Abstract
Ninety percent of cultured bacterial nitrate reducers with a 16S rRNA gene similarity of ≥97% had a narG or nosZ similarity of ≥67% or ≥80%, respectively, suggesting that 67% and 80% could be used as standardized, conservative threshold similarity values for narG and nosZ, respectively (i.e., any two sequences that are less similar than the threshold similarity value have a very high probability of belonging to different species), for estimating species-level operational taxonomic units. Genus-level tree topologies of narG and nosZ were generally similar to those of the corresponding 16S rRNA genes. Although some genomes contained multiple copies of narG, recent horizontal gene transfer of narG was not apparent.
Nitrate reducers (i.e., both dissimilatory nitrate reducers and denitrifiers) reduce nitrate to nitrite, which can then be reduced to ammonium by dissimilatory nitrate reducers or sequentially reduced to nitric oxide, nitrous oxide, and dinitrogen by denitrifiers (29). narG codes for the alpha subunit of the dissimilatory nitrate reductase, which reduces nitrate to nitrite and is thus common to both dissimilatory nitrate reducers and denitrifiers (29). nosZ codes for nitrous oxide reductase, which reduces nitrous oxide to dinitrogen and is common to denitrifiers but not dissimilatory nitrate reducers (29). Both narG and nosZ are commonly used as gene markers for community level analysis of nitrate reducers (2, 8, 9, 16, 18, 19, 20, 25). However, standardized criteria for assigning environmental narG and nosZ sequences to operational taxonomic units (OTUs) are required so that diverse data sets on nitrate-reducing communities can be normalized. The widespread ability of bacteria and archaea to denitrify (29) complicates the development of such criteria for genes involved in denitrification. Some closely related narG and closely related nosZ genes occur in distantly related taxa, and narG or nosZ phylogenies do not always reflect 16S rRNA phylogenies (17). However, nosZ-based phylogenies in general have a high degree of congruency with 16S rRNA gene-based phylogenies (3, 10, 30), and recent horizontal gene transfer of nosZ seems unlikely (10), indicating that denitrifier structural genes might be used for estimating the species-level novelty, as well as species-level diversity, of denitrifiers in environmental samples. The limited amount of data on horizontal gene transfer of narG (4, 24) identifies a need to extend such an approach to this gene. The limited number of studies that have compared 16S rRNA with narG or nosZ phylogenies accentuates the need for a more thorough analysis of the phylogenetic relatedness of these three genes (3, 4, 7). Thus, the main objectives of this study were to (i) resolve criteria for standardizing OTU assignment of environmental narG and nosZ sequences, (ii) determine whether those criteria can be used as indicators of novel species, and (iii) investigate the impact of horizontal gene transfer on narG.
Analysis of narG and nosZ sequence data.
One hundred fourteen narG and 85 nosZ sequences from pure cultures were retrieved from GenBank along with the associated 16S rRNA gene sequences (see Table S1 in the supplemental material). The fragments of narG, nosZ, and the 16S rRNA gene that were analyzed correspond to regions amplified by the commonly used primers narG1960f/-2650r (18), nosZF/-R (19), and 27F/1492R (14), respectively. Alignments of 16S rRNA gene fragments and in silico-translated narG and nosZ fragments were performed with ClustalW implemented in MEGA 4.0 (13) and manually refined. The number of base or amino acid differences per site (D) was calculated for pairwise comparisons of all narG, nosZ, and 16S rRNA gene fragments. The similarity (S) was expressed as S = 1 − D. The percent similarity of narG or nosZ from nucleic acid- and amino acid-based comparisons was plotted against the percent similarity of the corresponding 16S rRNA gene. This approach is similar to that used for derivation of 16S rRNA gene-based criteria for species delineation, which is based on DNA/DNA similarities (as a valid criterion to distinguish two species) (28).
Phylogenetic trees based on nucleotide sequences of 16S rRNA gene, narG, and nosZ fragments were calculated in MEGA 4.0 (13) using the neighbor-joining algorithm (21) and 500 bootstrap replications (5). Tree topologies were compared to evaluate the use of narG and nosZ as indicators of new species.
The impact of horizontal gene transfer on narG diversity was assessed by locating putatively horizontally transferred genes in whole-genome sequences of species using the score-based identification of genomic islands-hidden Markov model algorithm (SIGI-HMM) as implemented in Colombo (http://www.tcs.informatik.uni-goettingen.de/colombo-sigihmm), which identifies putatively alien genes based on differences in codon usage (27), and using SeqWord, which can be used to identify putatively alien genes based on oligonucleotide biases (6).
Correlations between structural gene sequences, species diversity, and horizontal gene transfer.
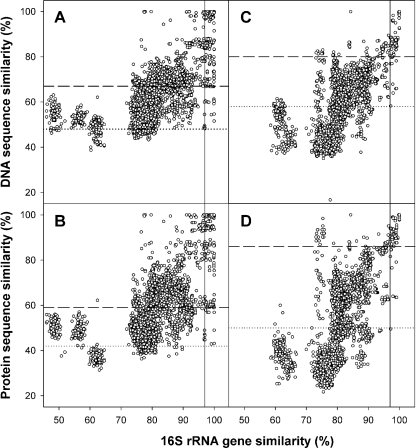
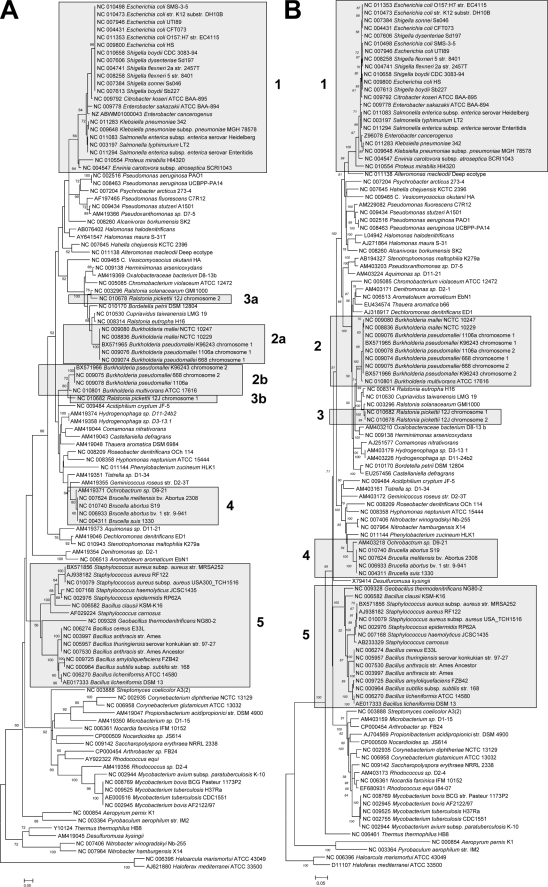
A 16S rRNA gene similarity of ≥97% (a conservative threshold similarity for assigning two organisms to different species [22, 23]) corresponded to an average narG sequence similarity of 85% (86% for translated amino acids). Nitrate reducers that had a <48% narG sequence similarity (or <42% for translated amino acids) always had a 16S rRNA gene similarity of <97% (Fig. 1) and therefore belonged to different species. Ninety percent of the nitrate reducers with a 16S rRNA gene similarity of ≥97% had a narG sequence similarity of ≥67% and a narG translated amino acid similarity of ≥59%. The same gene sequence and translated amino acid similarities were obtained for 90% of nitrate reducers with a 16S rRNA gene similarity of ≥98.7%, which has recently been suggested as a threshold similarity for species differentiation (22). The correlation analysis suggests that a narG sequence similarity of 67% and a narG amino acid similarity of 59% could be used as threshold similarities (i.e., any two sequences that are less similar than the threshold similarity value have a very high probability of belonging to different species) for estimating narG-derived species-level OTUs. These relationships are indicative of different evolutionary rates for narG and the 16S rRNA gene. Tree topologies of narG and 16S rRNA phylogenies were generally similar at the genus level, since species of the same genus mostly clustered together in both trees (Fig. 2). However, multiple copies of narG were present in some species. In such cases, the multiple narG copies clustered separately (Fig. 2), while the multiple copies of the 16S rRNA genes from the organisms analyzed clustered closely together (data not shown).
FIG. 1.
Correlation of DNA and in silico-translated amino acid sequence similarities of narG (A, B) and nosZ (C, D) versus 16S rRNA gene similarity. Dotted lines represent the similarity values, below which two sequences always had less than 97% 16S rRNA gene sequence similarity. The dashed lines represent the 90% quantile of pairwise sequence comparisons with a 16S rRNA gene sequence similarity of 97% (i.e., threshold similarity). The solid lines mark the 97% 16S rRNA gene similarities.
FIG. 2.
Comparison of narG (A) and 16S rRNA (B) phylogenies. Boxes 1, 4, and 5 display examples of closely related species clustering together in the narG tree. Boxes 2 and 3 illustrate that multiple copies of narG from the same organism cluster separately. Neighbor-joining trees were constructed from nucleotide sequences of narG and 16S fragments with approximate lengths of 650 and 1,400 bp, respectively. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. Bootstrap supports below 50% are not shown. The bars represent estimated sequence dissimilarities of 5% (A) and 0.1% (B).
The percentage of putatively alien genes found in whole-genome sequences ranged from 0% to 20.6% based on codon usage analysis, and the Nar operon was predicted to be putatively alien only in Pseudomonas stutzeri A1501 and was estimated to be transferred from the Gammaproteobacteria (see Table S1 in the supplemental material). In Nitrobacter winogradskyi Nb-255, narI and narJ were identified as putatively alien genes transferred from the Gammaproteobacteria (see Table S1 in the supplemental material). It appeared to be unlikely, based on oligonucleotide bias, that the Nar operon was horizontally transferred in any species (e.g., see Fig. S1 to S3 in the supplemental material).
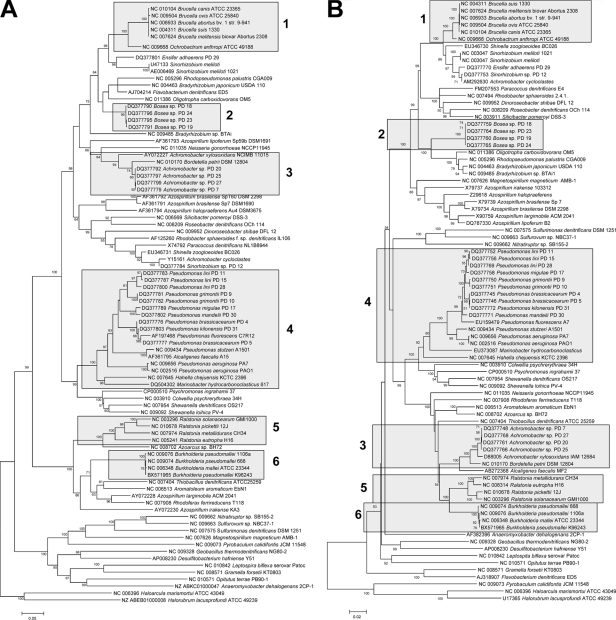
A 16S rRNA gene similarity of ≥97% (a conservative threshold similarity for assigning two organisms to different species [22, 23]) corresponded to an average nosZ sequence similarity of 89% (92% for translated amino acids). Denitrifiers with a <58% nosZ similarity (or <50% similarity of translated amino acids) always had a 16S rRNA gene similarity of <97% (Fig. 1) and therefore belonged to different species. Ninety percent of denitrifiers with a 16S rRNA gene similarity of ≥97% had a nosZ sequence similarity of ≥80% and a translated amino acid similarity of ≥86%. Ninety percent of denitrifiers with a 16S similarity of ≥98.7% had a nosZ sequence similarity of ≥85% and a translated amino acid similarity of ≥90%. The correlation analysis suggests that a nosZ sequence similarity of 80% and a nosZ amino acid similarity of 86% could be used as threshold similarities for estimating nosZ-derived species-level OTUs with a very high probability of estimating correctly. The calculated threshold similarities are in good agreement with those calculated for the nosZ fragment amplified by the primers nosZ661F/-1773R (9). Phylogenetic trees of nosZ and 16S rRNA genes had similar clustering at the genus level (Fig. 3). Multiple copies of nosZ per organism were not found. The different threshold similarities of nosZ and narG suggest that nosZ is more conserved than narG, indicating that species-level OTU assignment of nosZ might be more reliable than that of narG.
FIG. 3.
Comparison of nosZ (A) and 16S rRNA (B) phylogenies. Boxes 1 to 6 display examples of closely related species clustering together in the nosZ tree. Not all sequences of Azospirillum sp. cluster together in the nosZ tree. Neighbor-joining trees were constructed from nucleotide sequences of nosZ and 16S fragments with approximate lengths of 700 and 1,400 bp, respectively. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. Bootstrap supports below 50% are not shown. The bars represent estimated sequence dissimilarities of 5% (A) and 2% (B).
Conclusions.
The above considerations indicate that narG or nosZ analyses can be used to estimate species-level diversity of the associated bacteria on the basis of sequence similarities. Such analysis is considered to yield a minimum number of species in a sample (i.e., the true species-level diversity might be much higher). Analyses of gene markers of different functional groups revealed similar threshold similarities indicating species-level OTUs (e.g., for dsrAB and amoA gene fragments, they were 80 to 90%) (11, 12). Since narG or nosZ sequences from organisms of the same genus generally form coherent clusters in phylogenetic trees (Fig. 2 and 3), distinct clusters of environmental sequences could provide evidence for new genus-level diversity. Recent horizontal gene transfer does not appear to have occurred for either narG (as indicated by the results in this study) or nosZ (as indicated by the results in reference 10). The adaptation of genomic features of an alien gene to those of a host genome takes several hundred million years (15), and genes that were horizontally transferred 50 million years ago can be reliably detected by sequence analyses (26). Sequence differences might also be caused by gene duplication and diversification (e.g., in species in which multiple gene copies are present) rather than by horizontal gene transfer (1). However, analysis of codon usage and oligonucleotide bias does not always reveal the transfer of genes from closely related organisms with similar genomic features (27).
Supplementary Material
Acknowledgments
Support for this study was provided by the Deutsche Forschungsgemeinschaft (DFG HO 4020/2-2 and DR 310/3-3) and the University of Bayreuth.
Footnotes
Published ahead of print on 5 June 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bergthorsson, U., D. I. Andersson, and J. R. Roth. 2007. Ohno's dilemma: evolution of new genes under continuous selection. Proc. Natl. Acad. Sci. USA 104:17004-17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chèneby, D., S. Hallet, M. Mondon, F. Martin-Laurent, J. C. Germon, and L. Philippot. 2003. Genetic characterization of the nitrate reducing community based on narG nucleotide sequence analysis. Microb. Ecol. 46:113-121. [DOI] [PubMed] [Google Scholar]
- 3.Dandie, C. E., D. L. Burton, B. J. Zebarth, J. T. Trevors, and C. Goyer. 2007. Analysis of denitrification genes and comparison of nosZ, cnorB and 16S rDNA from culturable denitrifying bacteria in potato cropping systems. Syst. Appl. Microbiol. 30:128-138. [DOI] [PubMed] [Google Scholar]
- 4.Delorme, S., L. Philippot, V. Edel-Hermann, C. Deulvot, C. Mougel, and P. Lemanceau. 2003. Comparative genetic diversity of the narG, nosZ, and 16S rRNA genes in fluorescent pseudomonads. Appl. Environ. Microbiol. 69:1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felsenstein, J. 1985. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 6.Ganesan, H., A. S. Rakitianskaia, C. F. Davenport, B. Tummler, and O. N. Reva. 2008. The SeqWord genome browser: an online tool for the identification and visualization of atypical regions of bacterial genomes through oligonucleotide usage. BMC Bioinformatics 9:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregory, L. G., P. L. Bond, D. J. Richardson, and S. Spiro. 2003. Characterization of a nitrate-respiring bacterial community using the nitrate reductase gene (narG) as a functional marker. Microbiology 149:229-237. [DOI] [PubMed] [Google Scholar]
- 8.Henry, S., D. Bru, B. Stres, S. Hallet, and L. Philippot. 2006. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 72:5181-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horn, M. A., H. L. Drake, and A. Schramm. 2006. Nitrous oxide reductase genes (nosZ) of denitrifying microbial populations in soil and the earthworm gut are phylogenetically similar. Appl. Environ. Microbiol. 72:1019-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones, C. M., B. Stres, M. Rosenquist, and S. Hallin. 2008. Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol. Biol. Evol. 25:1955-1966. [DOI] [PubMed] [Google Scholar]
- 11.Kjeldsen, K. U., A. Loy, T. F. Jakobsen, T. R. Thomsen, M. Wagner, and K. Ingvorsen. 2007. Diversity of sulfate-reducing bacteria from an extreme hypersaline sediment, Great Salt Lake (Utah). FEMS Microbiol. Ecol. 60:287-298. [DOI] [PubMed] [Google Scholar]
- 12.Koops, H.-P., U. Purkhold, A. Pommerening-Röser, G. Timmermann, and M. Wagner. 2003. The lithoautotrophic ammonia-oxidizing bacteria, p. 778-811. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. Springer-Verlag, New York, NY.
- 13.Kumar, S., M. Nei, J. Dudley, and K. Tamura. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics, 1st ed. John Wiley and Sons, Ltd., Chichester, NY.
- 15.Lawrence, J. G., and H. Ochman. 1997. Amelioration of bacterial genomes: rates of change and exchange. J. Mol. Evol. 44:383-397. [DOI] [PubMed] [Google Scholar]
- 16.Mounier, E., S. Hallet, D. Cheneby, E. Benizri, Y. Gruet, C. Nguyen, S. Piutti, C. Robin, S. Slezack-Deschaumes, F. Martin-Laurent, J. C. Germon, and L. Philippot. 2004. Influence of maize mucilage on the diversity and activity of the denitrifying community. Environ. Microbiol. 6:301-312. [DOI] [PubMed] [Google Scholar]
- 17.Philippot, L. 2002. Denitrifying genes in bacterial and archaeal genomes. Biochim. Biophys. Acta 1577:355-376. [DOI] [PubMed] [Google Scholar]
- 18.Philippot, L., S. Piutti, F. Martin-Laurent, S. Hallet, and J. C. Germon. 2002. Molecular analysis of the nitrate-reducing community from unplanted and maize-planted soils. Appl. Environ. Microbiol. 68:6121-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rich, J. J., R. S. Heichen, P. J. Bottomley, K. Cromack, and D. D. Myrold. 2003. Community composition and functioning of denitrifying bacteria from adjacent meadow and forest soils. Appl. Environ. Microbiol. 69:5974-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rösch, C., A. Mergel, and H. Bothe. 2002. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68:3818-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou, N., and M. Nei. 1987. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 22.Stackebrandt, E., and J. Ebers. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 33:152-155. [Google Scholar]
- 23.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S ribosomal-RNA sequence-analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 24.Stolz, J. F., and P. Basu. 2002. Evolution of nitrate reductase: molecular and structural variations on a common function. ChemBioChem 3:198-206. [DOI] [PubMed] [Google Scholar]
- 25.Throback, I. N., K. Enwall, A. Jarvis, and S. Hallin. 2004. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49:401-417. [DOI] [PubMed] [Google Scholar]
- 26.Vernikos, G. S., N. R. Thomson, and J. Parkhill. 2007. Genetic flux over time in the Salmonella lineage. Genome Biol. 8:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waack, S., O. Keller, R. Asper, T. Brodag, C. Damm, W. F. Fricke, K. Surovcik, P. Meinicke, and R. Merkl. 2006. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics 7:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Trüper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 29.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zumft, W. G., and H. Körner. 2007. Nitrous oxide reductases, p. 67-81. In H. Bothe, S. J. Ferguson, and W. E. Newton (ed.), Biology of the nitrogen cycle, 1st ed. Elsevier Academic Press, Inc., Amsterdam, The Netherlands.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.