Abstract
Electron transfer dissociation (ETD) is evaluated as a technique to provide local information on higher order structure and dynamics of a whole protein molecule. Isotopic labeling of highly flexible segments of a model 18 kDa protein is carried out in solution under mildly denaturing conditions by means of hydrogen/deuterium exchange (HDX), followed by transfer of intact protein ions to the gas phase by means of electrospray ionization and mass-selection of a precursor ion for subsequent reactions with fluoranthene radical anions. The ETD process gives rise to abundant fragment ions, whose deuterium content can be measured as a function of duration of the HDX reaction in solution. No backbone protection is detected for all protein segments spanning the twenty-five residue long N-terminal part of the protein, which is known to lack structure in solution. At the same time, noticeable protection is evident for segments representing the structured regions of the protein. The results of this work suggest that ETD of intact protein ions is not accompanied by detectable hydrogen scrambling and can be used in tandem with HDX to probe protein conformation in solution.
Hydrogen/deuterium exchange in solution (HDX) combined with mass spectrometry (MS) has become a powerful tool to probe both conformation and dynamics of proteins under variety of conditions, where patterns of deuterium distribution across the polypeptide backbone can be obtained by fragmenting the protein molecule proteolytically in solution under the slow-exchange conditions (1) or in the gas phase prior to MS detection (2). Enzymatic fragmentation remains the most popular choice, as it can be implemented using a variety of platforms, although the methodology employing protein ion fragmentation in the gas phase (top-down HDX MS) is also gaining popularity. It offers several advantages, including a possibility to avoid back-exchange by excluding protein manipulation under the slow exchange conditions from the experimental scheme, and has been already applied to characterize dynamics and structural features of proteins in their native and non-native states (3–5). One unique feature of such experiments that is beginning to be exploited is its ability to correlate isotope distribution patterns with specific protein conformations using mass selection prior to ion activation as a means to obtain fragment ions originating from a particular conformer.
However, applications of top-down HDX MS are still limited due to concerns over the possibility of hydrogen scrambling accompanying collision-activated dissociation (CAD) of protein ions. Indeed, several reports have been published suggesting that proton mobility in the gas phase does influence the outcome of HDX CAD MS measurements for short peptides and under certain conditions for longer polypeptides (3, 6, 7). Some time ago we suggested that the specter of hydrogen scrambling in top-down HDX MS measurements may be alleviated by using non-ergodic fragmentation processes, where dissociation is induced by ion-electron interaction, rather than collisional activation (2). Indeed, the results of several recent studies suggest that only minimal rearrangement or labile hydrogen atoms occurs in the gas phase when electron capture dissociation (ECD) is used to probe local deuterium content of polypeptides (8, 9).
Another recently introduced fragmentation technique based on cation-electron interactions, electron transfer dissociation, ETD (10), apparently is even less prone to scrambling when applied to short peptides (11). While this report is very encouraging, it does not address a question of whether ETD can be used to probe protein conformation in top-down HDX MS experiments. Although ETD of intact protein ions is feasible (12), it is a multi-step process, which involves precursor ion interactions with the reagent anion followed by its charge reduction, subsequent formation of the radical site and, finally, radical-induced dissociation of an N-Cα bond. Rearrangement of labile hydrogen atoms that may occur within the protein ion prior to the dissociation events would result in hydrogen scrambling. Furthermore, increasing the yield of dissociation often requires that the ion-ion interactions in the gas phase be allowed to occur during an extended period of time, leading to “over-reaction” (13), which may also alter the isotopic distributions of fragment ions by inducing charge reduction. Although the fragment ion mass does not change upon charge reduction from n to n-1,1 the reduced fragment ion would carry one “extra” H⋅ atom compared to fragments at (n-1)+ charge state that were generated directly by protein ion dissociation, an apparent mass gain that may interfere with top-down HDX MS measurements.
In order to evaluate the occurrence and the extent of hydrogen scrambling that may accompany ETD of protein ions, we carried out HDX MS measurements using an 18 kDa model protein, pseudo-wild type Cellular Retinoic Acid Binding Protein I (wt*-CRABP I) containing a 21-residue His-tag at the N-terminus. While the protein itself is folded into a β-barrel conformation (Figure 1), which remains stable across a wide pH range, the His-tag portion of the protein (red segment in Figure 1) is entirely unstructured and its backbone, together with a short N-terminal segment of the wild type sequence (orange in Figure 1), and should lack any protection in solution (14). This makes the 25-residue N-terminal segment of the protein a convenient reporter of hydrogen scrambling, which has been used to detect occurrence of scrambling in HDX CAD MS (15).
Figure 1.
Tertiary structure and sequence of wt*-CRABP I showing the His-tag (red), the unstructured N-terminal segment of the wild-type protein (orange) and the first N-terminal segment forming a stable element of its secondary (strand β1, blue).
HDX was initiated by diluting a stock solution of fully deuterated wt*-CRABP I (120 µM in D2O) in H2O/10 mM ammonium acetate (1:15). HDX was carried out at pH 3.0 and 25°C and quenched by dropping the solution temperature to 0°C, followed by injection to the ESI source of a prototype 4.7T apeX-Ultra (Bruker Daltonics, Billerica, MA) FT ICR MS with ETD capability (12). Source conditions were adjusted to minimize collisional activation and exclude formation of CAD-generated fragment ions in the ESI interface. ETD of protein ions was carried out by isolating and trapping the precursor ion (intact protein, charge state +17), followed by introduction of fluoranthene anions. Overall, 63 fragment ions have been detected in these experiments. While a significant proportion of these ions were derived from the His-tag region (apparently due to the very high histidine content (16)), 39 fragments resulted from cleavages of N-Cα bonds in the protein backbone beyond the His-tag sequence.
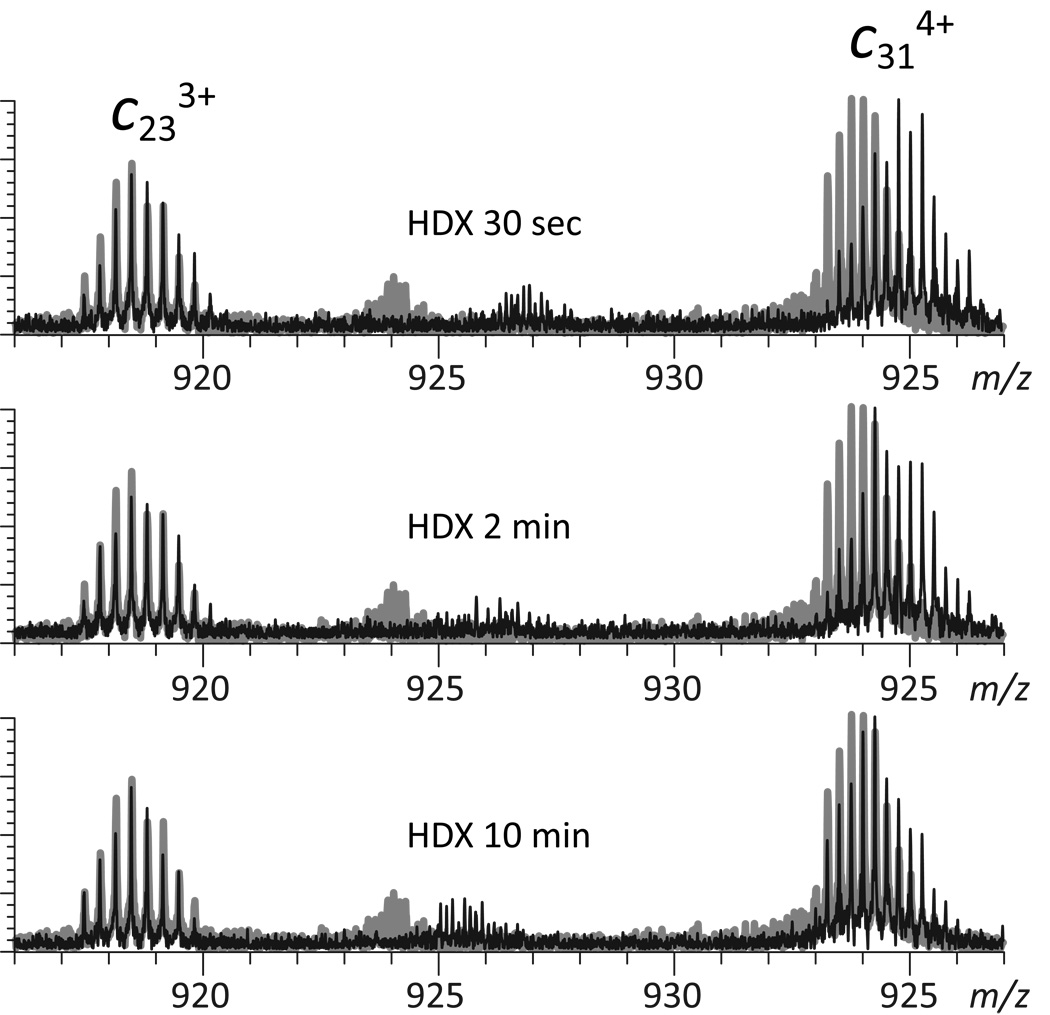
Although most fragmentation occurs during the initial 30 ms period, the ions were allowed to over-react for an additional 50–100 msec to evaluate the potential negative impact of fragment ions charge reduction on the quality of HDX MS measurements. Over-reaction manifests itself under these conditions not only by a significant reduction of the charge states of surviving precursor ions, but also by noticeable deviations of the isotopic distributions of fragment ions from the ones calculated using their empirical formulas (Supplementary Material, Figure S1). The magnitude of this bias towards higher masses is clearly dependent on the fragment ion charge state (Supplementary Material, Figure S2), consistent with the notion that charge reduction via electron gain in the gas phase does result in an apparent mass increase of fragment ions at lower charge states. For example, charge reduction of c31 5+ will generate (c31+H⋅)4+ ions, whose appearance in the mass spectrum is expected to distort the isotopic distribution of the entire c31 4+ ensemble. Such alterations of isotopic distributions of fragment ions under over-reaction conditions may certainly interfere with top-down HDX MS measurements by artificially inflating their deuterium content. However, we note that measurements of local deuterium content rely on comparing the amounts of labile deuterium atoms in two overlapping fragments (e.g., c31 4+ and c23 3+ in Figure 2) by either simple subtraction or deconvolution (17). Therefore, distortion of the isotopic distribution of c31 4+ is expected to be at least partially offset by that of c23 3+, minimizing the negative impact of fragment ion charge reduction on the ability to accurately measure deuterium content of the [Ala25→Ser32] segment. Indeed, the isotopic distribution of this segment calculated by deconvolution of experimentally measured distributions of c31 4+ and c23 3+ fragment ions is very close to the theoretical isotopic distribution for this segment calculated using its empirical formula (Figure S1).
Figure 2.
Evolution of isotopic distributions of c23 3+ and c31 4+ fragment ions in ETD mass spectra of wt*-CRABP I undergoing HDX in solution (black traces). The gray traces represent the end-point of the exchange reaction
Although fragment ion charge reduction is unlikely to have a significant negative impact on the quality of top-down HDX MS measurements even under the unfavorable over-reaction conditions (vide supra), extended reaction time may also increase the incidence of hydrogen scrambling in the gas phase as a result of intramolecular H/D exchange prior to fragmentation. In order to evaluate the extent of hydrogen scrambling, ETD was used to probe deuterium content of the unstructured N-terminal segment of the protein. Even after very brief exchange, isotopic distributions of all fragment ions derived from this region are nearly indistinguishable from the distributions representing the HDX endpoint, suggesting very fast exchange of labile deuterium atoms. The largest fragment ion exhibiting such behavior is c23 (Figure 3), which incorporates the entire unstructured N-terminal segment. Although the surviving intact protein ions retain 70 deuterons under these conditions (data not shown), no deuterium atoms are retained within c23 beyond the small number representing the HDX end-point (due to the small, but finite D2O/H2O dilution ratio). This suggests that no detectable intramolecular exchange of labile hydrogen atoms occurs during protein ion ETD.
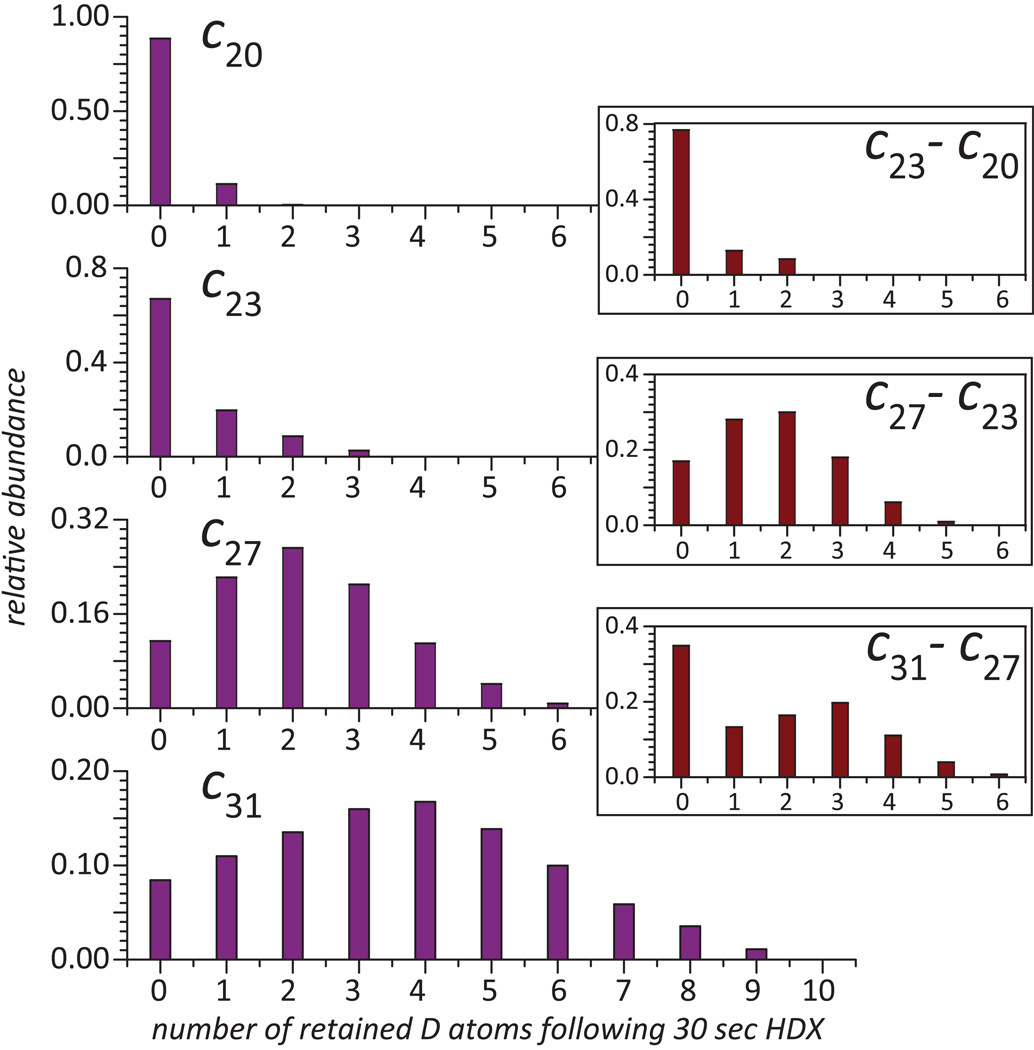
Figure 3.
Blue bars: distribution of deuterium atoms retained by ETD fragments of fully deuterated wt*-CRABP I exposed to protiated buffer (room temperature, pH 3.0) for 30 sec obtained by deconvoluting the isotopic distributions of each fragment ion at the end-point of the exchange reaction from that at following 30 sec of exchange. Brown bars (insets): distribution of retained deuterium atoms within short protein segments obtained by deconvoluting isotopic distributions of overlapping fragment ions.
Fragment ions derived from the protein beyond the [Gly1→Ala25] segment display a markedly different behavior. A noticeable retention of deuterium atoms is readily detected following brief exchange, and the deviation of such isotopic distributions from HDX end-point persists even following prolonged exchange in solution (e.g., c31 4+ in Figure 2). Deuterium content of each fragment ion can be obtained by removing contributions of naturally occurring isotopes and residual deuterium content (due to the finite D2O/H2O dilution ratio) by deconvolution using their isotopic distributions at HDX end-point (17). The results of these calculations for four adjacent fragment ions (c20, c23, c27 and c31) are shown in Figure 3. Clearly, retention of deuterium is correlated with the position of the cleavage site, as detectable retention of deuterium label is shown only by ions incorporating segments of the “native” sequence of CRABP I.
In order to present local protection patterns deduced from top-down HDX MS measurements and to minimize the influence of charge reduction on such measurements (vide supra), distributions of retained deuterium in partially overlapping fragments can be used to evaluate deuterium content of short protein segments (e.g., c27 and c31 to obtain deuterium content within the [Lys29→Ser32] segment). These calculations carried for several fragments using a deconvolution procedure developed in our laboratory (17) and the results clearly show that segments representing the unstructured region lack any protection, while noticeable protection is afforded to segments localized in the stable elements of the protein secondary structure (Figure 3).
Absence of deuterium retention within the protein segments representing the unstructured elements of wt*-CRABP I provides conclusive evidence that proton/deuteron mobility in protein ions undergoing ETD is very low and does not affect the outcome of local HDX MS measurements. This opens up a host of new exciting opportunities for extending the HDX MS armamentarium by including HDX ETD MS as a method to characterize protein conformation and dynamics at high spatial resolution without the need for proteolytic step in solution.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant R01 GM061666 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
we do not consider gain of an electron as a mass increase, since it would not be measurable using most commercial mass analyzers
References
- 1.Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol. Cell. Proteomics. 2002;1:132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Kaltashov IA, Eyles SJ. Crossing the phase boundary to study protein dynamics and function: combination of amide hydrogen exchange in solution and ion fragmentation in the gas phase. J. Mass Spectrom. 2002;37:557–565. doi: 10.1002/jms.338. [DOI] [PubMed] [Google Scholar]
- 3.Hoerner JK, Xiao H, Dobo A, Kaltashov IA. Is there hydrogen scrambling in the gas phase? Energetic and structural determinants of proton mobility within protein ions. J. Am. Chem. Soc. 2004;126:7709–7717. doi: 10.1021/ja049513m. [DOI] [PubMed] [Google Scholar]
- 4.Xiao H, Kaltashov IA. Transient structural disorder as a facilitator of protein-ligand binding: native H/D exchange-mass spectrometry study of cellular retinoic acid binding protein I. J. Am. Soc. Mass Spectrom. 2005;16:869–879. doi: 10.1016/j.jasms.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 5.Hoerner JK, Xiao H, Kaltashov IA. Structural and dynamic characteristics of a partially folded state of ubiquitin revealed by hydrogen exchange mass spectrometry. Biochemistry. 2005;44:11286–11294. doi: 10.1021/bi0509548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demmers JA, Rijkers DT, Haverkamp J, Killian JA, Heck AJ. Factors affecting gas-phase deuterium scrambling in peptide ions and their implications for protein structure determination. J. Am. Chem. Soc. 2002;124:11191–11198. doi: 10.1021/ja0125927. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson PL, Pan J, Wilson DJ, Dempsey B, Lajoie G, Shilton B, Konermann L. Hydrogen/deuterium scrambling during quadrupole time-of-flight MS/MS analysis of a zinc-binding protein domain. Anal. Chem. 2007;79:153–160. doi: 10.1021/ac061261f. [DOI] [PubMed] [Google Scholar]
- 8.Rand KD, Adams CM, Zubarev RA, Jørgensen TJD. Electron capture dissociation proceeds with a low degree of intramolecular migration of peptide amide hydrogens. J. Am. Chem. Soc. 2008;130:1341–1349. doi: 10.1021/ja076448i. [DOI] [PubMed] [Google Scholar]
- 9.Pan J, Han J, Borchers CH, Konermann L. Electron capture dissociation of electrosprayed protein ions for spatially resolved hydrogen exchange measurements. J. Am. Chem. Soc. 2008;130:11574–11575. doi: 10.1021/ja802871c. [DOI] [PubMed] [Google Scholar]
- 10.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zehl M, Rand KD, Jensen ON, Jorgensen TJ. Electron transfer dissociation facilitates the measurement of deuterium incorporation into selectively labeled peptides with single residue resolution. J. Am. Chem. Soc. 2008;130:17453–17459. doi: 10.1021/ja805573h. [DOI] [PubMed] [Google Scholar]
- 12.Kaplan DA, Hartmer R, Speir JP, Stoermer C, Gumerov D, Easterling ML, Brekenfeld A, Kim T, Laukien F, Park MA. Electron transfer dissociation in the hexapole collision cell of a hybrid quadrupole-hexapole Fourier transform ion cyclotron resonance mass spectrometer. Rapid Commun. Mass Spectrom. 2008;22:271–278. doi: 10.1002/rcm.3356. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan DA, Hartmer R, Easterling ML, Yang J, Kellersberger K, Park MA. Evaluation of electron transfer dissociation in a hybrid quadrupole-hexapole Fourier transform ion cyclotron resonance mass spectrometer; 56-th ASMS Conference on Mass Spectrometry and Allied Topics; Denver, CO.: ASMS; 2008. p TPC089. [DOI] [PubMed] [Google Scholar]
- 14.Eyles SJ, Speir P, Kruppa G, Gierasch LM, Kaltashov IA. Protein conformational stability probed by Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Chem. Soc. 2000;122:495–500. [Google Scholar]
- 15.Abzalimov RR, Kaltashov IA. Hydrogen exchange and tandem mass spectrometry; 18 Sanibel Conference on Mass Spectrometry; Sanibel Island, FL.: ASMS; 2006. [Google Scholar]
- 16.Xia Y, Gunawardena HP, Erickson DE, McLuckey SA. Effects of cation charge-site identity and position on electron-transfer dissociation of polypeptide cations. J. Am. Chem. Soc. 2007;129:12232–12243. doi: 10.1021/ja0736764. [DOI] [PubMed] [Google Scholar]
- 17.Abzalimov RR, Kaltashov IA. Extraction of local hydrogen exchange data from HDX CAD MS measurements by deconvolution of isotopic distributions of fragment ions. J. Am. Soc. Mass Spectrom. 2006;17:1543–1451. doi: 10.1016/j.jasms.2006.07.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.