450 involved in biotin biosynthesis in Bacillus subtilis. Arch. Biochem. Biophys. 384:351-360. [DOI] [PubMed] [Google Scholar]
- 66.Stone, P. J., K. J. O'Callaghan, M. R. Davey, and E. C. Cocking. 2001. Azorhizobium caulinodans ORS571 colonizes the xylem of Arabidopsis thaliana. Mol. Plant-Microbe Interact. 14:93-97. [DOI] [PubMed] [Google Scholar]
- 67.Suganuma, N., R. Shimokawa, T. Katoh, and T. Nagai. 1993. Presence of acetone and acetoacetate decarboxylase in soybean root-nodules. Soil Sci. Plant Nutr. 39:653-660. [Google Scholar]
- 68.Suzuki, S., T. Aono, K. B. Lee, T. Suzuki, C. T. Liu, H. Miwa, S. Wakao, T. Iki, and H. Oyaizu. 2007. Rhizobial factors required for stem nodule maturation and maintenance in Sesbania rostrata-Azorhizobium caulinodans ORS571 symbiosis. Appl. Environ. Microbiol. 73:6650-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taté, R., A. Riccio, M. Merrick, and E. J. Patriarca. 1998. The Rhizobium etli amtB gene coding for an NH4+ transporter is down-regulated early during bacteroid differentiation. Mol. Plant-Microbe Interact. 11:188-198. [DOI] [PubMed] [Google Scholar]
- 70.Tsien, H. C., B. L. Dreyfus, and E. L. Schmidt. 1983. Initial stages in the morphogenesis of nitrogen-fixing stem nodules of Sesbania rostrata. J. Bacteriol. 156:888-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Uchiumi, T., T. Ohwada, M. Itakura, H. Mitsui, N. Nukui, P. Dawadi, T. Kaneko, S. Tabata, T. Yokoyama, K. Tejima, K. Saeki, H. Omori, M. Hayashi, T. Maekawa, R. Sriprang, Y. Murooka, S. Tajima, K. Simomura, M. Nomura, A. Suzuki, Y. Shimoda, K. Sioya, M. Abe, and K. Minamisawa. 2004. Expression islands clustered on the symbiosis island of the Mesorhizobium loti genome. J. Bacteriol. 186:2439-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van der Ploeg, J. R., N. J. Cummings, T. Leisinger, and I. F. Connerton. 1998. Bacillus subtilis genes for the utilization of sulfur from aliphatic sulfonates. Microbiology 144:2555-2561. [DOI] [PubMed] [Google Scholar]
- 73.van der Ploeg, J. R., R. Iwanicka-Nowicka, T. Bykowski, M. M. Hryniewicz, and T. Leisinger. 1999. The Escherichia coli ssuEADCB gene cluster is required for the utilization of sulfur from aliphatic sulfonates and is regulated by the transcriptional activator Cbl. J. Biol. Chem. 274:29358-29365. [DOI] [PubMed] [Google Scholar]
- 74.van Ginkel, C. G., and J. A. M. Debont. 1986. Isolation and characterization of alkene-utilizing Xanthobacter spp. Arch. Microbiol. 145:403-407. [Google Scholar]
- 75.Webster, G., C. Gough, J. Vasse, C. Batchelor, K. O'Callaghan, S. Kothari, M. Davey, J. Dénarié, and E. Cocking. 1997. Interactions of rhizobia with rice and wheat. Plant Soil 194:115-122. [Google Scholar]
- 76.Webster, G., V. Jain, M. R. Davey, C. Gough, J. Vasse, J. Denarie, and E. C. Cocking. 1998. The flavonoid naringenin stimulates the intercellular colonization of wheat roots by Azorhizobium caulinodans. Plant Cell Environ. 21:373-383. [Google Scholar]
- 77.Yen, H. C., N. T. Hu, and B. L. Marrs. 1979. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J. Mol. Biol. 131:157-168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.