Abstract
An intI-targeted PCR assay was optimized to evaluate the frequency of partial class 2-like integrases relative to putative, environmental IntI elements in clone libraries generated from 17 samples that included various terrestrial, marine, and deep-sea habitats with different exposures to human influence. We identified 169 unique IntI phylotypes (≤98% amino acid identity) relative to themselves and with respect to those previously described. Among these, six variants showed an undescribed, extended, IntI-specific additional domain. A connection between human influence and the dominance of IntI-2-like variants was also observed. IntI phylotypes 80 to 99% identical to class 2 integrases comprised ∼70 to 100% (n = 65 to 87) of the IntI elements detected in samples with a high input of fecal waste, whereas IntI2-like sequences were undetected in undisturbed settings and poorly represented (1 to 10%; n = 40 to 79) in environments with moderate or no recent fecal or anthropogenic impact. Eleven partial IntI2-like sequences lacking the signature ochre 179 codon were found among samples of biosolids and agricultural soil supplemented with swine manure, indicating a wider distribution of potentially functional IntI2 variants than previously reported. To evaluate IntI2 distribution patterns beyond the usual hosts, namely, the Enterobacteriaceae, we coupled PCR assays targeted at intI and 16S rRNA loci to G+C fractionation of total DNA extracted from manured cropland. IntI2-like sequences and 16S rRNA phylotypes related to Firmicutes (Clostridium and Bacillus) and Bacteroidetes (Chitinophaga and Sphingobacterium) dominated a low-G+C fraction (∼40 to 45%), suggesting that these groups could be important IntI2 hosts in manured soil. Moreover, G+G fractionation uncovered an additional set of 36 novel IntI phylotypes (≤98% amino acid identity) undetected in bulk DNA and revealed the prevalence of potentially functional IntI2 variants in the low-G+C fraction.
Integrons are genetic modules described in pathogenic and commensal bacteria that confer the ability to capture and express promoterless DNA units, called gene cassettes, which encode a variety of adaptive functions including antibiotic resistance (9, 42, 64). The acquisition of gene cassettes occurs through a site-specific recombination mechanism catalyzed by an integron-encoded integrase (IntI). The integrative recombination reaction occurs primarily between an integron receptor site (attI) and a cassette-associated sequence known as the attC site or 59-base element (11). However, integron integrases are able to recognize and process nonspecific secondary targets as well as attI and attC sites with a high degree of sequence variation (20, 25). This versatility facilitates the exchange of exogenous genes between different integrons through various recombination reactions (attI × attC, attI × attI, and attC × attC) that propel the adaptability and evolution of bacterial genomes (8, 11, 31, 38, 55, 58). Although integrons can be chromosomally encoded, they also may be horizontally transferred via transduction or by transposons associated with conjugative plasmids (42, 61). Three major groups (classes 1 to 3) are known to be associated with laterally transferred elements and highly prevalent in the clinical scene. In most of the cases, these have also been reported to harbor almost exclusively gene cassettes encoding antibiotic resistance functions (42). All together, these traits have led to their designation as “mobile” (9) or “clinical” (22) integrons. Although integrons have been traditionally classified according to the percent identity of the nucleotide or predicted amino acid sequence of their respective intI genes (9, 43, 71), several structural features and differences in abundance patterns have been identified which distinguish classes 1 to 3 (9, 42).
Class 1 integrons are the most widely studied variant and are typically linked to replicative Tn21 transposons, which appears to contribute to their extensive distribution (48). A key feature commonly reported within the class 1 module is the presence of a highly conserved 3′ region comprised of a qacEΔ gene and a sul1 gene, which provide protection against quaternary ammonium compounds and sulfa drugs, respectively. In contrast, class 2 integrons are routinely associated with nonreplicative Tn7 transposons, are less frequently detected and, hence, remain an understudied group relative to their class 1 counterparts (42, 48, 65). Even less is known about the class 3 variants, which so far have been described in only three instances (71).
Except for the identical IntI2 elements recently reported in Providencia stuartii and Escherichia coli strains isolated from beef cattle sources and the human urinary tract, respectively, all known integrases encoded by class 2 integrons are considered nonfunctional due to the presence of the ochre 179 codon (6, 40, 42). Nevertheless, it has been argued that integrons with truncated class 2 integrases might be implicated in the transfer and high prevalence of antibiotic resistance genes among clinical isolates, possibly via the in trans activities of other functional integrases or the suppression of the stop codon (27). So far, class 2 integrons have been described in association with isolates affiliated to the gamma, beta, and epsilon subdivisions of the Proteobacteria but have been more frequently reported among members of the Gammaproteobacteria group, particularly the Enterobacteriaceae (1, 14, 19, 56, 57). However, most of these studies have focused on easily culturable, aerobic bacteria or those of clinical importance, leading to the exclusion of unculturable or difficult-to-grow commensals that could be inconspicuous but important reservoirs of class 2 elements in the environment. Although the occurrence and quantification of integrons and integron-associated genes by means of molecular, culture-independent methods are being increasingly documented outside the clinical scene (18, 22, 28, 48, 49, 51, 65, 70), the estimates of the extant diversity of the integron platform in nature are still rudimentary. Likewise, further work is needed for the identification of environmental hosts of integrons commonly found in clinical strains without the bias associated with culture techniques (48).
In order to provide a comprehensive view of integron integrase variation and prevalence patterns of IntI2 elements in the environment, we PCR amplified partial intI sequences from metagenomic DNA isolated from various terrestrial, marine, and deep-sea habitats exposed to various degrees of anthropogenic or fecal impact. Amplification conditions were optimized to facilitate the assessment of the frequency of IntI2-like sequences relative to that of environmental integron integrases. Additionally, since the guanine-plus-cytosine content of DNA corresponds to taxonomy (68), we coupled G+C fractionation of total DNA (4, 5, 29, 30) with PCR assays targeted at intI and 16S rRNA genes to identify potential, unconventional hosts of class 2 integrons in soil that had received swine manure.
MATERIALS AND METHODS
DNA extraction, PCR, and cloning experiments.
Metagenomic DNA was extracted from 17 environmental samples (Table 1; see also information on the sample sites in the supplemental material) with the FastDNA Spin kit (Qbiogene Inc., Carlsbad, CA). A single set of degenerate primers, hep35 (5′-TGCGGGTYAARGATBTKGATTT-3′) and hep36 (5′-CARCACATGCGTRTARAT-3′), complementary to nucleotide sequences corresponding to the Box I and Box II domains of integron integrases, was used to amplify a fragment of ∼491 bp (69). The amplification of integron-encoded integrases was optimized by conducting annealing temperature gradient experiments in a Robocycler Gradient 96 thermal cycler (Stratagene, La Jolla, CA). Optimization was carried out using three different amounts (5, 20, and 50 ng) of class 1 [E. coli SK1592(pDU202)] and class 2 (E. coli J53.3::Tn7) genomic templates and the following cycling conditions: initial denaturation at 94°C for 2 min followed by 35 cycles of melting at 94°C for 30 s, 1 min of an annealing temperature gradient ranging from 45 to 60°C, 45 s of extension at 72°C, and a final extension step of 7 min at 72°C. Since class 1 integrons are commonly detected among fecal bacteria, after optimization the PCR method was validated for the recovery of IntI2 elements and novel integron integrases in DNA extracted from a recently manured field (AMS4) and an adjacent nonagricultural site (NMS5). Both soils were of the same type (Capac loam). Previously described primers (8F, 5′-AGAGTTTGATCMTGGCTCAG-3′ [23]; 1392R, 5′-ACGGGCGGTGTGTACA-3′ [3]) were used for the amplification of 16S rRNA genes with the following cycling parameters: initial denaturation at 95°C for 3 min followed by 25 cycles of melting at 95°C for 45 s, 45 s of annealing at 57°C, 1 min 30 s of extension at 72°C, and a final extension step of 7 min at 72°C. All PCRs were carried out with the Robocycler Gradient 96 instrument.
TABLE 1.
Abundance and distribution of IntI phylotypes detected in total environmental DNA
| Clone or site code | Site description | No. of successfully sequenced clones | No. of clones encoding putative ORFsb | No. of clones encoding IntI-like ORFs | No. of unique (≤98% identical) IntI-like ORFsc | No. of nonclinical IntI-like clones | No. of ochre 179 IntI2-like clonesd | No. of potentially functional IntI2-like clonesd | No. of IntI1-like clonesd | No. of IntI3-like clonesd |
|---|---|---|---|---|---|---|---|---|---|---|
| BIO | Biosolids from sewage plant, MI | 71 | 6 | 65 | 6 | 1 | 54 | 4 | 0 | 6 |
| CM | Composted swine manure, OH | 83 | 0 | 83 | 4 | 0 | 81 | 0 | 2 | 0 |
| AMS4 | Agricultural soil, 3 days after swine manure application, MI | 68 | 4 | 64 | 7 | 19 | 42 | 3 | 0 | 0 |
| NMS5 | Nonagricultural soil adjacent to AMS4, MI | 90 | 5 | 85 | 5 | 85 | 0 | 0 | 0 | 0 |
| AMS23 | Agricultural soil, 1 wk after manure application, MI | 90 | 3 | 87 | 2 | 0 | 85 | 2 | 0 | 0 |
| AMS25 | AMS23, 4 wks after swine manure application, MI | 83 | 1 | 82 | 4 | 22 | 59 | 1 | 0 | 0 |
| NMS24 | Nonagricultural soil adjacent to AMS23, MI | 28 | 4 | 24 | 2 | 24 | 0 | 0 | 0 | 0 |
| MS8 | Barrow Canyon, Arctic Ocean sediment, 180 ma | 36 | 3 | 33 | 13 | 33 | 0 | 0 | 0 | 0 |
| MS7 | Barrow Canyon, Arctic Ocean sediment, 2,000 ma | 26 | 4 | 22 | 9 | 22 | 0 | 0 | 0 | 0 |
| MS6 | East Hannah Shoal, Arctic Ocean sediment, 160 ma | 25 | 14 | 11 | 4 | 11 | 0 | 0 | 0 | 0 |
| MS3 | Puget Sound, Carr Inlet sediment, Washington State, 84 ma | 47 | 10 | 37 | 13 | 37 | 0 | 0 | 0 | 0 |
| MS4 | Washington continental margin, Pacific Ocean, sediment, 1,000 ma | 15 | 9 | 6 | 3 | 6 | 0 | 0 | 0 | 0 |
| HI | HI, Laupahoehoe, native montane rainforest | 46 | 6 | 40 | 18 | 36 | 4 | 0 | 0 | 0 |
| MONA | PR, Mona Island, dry forest preserve | 86 | 1 | 85 | 20 | 85 | 0 | 0 | 0 | 0 |
| SAF | South Africa, Bien Donne, agricultural soil | 79 | 0 | 79 | 32 | 78 | 1 | 0 | 0 | 0 |
| UR | Uruguay, Progreso, agricultural (maize) soil | 70 | 0 | 70 | 8 | 70 | 0 | 0 | 0 | 0 |
| SLV | Slovenia, Ljubljana area, agricultural peat soil | 50 | 2 | 48 | 22 | 45 | 2 | 1 | 0 | 0 |
Water depth overlying the sediments sampled.
Unrelated to integron integrases.
With the exception of IntI1-, IntI2-, and IntI3-like phylotypes (which were also included), numbers refer to uninterrupted intI ORFs from the same source ≤98% identical relative to themselves and those from previously described IntI elements.
Over 80% identical relative to integrases from clinical strains.
Amplification reaction mixtures contained 19 pmol of each primer, 1× Taq buffer (Promega, Madison, WI), 3.5 mM MgCl2, 1 mM deoxynucleoside triphosphate mix (Invitrogen, Carlsbad, CA), 2.5 μg of bovine serum albumin (Roche, Indianapolis, IN), 0.6 U of Taq polymerase (Promega storage buffer B), and approximately 15 to 50 ng of template DNA in a total reaction volume of 12.5 μl. Approximately 10 to 15 ng of PCR product was ligated into the pCR4-TOPO plasmid vector for subsequent transformation of E. coli cells using the TOPO TA cloning for sequencing kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, with the exception that ligation reactions were conducted for 3 h and the entire transformation mix was spread on LB plates (∼75 μl/plate) containing 50 μg/ml of kanamycin.
Characterization of environmental intI sequences.
Ninety-six clones from each library were randomly chosen and sent to Macrogen Inc. (Gasan-dong, Seoul, Korea) for sequencing analysis. Among the successfully sequenced clones (Table 1) only those encoding an open reading frame (ORF) corresponding to at least 161 amino acids (with the exception of IntI1-, IntI2-, and IntI3-like phylotypes) and with hypothetical protein sequences ≤98% identical among themselves were chosen for further characterization. Sequences were analyzed with the BLAST (2) and the Conserved Domain Search (39) tools and compared against two well-known libraries of profile hidden Markov models (HMMs): the Pfam (7) and the TIGRfam (24) databases. The HMMER software package (16) was used to search the combined databases for the most likely models.
Environmental integron integrases were aligned against matching sequences in the GenBank database using the programs ClustalX (version 1.81) and BioEdit (version 5.0.9) (26, 67). The sequences from IntI1, IntI2, IntI3, and IntI4 (GenBank accession no. AAQ16665, AAT72891, AAO32355, and 99031763, respectively) were used as references for the identification of functional domains and amino acid residues (38, 44, 51). The numbering of the alignments was based on the amino acid sequence of the Vibrio cholerae IntI4 (VchIntI4), as its tridimensional structure is known (38). The amino acid sequences of XerC (GenBank accession no. P0A8P6) and XerD (GenBank accession no. P0A8P8) recombinases were used as an outgroup, since these are representatives of the closest relatives of integron integrases within the family of tyrosine recombinases (12, 44, 52). Comparative amino acid sequence analyses were carried out by constructing consensus neighbor-joining dendrograms using the MEGA software, version 4 (66). The dendrograms were generated using the p-distance model and tested by 1,000 bootstrap replications. All alignment positions containing gaps were excluded from the analysis.
G+C fractionation of metagenomic DNA extracted from manured soil and molecular screening for potential IntI2 hosts.
High-molecular-weight DNA was extracted from three replicate samples (5 g each) retrieved from an agricultural, manured field 4 weeks after manure application (AMS25) (Table 1; see also information on sample sites in the supplemental material). The samples were mixed with 2 g of sterile sand, ground in liquid nitrogen three times, and subsequently incubated for 30 min at 37°C after the addition of 5 ml of lysozyme (5 mg/ml, final concentration). The resulting suspensions were submitted to the sodium dodecyl sulfate-based lysis method described by Zhou et al. (72) except that samples were processed without shaking to minimize DNA shearing. To avoid background absorbance from humic acids usually associated with spectroscopy-based quantification, the extracted DNA was pooled and quantified by gel electrophoresis (0.8% agarose) relative to the 24-kb band of a lambda/HindIII DNA marker (Invitrogen, Carlsbad, CA). An amount corresponding to 50 μg was precipitated and sent to Alimetrics Ltd. (Helsinki, Finland) for G+C fractionation analysis (29, 30). The processed DNA was recovered in 16 fractions representing G+C content increments of ∼5% ranging from 20 to 80%. Two fractions, one corresponding to constituent populations with a G+C content of ∼40 to 45% (F7) and another representative of populations with a G+C content of ∼60 to 65% (F12), were chosen for subsequent PCR analyses. Prior to PCR amplification these fractions were desalted with 10 mM Tris, pH 7, using PD-10 Sephadex columns (GE Healthcare Biosciences Corp., Piscataway, NJ) according to the manufacturer's instructions. To avoid excessive dilution of PCR template, the eluted DNA was collected in 13 aliquots of ∼250 μl each. The DNA in each aliquot was quantified by spectroscopy (the optical density at 260 nm), and ∼15 ng of DNA from the aliquot having the highest DNA concentration was added as template for PCRs.
Small-subunit 16S rRNA and intI genes were PCR amplified, cloned, and sequenced for each fraction in order to evaluate the distribution of class 2 integrases across bacterial populations associated with manured soil. The 16S rRNA sequences were processed and analyzed through the Ribosomal Database Project's (RDP) Pipeline tool (10). The library compare tool was used to evaluate the microbial community composition from both fractions, whereas the sequences were characterized at the phylum level with the RDP-Classifier tool using a bootstrap confidence threshold of 80%. Additionally, the SeqMatch tool was used to estimate the taxonomic distance of the retrieved sequences based on Sab scores. Previous applications of the G+C fractionation method have used Sab scores of >0.95 to identify 16S rRNA phylotypes at the species level relative to type strains, whereas Sab scores ranging from 0.70 to 0.94 have been applied for characterization at the genus level (4, 30). Those sequences that could not be characterized with the SeqMatch tool were described in terms of their percent identity relative to their respective closest matches published in GenBank.
Nucleotide sequence accession numbers.
Representative sequences of IntI detected in total DNA were deposited in GenBank under the following accession numbers: DQ282194 to DQ282201 (biosolids, MI), DQ282202 to DQ282205 (composted manure, OH), DQ282206 to DQ282223 (HI), GQ161094, DQ282224 to DQ282228, and DQ287857 to DQ287863 (manured soils, MI), DQ282229 to DQ282248 (PR), DQ282249 to DQ282261 (marine sediments, Puget Sound, Carr Inlet, WA, 84 m), DQ282262 to DQ282264 (Washington coast site, 1,000 m), DQ282265 to DQ282268 (Arctic marine sediments, East Hanna Shoal site, 160 m), DQ282269 to DQ282277 (Arctic marine sediments, Barrow Canyon site, 2,000 m), DQ282278 to DQ282290 (Arctic marine sediments, Barrow Canyon site, 180 m), DQ282291 to DQ282297 (nonagricultural soil, MI), DQ282298 to DQ282345 (South Africa), DQ282346 to DQ282367 (Slovenia), and DQ282368 to DQ282376 (Uruguay). The sequences of intI and 16S rRNA phylotypes identified in G+C fractions were deposited in GenBank under accession numbers FJ615774 to FJ615799, FJ615800 to FJ615829, FJ615830 to FJ615920, and FJ615921 to FJ616000.
RESULTS AND DISCUSSION
Optimization and validation of the intI-targeted PCR assay.
The PCR method used was optimized to facilitate the detection of class 2 integrases as well as that of novel integrases relative to class 1 elements (see Fig. S1 in the supplemental material). The lowest annealing temperature for the specific amplification of the class 1 template was 56°C, but the yield was poor relative to the amplification of the class 2 template at the same temperature. Amplification at annealing temperatures of <56°C generated unspecific banding patterns that in several instances lacked the targeted fragment. In contrast, discrete amplicons (∼491 bp) were consistently obtained with the class 2 control template when the annealing temperature ranged from 50 to 56°C, while artifacts of >500 bp were observed at temperatures ranging from 45 to 49°C. We thus chose an annealing temperature of 52°C for the recovery of environmental integrases, since it suggested a greater potential for the amplification of novel intI genotypes from community DNA and facilitated the amplification of class 2 elements over class 1 intI (see Fig. S1 in the supplemental material).
Since class 2 integrons have been associated primarily with enteric bacteria, DNA from an agricultural field (AMS4, MI) (Table 1) sampled 3 days after manure application was used to test the PCR method for the recovery of class 2 integrons. In contrast, DNA from an adjacent, nonagricultural field (NMS5, MI) (Table 1) was used to evaluate the efficiency of the PCR assay for the retrieval of novel intI sequences distantly related to those of clinical integrons, since this site had no history of manure application. An amplicon of the expected size (∼491 bp) was cloned from both of these habitats. Analyses of predicted ORFs identified the family of integron-encoded integrases as the closest match for 64 and 85 of the total clones sequenced from sites AMS4 (n = 68) and NMS5 (n = 90), respectively (Table 1). These coded for a protein sequence that was in frame with the third position of the forward primer, which corresponded to a codon for a conserved arginine residue (R143, IntI4 numbering system) present in the Box I region of integron integrases. About 70% of the intI sequences recovered from the manured field (AMS4) had predicted amino acid sequences over 80% identical to that of a class 2 integrase (GenBank accession no. AY639870), while ∼94% of the sequences retrieved from the NMS5 site were 58 to 83% identical to IntI sequences unrelated to clinical integrons previously recovered from environmental, uncultured bacteria (49, 51) or reported in sequenced genomes of commensal species (GenBank accession no. ZP_00299196, ZP_00172930, and NP_842193). The dominance of IntI2-like sequences in the manured field suggested that fecal bacteria could be important reservoirs of class 2 integrons and that large loads of animal waste may increase the abundance of IntI2 elements in fertilized cropland to a threshold level that facilitates their detection relative to soil not exposed to agricultural impact. Besides being appropriate for surveying IntI2 sequences, the primers and amplification conditions used were suitable for detecting novel, “nonclinical” IntI elements, particularly in a setting with no history of fecal input or agricultural use. These findings were further evaluated by applying the same approach to community DNA from biosolids, composted swine manure, soils with different histories of fecal or anthropogenic impact, and marine sediments from the Pacific and Arctic Oceans collected from 84 to 2,000 m below sea level (Table 1; see also the information on sampling sites in the supplemental material).
Characterization of environmental IntI elements.
A threshold of ≤98% identity at the amino acid sequence level was used for the designation of a novel phylotype. Based on this criterion, 169 partial intI genes were detected. All were identified as putative integron integrases on the basis of analyses conducted with the BLAST and the CD-Search tools. These results were verified by comparing each sequence against HMM profiles from the Pfam and TIGRfam databases. For all 169 IntI sequences, the TIGRfam integron integrase family model produced the most statistically significant scores which corresponded to the lowest maximum (4.7 × 10−17), median (2.2 × 10−31), and minimum (4.7 × 10−59) E-values establishing them as members of the integron integrase group.
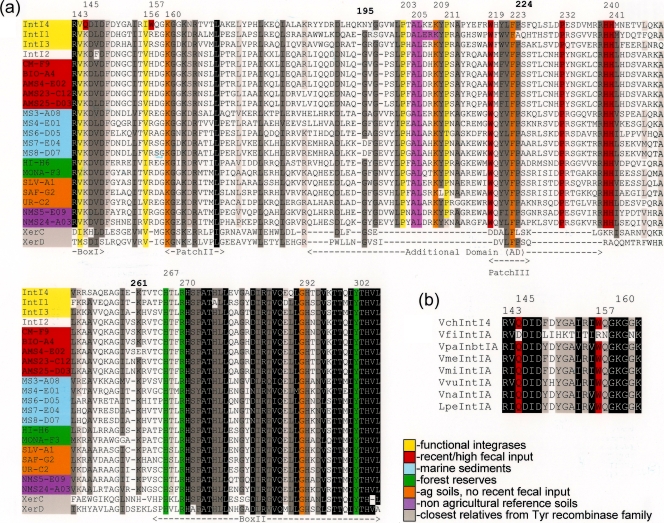
Comparative sequence alignments against functional integron integrases were carried out to further characterize the inferred IntI sequences. Although putative sequences ∼50% of a full-length IntI element were retrieved, the high conservation of motifs and residues critical for binding and recombination activity suggested that (with the exception of ochre 179 IntI2-like elements) most of the recovered integrases might represent novel integrons that could be functional. The IntI-specific additional domain was detected in each of the environmental IntI sequences. The motifs designated as Boxes I and II and Patches II and III, which are characteristic of members of the tyrosine recombinase family, were also found (44, 51, 52) (Fig. 1a). The aligned sequences showed 56 conserved residues; 43 of them were ≥95% identical while the remaining 13 were ≥95% similar. These were distributed mainly across the above-mentioned regions. Furthermore, the following amino acids which are involved in catalytic and DNA-binding functions were detected: K160 (∼99% conserved, Patch II), A205 (∼95% conserved, additional domain), L206 (∼93% conserved, additional domain), K209 (∼99% conserved, additional domain), F223 (∼98% conserved, Patch III), and G292 (∼99% conserved, Box II), as well as H267 (∼99% conserved, Box II), R270 (100% conserved, Box II), and Y302 (100% conserved, Box II) from the signature RHRY tetrad of the tyrosine recombinase family (44, 51, 55).
FIG. 1.
(a) Alignment of partial amino acid sequences of environmental integrases representative of each sampled site against functional integrase classes (1, 3, and 4) and tyrosine recombinases XerC and XerD. Numbering is based on the VchIntI4 sequence (38). The position of conserved motifs among integron-encoded integrases is indicated by segments labeled as Boxes I and II and Patches II and III and as additional domains (AD). The degree of conservation (percent identity) among equivalent residues is indicated as follows: black (100%), dark gray (95%), and light gray (85%). Conserved amino acids (93 to 99% identical) with functions related to protein folding, DNA binding, and recombination activity are highlighted in orange, pink (44), and red (38), respectively. Yellow columns show conserved proline residues (≥85% identical) detected within the AD region and conserved hydrophobic amino acids preceding functional residues exclusive of the VchIntI sequence. Green columns correspond to H, R, and Y residues from the conserved RHRY tetrad, which is characteristic of the entire tyrosine recombinase family (52). Numbers in boldface indicate the position of insertions and substitutions in functional integrases. GenBank accession numbers for the protein sequences of reference integrases (classes 1 to 4) and tyrosine recombinases XerC and XerD are AAQ16665, AAT72891, AAO32355, 99031763, P0A8P6, and P0A8P8, respectively. (b) Partial sequence alignment showing conserved residues (Q145 and W157; red columns) that seem to be exclusive of the Vibrio IntI clade.
The crystal structure of VchIntI4 was recently determined under conditions reminiscent of an attC × attC recombination reaction (38). This model revealed that the VchIntI4-attC assemblage requires binding of four IntI molecules arranged in dimers consisting of an attacking and a nonattacking subunit attached to each attC target. According to this structure, residues Q145 (Box I), W157 (downstream Box I), and W219 (patch III) are implicated in the attachment of the attacking IntI molecules, whereas residues P232 (additional domain), H240, and H241 (downstream of the additional domain) facilitate binding of the nonattacking monomers, generating a stable synaptic complex that undergoes subsequent recombination. This study also reports that K160 (Patch II) creates important base contacts related to cleavage activity.
Sequence alignments also demonstrated that functional residues W219, P232, H240, and H241 were nearly 99, 92, 94, and 98% conserved, respectively, across the environmental IntI sequences (Fig. 1a). However, the presence of the functional residues Q145 and W157 appeared to be exclusive of IntI sequences from the Vibrio clade, as these were replaced across “clinical” and environmental IntI by K (100% conserved) and R (85% conderved) or H (10% conserved) residues, respectively. Relative to the structural model proposed for IntI4, these substitutions suggest the possibility of structural variations within the integrative recombination mechanism among different integrases. The R157 residue was also conserved in class 1 and class 3 integrases, whereas H157 was conserved among class 2-like sequences. Moreover, positions corresponding to Q145 and W157 were both preceded by hydrophobic amino acids (V, L, and I) which were conserved among all the aligned IntI sequences.
Extended additional domain identified in environmental IntI phylotypes.
We also found that six clones (55 to 77% identity) recovered across remote terrestrial locations (HI-G2, MONA-G2, SAF-A8, SAF-F5, and UR-C9) and marine sediments (MS8-F05) possessed a longer (1 to 8 amino acids) integron-specific additional domain relative to previously described IntI elements (see Fig. S2 in the supplemental material). In all instances the insertions were located downstream of the Patch III motif after a 75% conserved glycine residue (position 235 [VchIntI4 numbering]). Insertions and deletions associated with the Patch I and Box I domains, respectively, have been reported in an integrase phylotype detected in total DNA recovered from deep-sea hydrothermal vent fluids (18). To our knowledge, the above-mentioned, extended additional domain is the first description of this characteristic for integron integrases. Whether this trait allows function or represents a disabling insertion is unknown. However, according to the VchIntI4 crystal structure the IntI additional domain contains an α-helix motif that is critical for synapse formation. (38).
Diversity and prevalence patterns of IntI2-like elements and environmental IntI sequences.
Phylogenetic analysis demonstrated that all unique clones identified as integron integrases formed a clade that excluded representatives of the XerC and XerD recombinases, the closest relatives of integron integrases within the tyrosine recombinase family (Fig. 2) (59). This trend was supported by values corresponding to 99% of 1,000 bootstrap replications (Fig. 2). In agreement with previous phylogenetic analyses of integron integrases (59), we found that class 2 and class 3 integrases formed sister groups, whereas the cluster comprised of the Vibrio IntI group along with integrases retrieved from marine sediments were the most distantly related to nearly all other IntI phylotypes, followed by the IntI2-like group (Fig. 2, clusters I, II, III, and G). Mutational hot spots have been identified among intI genes from the Vibrio clade relative to other intI sequences, suggesting that this group of integrases is under positive selection (50), which is also consistent with the clustering of Vibrio IntI elements near the outgroup sequences. In accordance with these previous findings, again, relative to other IntI sequences, we identified conserved, nonsynonymous substitutions among Vibrio integrases at positions corresponding to amino acids implicated in DNA binding functions (Q145 and W157 [VchIntI4 numbering]) (Fig. 1a and b).
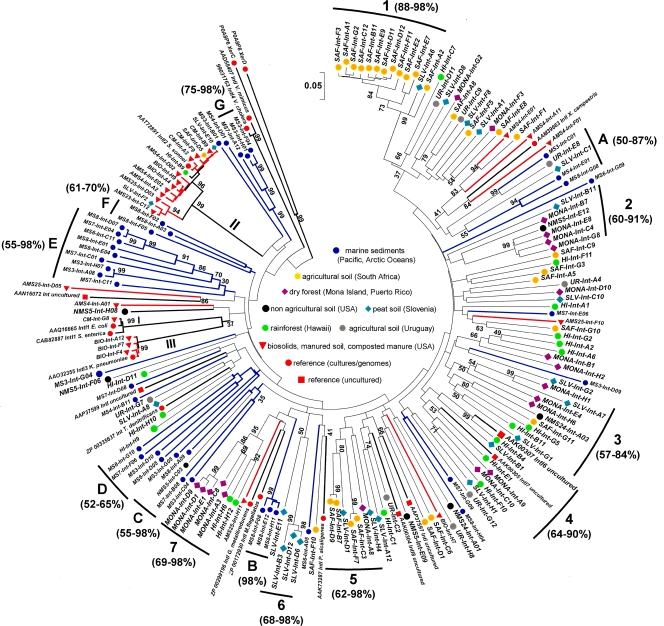
FIG. 2.
Consensus neighbor-joining tree (compared positions for 108 amino acid residues), illustrating the relationship of representative IntI sequences recovered from a variety of marine and terrestrial habitats. Values represent the percentage of 1,000 bootstrap replications that supported the branching order. Roman numbers (I to III) designate clusters containing “clinical” integrases (classes 1 to 3). Red branches point out the arrangement of IntI sequences detected in environments with high fecal impact, while thicker black branches highlight the position of outgroup sequences (XerC and XerD recombinases) and reference sequences (best database matches). Numbers 1 to 7 denote clusters dominated by at least three soil IntI phylotypes originating from the same sample, whereas letters (A to G) indicate the location of clusters dominated by IntI phylotypes recovered from marine and deep-sea sediments (blue branches). The percent identities of IntI sequences associated with the highlighted clusters are shown in parentheses. Values of bootstrap support below 30% are not shown. GenBank accession numbers follow the designation of the reference and outgroup sequences.
In agreement with optimization and validation of the intI-targeted PCR assay, IntI sequences closely related to that of class 2 integrons were dominant in libraries generated from samples with high fecal impact from human and animal sources (biosolids, composted swine manure, and recently manured [swine] soils) (Table 1). Nearly 98% of the predicted amino acid sequences recovered from composted swine manure (n = 83) and all of those retrieved from soil sampled after 1 week of manure application (AMS23; n = 87) were 80 to 100% identical to class 2 integrases. Likewise, about 89% of the sequences retrieved from biosolids (n = 65) and ∼73% of those from soil sampled after 1 month of manure application (AMS25; n = 82) were 80 to 99% identical to class 2 IntIs. Among this particular set of samples only two class 1 IntIs were detected in composted manure, while six IntI3 phylotypes were identified in biosolids, confirming that PCR conditions favored the detection of IntI2 among clinical integrons. Interestingly, four sequences ∼97 to 98% identical to IntI2 which lacked the distinctive internal stop codon of this integrase type (ochre 179) were detected in biosolids (clones BIO-Int-A1, A4, A10, and H9), while two were recovered from cropland fertilized with swine manure (AMS4-Int-A2 and E02 [see Fig. S3 in the supplemental material]). Additionally, five IntI sequences 89 to 95% identical to IntI2 (AMS23-Int-C12, E01, AMS25-Int-D03, SLV-Int-F2, and AMS4-Int-D02) also lacked the ochre 179 codon and were detected in manured agricultural fields (see Fig. S3 in the supplemental material).
Mutagenesis studies have proved that the replacement of the internal TAA stop codon by a GAG triplet coding for a hydrophilic, negatively charged Glu (the equivalent residue in IntI1) restores recombination activity (27). In contrast, the full-length IntI2 from Providencia stuartii and E. coli as well as the IntI2-like sequences described herein share a hydrophilic, neutral Gln residue at the ochre 179 position which is encoded by a CAA or a CAG triplet, indicating that functional IntI2s are more common and widely distributed than previously assessed with culture techniques (6, 40). Furthermore, the detection of a potentially functional IntI2-like genotype (AMS25-Int-D03) at the agricultural field AMS25 (Table 1), as well as the prevalence of class 2 elements at this site (∼73% of the retrieved sequences), suggested that fecal bacteria serving as IntI2 hosts were persistent under field conditions after 1 month of manure application. Nevertheless, only three IntI2-like clones were also detected in an agricultural soil from Slovenia (Table 1) that was sampled 3 years after animal waste application, suggesting that the dominance of class 2 integrases is a transient effect in manure-supplemented cropland. Similarly, one and four IntI2-like sequences were recovered from an agricultural field in South Africa and a forest preserve in Hawaii, respectively (Table 1). The presence of these IntI2 elements could be attributed to background levels or the use of animal manures at the South Africa site. As for the Hawaiian sample, which was collected at Laupahoehoe National Forest (54), it is also possible that intestinal strains with class 2 elements are being dispersed by feral pigs, as their local populations are large enough to turn over the soil surface on a 3-year average (P. Vitosuek, Stanford University, personal communication). Hence, it is possible that large numbers of wild pigs may also influence the spread and persistence of IntI2 through the fecal-oral route independent of human activity.
Except for clones SLV-Int-E10, SLV-Int-F2, SAF-Int-D5, and HI-Int-B9 (Fig. 2, cluster II) the screening of undisturbed habitats and those with no recent fecal impact allowed the detection of a varied set of novel integrases unrelated to those from clinical strains (39 to 75% identical), suggesting that fecal environments select for a much more restricted set of integrons (Fig. 2 and Table 1; see also the information on the sites provided in the supplemental material). The largest numbers of unique IntI sequences were recovered from soil samples collected in South Africa, Slovenia, Mona Island, and Hawaii and corresponded to 32, 22, 20, and 18 phylotypes, respectively. These totals were greater than that reported by a previous integrase survey which detected 15 phylotypes representing ∼82% of the predicted intI genes from a mine tailing sample (49). However, as few as two to five unique phylotypes were recovered from nonagricultural soil in Michigan (NMS24 and NMS5), whereas eight were detected in agricultural soil from Uruguay. Defined patterns of phylogenetic clustering were not prevalent among the above-mentioned soil integrases, reflecting the broad range of amino acid identity observed in these sequences. In only a few instances, some representative, soil IntI from Mona Island, Hawaii, Slovenia, and South Africa formed small, distantly related groups dominated by at least three phylotypes of common origin (Fig. 2, clusters 1 to 7). Remarkably, 12 of the clones recovered from South Africa grouped at the most distant branch relative to the XerC/XerD outgroup (Fig. 2, cluster 1).
Forty-two integron integrases were retrieved across marine and deep-sea sediments from the Pacific and Arctic Oceans. The sequence identity among them ranged from 43 to 98%. Thirteen phylotypes were recovered from the Puget Sound site (Carr Inlet, WA) at a depth of 84 m (MS3 series), while three were obtained from the Washington continental margin under a water depth of 1,000 m (MS4 series). Thirteen and nine phylotypes were detected at the Barrow Canyon site (Arctic Ocean) under water depths of 180 (MS8 series) and 2,000 m (MS7 series), respectively. Additionally, four IntI types were retrieved from the East Hanna Shoal station (Arctic Ocean) from a water depth of 160 m (MS6 series). Phylogenetic analysis revealed that marine IntIs were distributed mainly across distantly related groups dominated by sequences from oceanic and deep-sea sediments (Fig. 2, clusters A to G). Integrons have been described in culturable marine bacteria associated with the water column (1, 59) as well as in endosymbionts of mussels and fluids recovered from deep-sea hydrothermal vent sites (18). However, each of the marine and deep-sea phylotypes that we detected probably represent a new integron class, since they were ≤67% identical with respect to previously described IntI sequences. Relative to fecal and soil-related environments the numbers of successfully sequenced clones obtained from sediments were lower, presumably due to a low yield of PCR products and the instability of the resulting transformants during propagation. Nevertheless, we consider that class 2 integrases were not detected in sediments due to the lack of significant anthropogenic impact, as we have confirmed the association of intI2 prevalence with human influence by using intI2-specific primers in other samples (unpublished results).
Finally, intI pseudogenes (as judged by the presence of premature stop codons) distantly related to “clinical” integrases were sequenced in several instances from both soil and marine environments and indicated the existence of a widespread pool of nonfunctional intI genotypes in nature. Nearly one-third of the intI genes detected in a recent survey that included 103 bacterial genomes were pseudogenes (50), suggesting, as argued earlier (21), that selection in favor of nonfunctional integrases could be beneficial, since cassettes encoding traits critical for fitness under specific scenarios are less likely to be lost through the inactivation of the native IntI.
Application of the G+C fractionation technique for the detection of potential IntI2 reservoirs.
DNA from a manured soil (AMS25) sampled 4 weeks after manure application was fractionated based on its G+C content to reduce the complexity of the microbial community and facilitate the evaluation of patterns of IntI sequences relative to the bacterial community composition. We chose fraction F7 (∼40 to 45% G+C) and fraction F12 (∼60 to 65% G+C) for three reasons: (i) most of the soil-extracted DNA was in the range of 40 to 65% G+C; (ii) they provided a greater potential for separating distinct bacterial groups, as the G+C content of dominant intestinal bacteria with fermentative or anaerobic metabolism typically ranges from 30 to 45% while that of aerobic, heterotrophic species prevalent in soil has been reported to be over 60% (5, 29, 53, 54); and (iii) our choice of fractions was also promising for excluding common carriers of class 2 integrons such as the enterics, as several of the key representatives from this group have a G+C content ranging from 46 to 60% (5). Therefore, we hypothesized that this culture-independent approach would facilitate the detection of potential, inconspicuous reservoirs of class 2 elements.
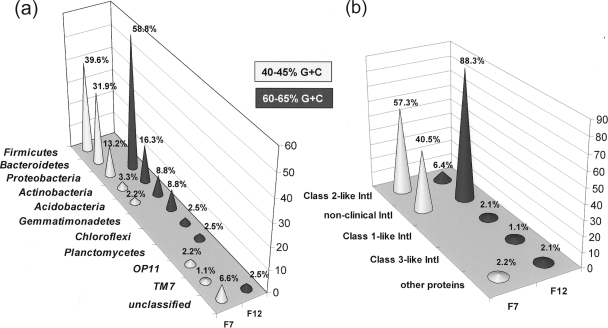
The method of G+C fractionation was capable of reducing the complexity of the analyzed metagenome and allowed us to detect patterns in the abundance of IntI phylotypes in manured soil with respect to the community structure of each individual fraction. Analysis of clone libraries of 16S rRNA genes revealed that both fractions had different community structures comprised by unique phylotypes (≤97% identical). According to the RDP-II Classifier tool, the high G+C fraction (F12) showed a community structure consistent with that typically described in soil (32), as it was dominated by representatives from the Proteobacteria (58.8%; n = 81) Actinobacteria (16.3%), and Acidobacteria (8.8%) (Fig. 3a). In contrast, sequences related to the Firmicutes (39.6%; n = 91) and Bacteroidetes (31.9%) comprised the most prevalent lineages detected in the low-G+C fraction (F7). Remarkably, Firmicutes-like phylotypes very similar (Sab scores, 0.94 to 0.99) to those found in intestinal or fecal niches (Clostridia and Bacilli) (13, 33, 34, 35, 37, 63) not only were exclusive of this fraction but their relative frequency (∼21%) was nearly 10 times higher than that of “fecal” Rhodococcus phylotypes detected in fraction F12 (∼2%), indicating a higher prevalence of fecal bacteria in fraction F7 (Fig. 3a; see also Tables S1 and S2 in the supplemental material). These data confirmed that fecal porcine species were persistent in manured soil after a month of manure incorporation. Neither fraction showed the presence of phylotypes related to the Enterobacteriaceae.
FIG. 3.
Relative frequency distribution of 16S rRNA (a) and IntI phylotypes (b) in DNA fractions corresponding to 40 to 45% and 60 to 65% G+C contents, recovered from an agricultural soil from Michigan (AMS25) sampled after 1 month of manure application.
Total DNA fractionation also revealed that the relative abundance of IntI2-like elements was nearly 9 times higher in fraction F7 (∼57%; n = 91) than in fraction F12 (∼6.4%; n = 96), supporting the notion of Firmicutes and Bacteroidetes as potential sources of class 2 integrases (Fig. 3b). Although this study does not provide experimental evidence in support of an absolute linkage between these two phyla and class 2 integrases, results from G+C experiments and from our culture-independent survey of IntI2-like variants are in agreement with the prevalence of class 1 integrons in Firmicutes from fecal environments (48) as well as with the recent description in public databases (47) of a class 2 integron detected in a Firmicutes (Aerococcus viridans) isolated from agricultural slurry from tylosin-fed pigs (K. G. Byrne-Bailey et al., unpublished data; GenBank accession no. FJ474094).
Clones similar to aerobic, soil-borne Bacteroidetes were the second most prevalent group in the low-G+C fraction, suggesting a role as potential IntI2 hosts (Fig. 3). However, to our knowledge “clinical integrons” have not been described among environmental Bacteroidetes, and information on the distribution of class 2 IntIs among clinical representatives is limited. In a recent study, classes 1 to 3 were not detected among clinical Bacteroides species (n = 69), whereas these were common among medically important isolates of the Gammaproteobacteria (15). Class 2 integrons have been reported among members of the Gammaproteobacteria, particularly within representatives from the orders Enterobacteriales, Pseudomonadales, Aeromonadales, and Vibrionales (9, 46). Nevertheless, only a single clone similar (Sab score, 0.92) to an uncultured, unclassified phylotype related to the Pseudomonadaceae was detected in fraction F7, corroborating that the G+C fractionation was effective in excluding canonical IntI2 hosts (see Table S1 in the supplemental material).
The detection of potentially functional IntI2 phylotypes was also enhanced by the resolving capacity of the G+C fractionation technique. Twenty-three IntI2-like phylotypes lacking the ochre 179 codon were identified in the low-G+C fraction (IntI-F7 series), revealing an unsuspected higher frequency of these variants relative to bulk DNA from the AMS25 site and to that from any other sample where IntI2 elements were prevalent (agricultural soils and biosolids, one to four IntI2 phylotypes) (see Fig. S3 in the supplemental material). Within this group, clone IntI-F7-E8 showed a proline residue at position 179, whereas two others had unique, premature termination codons at positions 214 (IntI-F7-C11) and 280 (IntI-F7-E5), which are located at the additional and Box II domains of integron integrases, respectively (see Fig. S4 in the supplemental material). A total of six unique (77 to 98% identical among themselves) potentially functional IntI2-like sequences were identified (IntI-F7-C11, D12, E6, E8, G6, and H5). Additionally, five unique phylotypes distantly related to “clinical” integrases were found, which represented ∼41% of the total sequences (see Fig. S5 in the supplemental material). Class 1 and class 3-like elements were not detected in fraction F7.
Likewise, an additional set of 25 unique phylotypes (45 to 98% identical) were identified in the high-G+C fraction which were not detected in total DNA from any of the other samples tested (see Fig. S5 in the supplemental material). These were also different compared to previously described integrases. Only two “nonclinical,” IntI phylotypes from fraction F12 were closely related (88 to 99% identical) to those from the F7 fraction (see Fig. S5 in the supplemental material). The functional residues K160, A205, K209, F223, H267, Y302 (100% conserved) R270, G292 (∼97% conserved), and L206 (∼94% conserved) were also identified among these new sequences. As observed earlier, K and R residues were 100 and 97% conserved at positions corresponding to the functional amino acids Q145 and W157 of the VchIntI4 sequence. Six of the IntI variants recovered from fraction F12 were closely related (88 to 98% identical) to class 2 integrases, and four of them also lacked the ochre 179 codon. These corresponded to three IntI2-like phylotypes (IntI-F12-C4, E7, and F12) 90 to 99% identical relative to themselves and over 99% identical to IntI2-like sequences found in the low-G+C fraction. Only three clones were found to be 72 to 73% identical to class 1 and class 3 integrases, while two were distant matches to protein sequences unrelated to integron integrases. The recovery of higher numbers of new IntI elements, particularly those different from “clinical” IntIs, emphasizes the value of the G+C fractionation process for the detection of novel IntI phylotypes swamped by those dominant in bulk DNA.
Novel integron integrases and class 2 integrases in the environment.
Overall, our extensive environmental survey of IntI elements uncovered 205 partial sequences of novel and distinct IntI phylotypes (≤98% identical). These corresponded to 169 detected in bulk DNA from different terrestrial and marine ecosystems, while 11 and 25, respectively, were found in total DNA fractions with ∼40 to 45 and ∼60 to 65% G+C contents. Here we have documented the first report of IntI elements in marine and deep-sea sediments from the Arctic Ocean and the Washington continental margin, as well as in soil-related environments from the northeast area of the Caribbean (Mona Island, PR). The recovery of such a widespread and diverse set of integron integrases from natural environments, in conjunction with previous reports on the existence of varied gene cassette pools across different habitats and the versatility of the recombination function of integron integrases, reinforces the notion of the ubiquitous distribution of the integron module and the evolutionary significance of this gene capture and expression system (20, 45, 49, 51). Additionally, two important findings arising from the comparative analysis of our environmental sequences with respect to that of functional integrases were (i) the presence of a previously undescribed insertion at the IntI-specific additional domain and (ii) the lack of conservation of VchIntI4-specific, functional residues (Q145 and W157) among environmental integrases and those from clinical strains. The former feature may have functional implications, as this region is involved in synapse formation during the integrative recombination reaction (38) while the latter suggests potential alternative structural arrangements to those described for VchIntI4.
The connection between the presence of IntI2-like elements in environments impacted by fecal waste from animals, including humans, allows us to suggest that class 2 integrons may have been extracted from the extant environmental pool, presumably through the food chain and in the absence of antibiotic selection, and then laterally transferred and enriched in the gastrointestinal tract. These events seem feasible, as the ability of exogenous bacteria to survive transit in the human and animal gut environment and the occurrence of bacterial, lateral gene transfer in the intestinal setting are well documented (17, 36, 41, 60, 62). Collectively, the results from our global, molecular survey and G+C experiments also present new insight into the prevalence and dispersal of potentially functional IntI2-like phylotypes, which showed an unsuspected wider distribution and diversity than those previously described with culture-based techniques (6, 40). Furthermore, the use of the G+C fractionation method allowed inferences of dispersal of class 2 intI genes in potential hosts that have not been identified before, which may guide future isolation efforts of IntI2 carriers. Hence, risks to human health rising from the manipulation of biosolids and animal manures and the possible dissemination of inconspicuous functional variants of the IntI2 platform as well as antibiotic resistance determinants encoded by integrons from fecal niches should be evaluated from a broader perspective.
Supplementary Material
Acknowledgments
This work was supported by the Reservoirs of Antibiotic Resistance Network under support from NIAID, by MSU's Emerging Initiatives on Pharmaceuticals in the Environment, and by the Center for Microbial Ecology.
We thank those who provided the samples and the site information: Lee Jacobs, Verónica Grüntzig, Allan Devoll, Alban Ramette, Klaus Nüsslein, Blaz Stres, Enríque Estramil, and Lucienne E. Mansvelt. We also thank Konstantinos Konstantinidis for assistance with high-throughput analysis of sequences and Anne O. Summers for control strains used in PCR experiments.
Footnotes
Published ahead of print on 5 June 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahmed, A. M., F. Kawaguchi, and T. Shimamoto. 2006. Class 2 integrons in Vibrio cholerae. J. Med. Microbiol. 55:643-644. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amman, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apajalahti, J. H. A., H. Kettunen, A. Kettunen, W. E. Holben, P. H. Nurminen, N. Rautonen, and M. Mutanen. 2002. Culture-independent microbial community analysis reveals that inulin in the diet primarily affects previously unknown bacteria in the mouse cecum. Appl. Environ. Microbiol. 68:4986-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apajalahti, J. H., L. K. Särkilahti, B. R. Mäki, J. P. Heikkinen, P. H. Nurminen, and W. E. Holben. 1998. Effective recovery of bacterial DNA and percent-guanine-plus-cytosine-based analysis of community structure in the gastrointestinal tract of broiler chickens. Appl. Environ. Microbiol. 64:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barlow, R. S., and K. S. Gobius. 2006. Diverse class 2 integrons in bacteria from beef cattle sources. J. Antimicrob. Chemother. 58:1133-1138. [DOI] [PubMed] [Google Scholar]
- 7.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biskri, L., M. Bouvier, A.-M. Guérout, S. Boisnard, and D. Mazel. 2005. Comparative study of class 1 integron and Vibrio cholerae superintegron integrase activities. J. Bacteriol. 187:1740-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boucher, Y., M. Labbate, J. E. Koenig, and H. W. Stokes. 2007. Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol. 15:301-309. [DOI] [PubMed] [Google Scholar]
- 10.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collis, C. M., G. D. Recchia, M.-J. Kim, H. W. Stokes, and R. M. Hall. 2001. Efficiency of recombination reactions catalyzed by class 1 integron integrase IntI1. J. Bacteriol. 183:2535-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornet, F., B. Hallet, and D. J. Sherratt. 1997. Xer recombination in Escherichia coli. Site-specific DNA topoisomerase activity of the XerC and XerD recombinases. J. Biol. Chem. 272:21927-21931. [DOI] [PubMed] [Google Scholar]
- 13.Cotta, M. A., T. R. Whitehead, and R. L. Zeltwanger. 2003. Isolation, characterization and comparison of bacteria from swine faeces and manure storage pits. Environ. Microbiol. 5:737-745. [DOI] [PubMed] [Google Scholar]
- 14.Crespo, O., M. Catalano, S. Piñeiro, M. Matteo, A. Leanza, and D. Centrón. 2005. Tn7 distribution in Helicobacter pylori: a selective paradox. Int. J. Antimicrob. Agents 25:341-344. [DOI] [PubMed] [Google Scholar]
- 15.Dillon, B., L. Thomas, G. Mohmand, A. Zelynski, and J. Iredell. 2005. Multiplex PCR for screening of integrons in bacterial lysates. J. Microbiol. Methods 62:221-232. [DOI] [PubMed] [Google Scholar]
- 16.Eddy, S. 1998. HMMER user's guide: biological sequence analysis using profile hidden Markov models. http://www.csb.yale.edu/userguides/seq/hmmer/docs/index.html.
- 17.Elli, M., M. L. Callegari, S. Ferrari, E. Bessi, D. Cattivelli, S. Soldi, L. Morelli, N. Goupil Feuillerat, and J. M. Antoine. 2006. Survival of yogurt bacteria in the human gut. Appl. Environ. Microbiol. 72:5113-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elsaied, H., H. W., Stokes, T. Nakamura, K. Kitamura, H. Fuse, and A. Maruyama. 2007. Novel and diverse integron integrase genes and integron-like gene cassettes are prevalent in deep-sea hydrothermal vents. Environ. Microbiol. 9:2298-2312. [DOI] [PubMed] [Google Scholar]
- 19.Fluit, A. C., and F. J. Schmitz. 2004. Resistance integrons and super-integrons. Clin. Microbiol. Infect. 10:272-288. [DOI] [PubMed] [Google Scholar]
- 20.Francia, M. V., F. de la Cruz, and J. M. García Lobo. 1993. Secondary-sites for integration mediated by the Tn21 integrase. Mol. Microbiol. 10:823-828. [DOI] [PubMed] [Google Scholar]
- 21.Gillings, M. R., M. P. Holley, H. W. Stokes, and A. J. Holmes. 2005. Integrons in Xanthomonas: a source of species genome diversity. Proc. Natl. Acad. Sci. USA 102:4419-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillings, M. R., S. Krishnan, P. J. Worden, and S. A. Hardwick. 2008. Recovery of diverse genes for class 1 integron-integrases from environmental DNA samples. FEMS Microbiol. Lett. 287:56-62. [DOI] [PubMed] [Google Scholar]
- 23.Giovannoni, S. J. 1991. The polymerase chain reaction, p. 177-201. In E. Stackenbrandt (ed.), Nucleic acids techniques in bacterial systematics. John Wiley & Sons, New York, NY.
- 24.Haft, D. H., B. J. Loftus, D. L. Richardson, F. Yang, J. A. Eisen, I. T. Paulsen, and O. White. 2001. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res. 29:41-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall, R. M., D. E. Brookes, and H. W. Stokes. 1991. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol. Microbiol. 15:1941-1959. [DOI] [PubMed] [Google Scholar]
- 26.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 27.Hansson, K., L. Sundström, A. Pelletier, and P. H. Roy. 2002. IntI2 integron integrase in Tn7. J. Bacteriol. 184:1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardwick, S. A., H. W. Stokes, S. Findlay, M. Taylor, and M. R. Gillings. 2008. Quantification of class 1 integron abundance in natural environments using real-time quantitative PCR. FEMS Microbiol. Lett. 278:207-212. [DOI] [PubMed] [Google Scholar]
- 29.Holben, W. E., and D. Harris. 1995. DNA-based monitoring of total bacterial community structure in environmental samples. Mol. Ecol. 4:627-631. [DOI] [PubMed] [Google Scholar]
- 30.Holben, W. E., K. P. Feris, A. Kettunen, and J. H. A. Apajalahti. 2004. GC fractionation enhances microbial community diversity assessment and detection of minority populations of bacteria by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:2263-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmes, A. J., M. P. Holley, A. Mahon, B. Nield, M. Gillings, and H. W. Stokes. 2003. Recombination activity of a distinctive integron-gene cassette system associated with Pseudomonas stutzeri populations in soil. J. Bacteriol. 185:918-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janssen, P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Møller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leung, K., and E. Topp. 2001. Bacterial community dynamics in liquid swine manure during storage: molecular analysis using DGGE/PCR of 16S rDNA. FEMS Microbiol. Ecol. 38:169-177. [Google Scholar]
- 35.Ley, R. E., P. J. Turnbaugh, S. Klein, and J. I. Gordon. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022-1023. [DOI] [PubMed] [Google Scholar]
- 36.Lick, S., K. Drescher, and K. J. Heller. 2001. Survival of Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus in the terminal ileum of fistulated Göttingen minipigs. Appl. Environ. Microbiol. 67:4137-4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu, J., S. Sanchez, C. Hofacre, J. J. Maurer, B. G. Harmon, and M. D. Lee. 2003. Evaluation of broiler litter with reference to the microbial composition as assessed by using 16S rRNA and functional gene markers. Appl. Environ. Microbiol. 69:901-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacDonald, D., G. Demarre, M. Bouvier, D. Mazel, and D. N. Gopaul. 2006. Structural basis for broad DNA-specificity in integron recombination. Nature 440:1157-1162. [DOI] [PubMed] [Google Scholar]
- 39.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Márquez, C., M. Labbate, A. J. Ingold, P. R. Chowdhury, M. S. Ramírez, D. Centrón, G. Borthagaray, and H. W. Stokes. 2008. Recovery of a functional class 2 integron from an Escherichia coli strain mediating a urinary tract infection. Antimicrob. Agents Chemother. 52:4153-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mater, D. D., L. Bretigny, O. Firmesse, M. J. Flores, A. Mogenet, J. L. Bresson, and G. Corthier. 2005. Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus survive gastrointestinal transit of healthy volunteers consuming yogurt. FEMS Microbiol. Lett. 250:185-187. [DOI] [PubMed] [Google Scholar]
- 42.Mazel, D. 2006. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4:608-620. [DOI] [PubMed] [Google Scholar]
- 43.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 1998. A distinctive class of integron in the Vibrio cholerae genome. Science 280:605-608. [DOI] [PubMed] [Google Scholar]
- 44.Messier, N., and P. H. Roy. 2001. Integron integrases possess a unique additional domain necessary for activity. J. Bacteriol. 183:6699-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michael, C. A., M. R. Gillings, A. J. Holmes, L. Hughes, N. R. Andrew, M. P. Holley, and H. W. Stokes. 2004. Mobile gene cassettes: a fundamental resource for bacterial evolution. Am. Nat. 164:1-12. [DOI] [PubMed] [Google Scholar]
- 46.Moura, A., I. Henriques, R. Ribeiro, and A. Correia. 2007. Prevalence and characterization of integrons from bacteria isolated from a slaughterhouse wastewater treatment plant. J. Antimicrob. Chemother. 60:1243-1250. [DOI] [PubMed] [Google Scholar]
- 47.Moura, A., M. Soares, C. Pereira, N. Leitão, I. Henriques, and A. Correia. 2009. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25:1096-1098. [DOI] [PubMed] [Google Scholar]
- 48.Nandi, S., J. J. Maurer, C. Hofacre, and A. O. Summers. 2004. Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. Proc. Natl. Acad. Sci. USA 101:7118-7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nemergut, D. R., A. P. Martin, and S. K. Schmidt. 2004. Integron diversity in heavy-metal-contaminated mine tailings and inferences about integron evolution. Appl. Environ. Microbiol. 70:1160-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nemergut, D. R., M. S. Robeson, R. F. Kysela, A. P. Martin, S. K. Schmidt, and R. Knight. 2008. Insights and inferences about integron evolution from genomic data. BMC Genomics 9:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nield, B. S., A. J. Holmes, M. R. Gillings, G. D. Recchia, B. C. Mabbutt, K. M. Nevalainen, and H. W. Stokes. 2001. Recovery of new integron classes from environmental DNA. FEMS Microbiol. Lett. 195:59-65. [DOI] [PubMed] [Google Scholar]
- 52.Nunes-Düby, S. E., H. J. Kwon, R. S. Tirumalai, T. Ellenberger, and A. Landy. 1998. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26:391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nüsslein, K., and J. M. Tiedje. 1998. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl. Environ. Microbiol. 64:1283-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nüsslein, K., and J. M. Tiedje. 1999. Soil bacterial community shift correlated with change from forest to pasture vegetation in a tropical soil. Appl. Environ. Microbiol. 65:3622-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Partridge, S. R., G. D. Recchia, C. Scaramuzzi, C. M. Collis, H. W. Stokes, and R. M. Hall. 2000. Definition of the attI1 site of class 1 integrons. Microbiology 146:2855-2864. [DOI] [PubMed] [Google Scholar]
- 56.Ramírez, M. S., C. Quiroga, and D. Centrón. 2005. Novel rearrangement of a class 2 integron in two non-epidemiologically related isolates of Acinetobacter baumanni. Antimicrob. Agents Chemother. 49:5179-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramírez, M. S., L. J. Vargas, V. Cagnoni, M. Tokumoto, and D. Centrón. 2005. Class 2 integron with a novel cassette array in a Burkholderia cenocepacia isolate. Antimicrob. Agents Chemother. 49:4418-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowe-Magnus, D. A., A. M. Guerout, and D. Mazel. 2002. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol. Microbiol. 43:1657-1669. [DOI] [PubMed] [Google Scholar]
- 59.Rowe-Magnus, D. A., and D. Mazel. 2001. Integrons: natural tools for bacterial genome evolution. Curr. Opin. Microbiol. 4:565-569. [DOI] [PubMed] [Google Scholar]
- 60.Salyers, A. A., A. Gupta, and Y. Wang. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12:412-416. [DOI] [PubMed] [Google Scholar]
- 61.Schmieger, H., and P. Schicklmaier. 1999. Transduction of multiple drug resistance of Salmonella enterica serovar typhimurium DT104. FEMS Microbiol. Lett. 170:251-256. [DOI] [PubMed] [Google Scholar]
- 62.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Snell-Castro, R., J. J. Godon, J. P. Delgenès, and P. Dabert. 2005. Characterisation of the microbial diversity in a pig manure storage pit using small subunit rDNA sequence analysis. FEMS Microbiol. Ecol. 52:229-242. [DOI] [PubMed] [Google Scholar]
- 64.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 65.Stokes, H. W., C. L. Nesbø, M. Holley, M. I. Bahl, M. R. Gillings, and Y. Boucher. 2006. Class 1 integrons potentially predating the association with Tn402-like transposition genes are present in a sediment microbial community. J. Bacteriol. 188:5722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 67.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vandamme, P., B. Pot, M. Gillis, P. de Vos, K. Kersters, and J. Swings. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.White, P. A., C. J. McIver, Y. Deng, and W. D. Rawlinson. 2000. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182:265-269. [DOI] [PubMed] [Google Scholar]
- 70.Wright, M. S., C. Baker-Austin, A. H. Lindell, R. Stepanauskas, H. W. Stokes, and J. V. McArthur. 2008. Influence of industrial contamination on mobile genetic elements: class 1 integron abundance and gene cassette structure in aquatic bacterial communities. ISME J. 2:417-428 [DOI] [PubMed] [Google Scholar]
- 71.Xu, H., J. Davies, and V. Miao. 2007. Molecular characterization of class 3 integrons from Delftia spp. J. Bacteriol. 17:6276-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.