Abstract
The soluble N-ethylmaleimide sensitive factor attachment protein receptor (SNARE) family of proteins is required for eukaryotic intracellular membrane fusions. Vesicle fusion for formation of the prospore membrane (PSM), a membrane compartment that forms de novo during yeast sporulation, requires SNARE function, phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2], and the activity of the phospholipase D (PLD) Spo14p, which generates phosphatidic acid (PA). The SNARE syntaxin Sso1p is essential for PSM production while the functionally redundant homolog in vegetative growth, Sso2p, is not. We demonstrate that Sso1p and Sso2p bind similarly in vitro to PA or phosphoinositide-containing liposomes and that the conserved SNARE (H3) domain largely mediates PA-binding. Both green fluorescent protein-Sso fusion proteins localize to the developing PSM in wild-type cells and to the spindle pole body in spo14Δ cells induced to sporulate. However, the autoregulatory region of Sso1p binds PI(4,5)P2-containing liposomes in vitro with a greater ability than the equivalent region of Sso2p. Overexpression of the phosphatidylinositol-4-phosphate 5-kinase MSS4 in sso1Δ cells induced to sporulate stimulates PSM production; PLD activity is not increased under these conditions, indicating that PI(4,5)P2 has roles in addition to stimulating PLD in PSM formation. These data suggest that PLD-generated PA and PI(4,5)P2 collaborate at multiple levels to promote SNARE-mediated fusion for PSM formation.
The soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) family of proteins is required for the fusion of vesicles to target membranes in eukaryotic cells (53). The process of SNARE-mediated fusion is both structurally and mechanistically similar in different intracellular transport pathways and is evolutionarily conserved from yeast to human (18, 31, 34). In vitro experiments demonstrated that SNAREs have the ability to effect fusions of liposomes in the absence of other components, indicating that these proteins directly mediate the fusion event (56). SNAREs can be broadly categorized as either vesicle SNAREs (v-SNAREs) or target membrane SNAREs (t-SNAREs), respectively. The interaction of SNAREs on apposed membranes can overcome the energy barrier generated by charged headgroups of lipids comprising the bilayers. As an incoming vesicle approaches its target membrane, the v-SNAREs and t-SNAREs assemble via their SNARE domains into a four-helix bundle termed a SNAREpin, bringing the two bilayers into closer proximity (3, 55, 56). The outer membrane layers of both the vesicle and target membrane mix, forming a hemifusion intermediate before full fusion of the membranes occurs (23, 24, 29, 58).
The helices comprising the SNAREpin are supplied by three different SNARE subfamilies. Two of these subfamily members, syntaxin and SNAP-25, are t-SNAREs; the former contributes one helix while the latter contributes two helices (16). The syntaxin and SNAP-25 homologs heterodimerize to form the t-SNARE complex before the trans-interaction with the helix of vesicle-associated membrane protein/synaptobrevin v-SNARE (42). Discrete intracellular fusion events are mediated by SNAREpins comprising different constituent syntaxin, SNAP-25, and vesicle-associated membrane protein homologs (18, 53).
In addition to SNAREs, lipids facilitate membrane fusion events for both membrane curvature induction required for procession through intermediate states of fusion and direct regulation of SNARE molecules (32, 33). Cone-shaped lipids such as diacylglycerol and phosphatidic acid (PA) induce negative (concave) curvature while inverted cone shapes, such as lysophosphatidic acid (LPA), have the opposite effect (26, 27). The assembly of SNARE complexes requires correct lipid composition at the fusion site; addition of inverted cone-shaped lipids antagonized in vitro SNARE complex assembly (35). Recent studies have shown that phosphatidylinositides also play roles in SNARE-mediated fusions. Phosphatidylinositol-3-phosphate [PI(3)P] interacts with the Saccharomyces cerevisiae SNARE Vam7p via its phox homology domain and appears to facilitate targeting to the vacuole (15). Additionally, phosphoinositides increased the rates of in vitro fusion of proteoliposomes that approximated physiological protein and lipids in vivo (36). Phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] was shown to bind to the juxtamembrane region of syntaxin-1 in PC12 cells and has both stimulatory and inhibitory effects on in vitro fusion rates (20).
The activity of the lipid-modifying enzyme phospholipase D (PLD) also appears to be important for vesicle fusions. PLD catalyzes the hydrolysis of phosphatidylcholine (PC) to PA in a PI(4,5)P2-dependent manner (22, 49). In S. cerevisiae, PLD activity is required for the de novo formation of a novel compartment, the prospore membrane (PSM), during sporulation (48). Vesicles trafficked from the Golgi and endosomal compartments dock at the spindle pole body (SPB) and participate in SNARE-mediated fusions for PSM formation (38, 40, 41). Cells induced to sporulate that lack the yeast PLD Spo14p show docked but unfused vesicles at the SPB (40, 44). Interestingly, cells lacking Sso1p, a syntaxin functionally redundant with Sso2p at the plasma membrane (PM), display a similar phenotype while sso2Δ cells display no sporulation defect (2, 21, 40). The specific requirement for Sso1p in sporulation is not fully understood although the Sso1p autoregulatory Habc motif is important (43).
In this study, we demonstrate that Sso1p acts downstream of Spo14p (PLD)-generated PA during PSM formation. Sso1p and Sso2p bind PA and additional phosphoinositide species; PA binding is mediated by the conserved H3 motif. Additionally, the Sso1p Habc domain shows a greater ability to interact with PIP2-containing liposomes in vitro than the equivalent region of Sso2p. Overexpression of the PI(4)P 5-kinase Mss4p results in PSM formation in sso1Δ cells induced to sporulate. Together, these data indicate that both PA and PI(4,5)P2 are required for efficient fusion and furthermore suggest a novel role for PI(4,5)P2 in the regulation of specialized SNARE fusion events.
MATERIALS AND METHODS
Yeast strains, media, and genetic manipulation.
Strains used in this study are derived from the SK-1 background and are listed in Table 1. DNA-mediated transformation of yeast cells was performed using the lithium acetate method (19). Y7795 and Y7799 were transformed with EcoRI-linearized URA3-MPC54-red fluorescent protein (RFP) for integration at MPC54 (40). Y4459 was generated by crossing sso1Δ haploids transformed with pKR466 linearized with XbaI and ClaI for integration at SPO14 (45). Integrations were verified by PCR. Growth and sporulation on solid and liquid media was performed as described previously (37). For green fluorescent protein (GFP) and RFP fusion protein analyses, live cells were examined on a Zeiss Axioskop 2 fluorescent microscope during mitotic growth and at various times postinduction of sporulation. Temperature-sensitive strains were induced on solid sporulation medium for 8 h at room temperature (∼23°C) before temperature shift to 34.5°C overnight; cells were examined at ∼16 h postshift for PSM formation.
TABLE 1.
Yeast strains
| Strain | Genotype or description |
|---|---|
| AN120a | MATa/α ura3/ura3his3/his3leu2/leu2 trp1/trp1 arg4/ARG4 rme1::LEU2/RME1 |
| Y4350 | MATa/α ura3/ura3 leu2/leu2 TRP1/trp1 mss4::HIS3/mss4::HIS3 plus mss4-2LEU2 |
| Y4451b | AN120 but spo14::URA3/spo14::URA3 |
| Y4454b | AN120 but sso1::his5+/sso1::his5+ |
| Y4459 | Y4454 but spo14::URA3/spo14::URA3 |
| Y7795 | Y4451 but URA3-MPC54-RFP |
| Y7799 | AN120 but URA3-MPC54-RFP |
| Y7834 | Y7799 plus pRS424-TEF2pr-GFP-SSO1 |
| Y7836 | Y7799 plus pRS424-SPO20pr-GFP-SSO1 |
| Y7852 | Y7795 plus pRS424-TEF2pr-GFP-SSO1 |
| Y7854 | Y7795 plus pRS424-SPO20pr-GFP-SSO1 |
| Y7945 | Y7799 plus pRS424-SPO20pr-GFP-SSO2 |
| Y7953 | Y7795 plus pRS424-SPO20pr-GFP-SSO2 |
| Y8026 | Y7799 plus pRS424-TEF2pr-GFP-SSO2 |
| Y8034 | Y7795 plus pRS424-TEF2pr-GFP-SSO2 |
| Y8116 | Y4454 plus YEp352-SSO2 |
| Y8119 | Y4454 plus YEp352 |
| Y8121 | AN120 plus YEp352-SSO1 |
| Y8124 | AN120 plus YEp352-SSO2 |
| Y8127 | AN120 plus YEp352 |
| Y8159 | Y4454 plus YEp-MSS4 and pUN40-SSO2 |
| Y8208 | AN120 plus YEp-MSS4 and pRS424-G20 |
| Y8211 | AN120 plus YEp352 and pRS424-G20 |
| Y8214 | Y4454 plus YEp-MSS4 and pRS424-G20 |
| Y8217 | Y4454 plus YEp352 and pRS424-G20 |
| Y8220 | AN120 plus YEp-MSS4 and YEp-SPO14 |
| Y8222 | AN120 plus YEp352 and YEp-SPO14 |
| Y8224 | Y4454 plus YEp-MSS4 and YEp-SPO14 |
| Y8226 | Y8226 plus YEp352 and YEp-SPO14 |
| Y8232 | AN120 plus pUN40-SSO2 |
| Y8234 | AN120 plus pUN40 |
| Y8238 | Y4454 plus pUN40-SSO2 |
| Y8240 | Y4454 plus pUN40 |
| Y8262 | Y4454 plus pUN30-SSO1 |
| Y8265 | Y4454 plus pRS424-SSO1 |
| Y8298 | Y4454 plus YEp-MSS4 plus YEp-SSO2 |
| Y8319 | Y4454 plus pUN50-SSO1R45A R63D |
| Y8335 | Y4454 plus pUN40-SSO2A49R D67R |
| Y8347 | Y4454 plus YEp-SSO2A49R D67R |
Plasmids.
All plasmids used in this study are listed in Table 2; PCR primer oligonucleotides are listed in Table 3. All constructs were verified by nucleotide sequencing. PCR amplification of SSO1, including 1,000 bp 5′ of and 500 bp 3′ of the open reading frame (ORF), using primers P630 and P631 was performed. The resulting product was digested with EcoRI and BamHI and inserted into similarly digested pUN30 and pRS424 to generate both centromeric and 2μ SSO1 plasmids. PCR with P591 and P592 was performed to amplify SSO2, including 5′ and 3′ flanking sequences; the product was digested with SphI and EcoRI and then inserted into similarly digested pUN40 and YEp352. YEp-SPO14 and YEp-MSS4 have been described previously (47, 48). Plasmids RS424-TEF2pr-GFP-SSO1, RS424-SPO20pr-GFP-SSO1, RS424-G20, and RS306-MPC54-RFP are from A. Neiman (40). To generate the equivalent plasmids for SSO2, PCR amplification with P610 and P603 was performed to amplify the SSO2 ORF lacking the ATG codon and including 500 bp 3′ of the ORF; the product was then digested with XbaI and XhoI. pRS424-TEF2pr-GFP-SSO1 was digested with EcoRI and XbaI to recover the GFP fragment. A three-way ligation was performed with the above fragments along with pRS424-SPO20pr isolated by digestion with EcoRI and XhoI to generate pRS424-SPO20pr-GFP-SSO2. This construct was then digested with EcoRI and XhoI to recover the combined GFP-SSO2 fragment, which was inserted into similarly digested pRS424-TEF2pr to generate pRS424-TEF2pr-GFP-SSO2. The mutant SSO plasmids (pUN50 carrying SSO1 with the R45A R63D mutation [pUN50-SSO1R45A R63D] and pUN40-SSO2A49R D67R) were generated with a QuikChange Lightning Multi Site-Directed Mutagenesis Kit according to the manufacturer's protocol (Stratagene, La Jolla, CA) using primers P633 and P634 for SSO1 and P635 and P636 for SSO2 and the corresponding wild-type plasmids as templates. The wild-type SSO plasmids expressed in vegetative cells were able to complement the growth defect of an sso1Δ sso2Δ strain (data not shown).
TABLE 2.
Plasmids used in this study
| Name | Yeast marker(s) | Promoter | Cloned gene | Source or reference |
|---|---|---|---|---|
| pUN40 | TRP1, CEN | 11 | ||
| pUN30-SSO1 | TRP1, CEN | SSO1 | SSO1plus 500 bp 3′ | This study |
| pUN50-SSO1 | URA3, CEN | SSO1 | SSO1plus 500 bp 3′ | This study |
| pUN50-SSO1R45A R63D | URA3, CEN | SSO1 | SSO1plus 500 bp 3′ | This study |
| pUN40-SSO2 | TRP1, CEN | SSO2 | SSO2plus 300 bp 3′ | This study |
| pUN40-SSO2A49R D67R | TRP1, CEN | SSO2 | SSO2plus 300 bp 3′ | This study |
| pRS424 | TRP1, 2μ | 51 | ||
| pRS424-SSO1 | TRP1, 2μ | SSO1 | SSO1plus 500 bp 3′ | This study |
| YEp352 | URA3, 2μ | 17 | ||
| YEp352-SSO1 | URA3, 2μ | SSO1 | SSO1plus 500 bp 3′ | This study |
| YEp352-SSO2 | URA3, 2μ | SSO2 | SSO2plus 300 bp 3′ | This study |
| YEp352-SSO2A49R D67R | URA3, 2μ | SSO2 | SSO2plus 300 bp 3′ | This study |
| YEp-MSS4-URA | URA3, 2μ | MSS4 | MSS4 | 48 |
| YEp-MSS4-TRP | TRP1, 2μ | MSS4 | MSS4 | 48 |
| YEp-SPO14 | TRP1, 2μ | SPO14 | SPO14 | 47 |
| pRS424-TEF2pr-GFP-SSO1 | TRP1, 2μ | TEF2 | GFP-SSO1 | 40 |
| pRS424-SPO20pr-GFP-SSO1 | TRP1, 2μ | SPO20 | GFP-SSO1plus 500 bp 3′ | 40 |
| pRS424-TEF2pr-GFP-SSO2 | TRP1, 2μ | TEF2 | GFP-SSO2 | This study |
| pRS424-SPO20pr-GFP-SSO2 | TRP1, 2μ | SPO20 | GFP-SSO2plus 500 bp 3′ | This study |
| pRS424-G20 | TRP1, 2μ | DTR1 | GFP-SPO2051-91 | 40 |
| pRS306-MPC54-RFP | URA3, integration | MPC54 | RFP-MPC54 | 40 |
| pKR466 | URA3, integration | 45 | ||
| pGEX-5X-1 | GST | 52 | ||
| pGEX-SSO1-TMD | GST-SSO11-265 | J. Jäntti | ||
| pGEX-SSO2-TMD | GST-SSO21-269 | J. Jäntti | ||
| pGEX-SSO1-start-HLH | GST-SSO11-148 | This study | ||
| pGEX-SSO1-Habc | GST-SSO132-148 | This study | ||
| pGEX-SSO1-HabcR45A R63D | GST-SSO132-148 | This study | ||
| pGEX-SSO1-HLH | GST-SSO1149-192 | This study | ||
| pGEX-SSO1-HLH-SNARE | GST-SSO1149-264 | This study | ||
| pGEX-SSO1-SNARE | GST-SSO1194-264 | This study | ||
| pGEX-SSO2-Habc | GST-SSO235-152 | This study | ||
| pGEX-SSO2-HabcA49R D67R | GST-SSO235-152 | This study | ||
| pGEX-SSO2-SNARE | GST-SSO2196-268 | This study |
TABLE 3.
Primers
| Name | Target | Sequence |
|---|---|---|
| P294 | URA3-3′ | 5′-CCTTGGTGGTACGAACATCC-3′ |
| P295 | URA3-5′ | 5′-AGAAAAGCAGGCTGGGAAGC-3′ |
| P300 | MPC54-5′ | 5′-GTAACTTCTCGTATAAGGCC-3′ |
| P493 | SPO14-5′ | 5′-ACAGTGGCTAATGGACCCG-3′ |
| P630 | SSO1-5′ | 5′-GATCGATCACGGATCCTACCGAAGAAAGAATACG-3′ |
| P631 | SSO1-3′ | 5′-GATCGATCACGAATTCAAACGTCTAATATAGCGG-3′ |
| P591 | SSO2-5′ | 5′-TAGAATTCGTGAATAAAGAAGCCAGC-3′ |
| P592 | SSO2-3′ | 5′-CCTTGGAACCGCATGCTAAGGGCGACAGTTTTTC-3′ |
| P610 | SSO2-ORF-5′ | 5′-GCTCTAGAATGAGCAACGCTAATCCT-3′ |
| P603 | SSO2-ORF-3′ | 5′-CGCGCTCGAGACACATAATAGGAATTGG-3′ |
| P558 | SSO1-start-5′ | 5′-CGGGATCCCCAGTTATAATAATCCGTAC-3′ |
| P559 | SSO1-HLH-3′ | 5′-CGGAATTCTTCCGCAAGAGCAGTCTT-3′ |
| P517 | SSO1-Habc-5′ | 5′-TTGGATCCCCGATTTCGTGGGCTTCATG-3′ |
| P518 | SSO1-Habc 3′ | 5′-TTGAATTCATACTGCCTCTTGGCTTG-3′ |
| P550 | SSO1-HLH-5′ | 5′-TTGGATCCCCATGATCATTCAACCAGAG-3′ |
| P587 | SSO1-H3-5′ | 5′-TTGGATCCCCGTCCAGGCAAGGCACCAA-3′ |
| P556 | SSO1-H3-3′ | 5′-GCGCGCGGAATTCTCTAATCTTGTTCTTTCT-3′ |
| P520 | SSO2-Habc-5′ | 5′-TTGGATCCCCGACGATTTCGTAGCTTTT-3′ |
| P519 | SSO2-Habc-3′ | 5′-TTGAATTCGTACTGTCTCTTCGCCTG-3′ |
| P588 | SSO2-H3-5′ | 5′-TTGGATCCCCGTACAGGCTAGACATCAA-3′ |
| P557 | SSO2-H3-3′ | 5′-GCGCGCGGAATTCTCTTATTTTGTTTTTTCT-3′ |
| P633 | SSO1 RtoA | 5′-AAGATCAGTCAAATCAATGCCGATCTCGATAAGTACGACCATACC-3′ |
| P634 | SSO1 RtoD | 5′-CAGGTCGATTCTTTGCATAAGGACCTACTGACCGAAGTTAATGAG-3′ |
| P635 | SSO2 AtoR | 5′-AACAAGATCAACTCAATAAATCGTAACTTGTCCAGGTACGAAAAC-3′ |
| P636 | SSO2 DtoR | 5′-CAAATTGATGCGCAACACAAACGCCTACTTACTCAAGTGAGTGAG-3′ |
The pGEX-SSO1-TMD (where TMD is the transmembrane domain) and pGEX-SSO2-TMD plasmids are from J. Jäntti. All PCR products used to generate glutathione S-transferase (GST)-SSO fusions were digested with BamHI and EcoRI and inserted into similarly digested pGEX-5X-1. To generate pGEX-SSO1-start-HLH (where HLH is hinge-loop-helix), PCR with P558 and P559 was performed to amplify the indicated SSO1 region. To generate the other pGEX-SSO constructs, PCR products using primers listed in Table 3 were used as inserts. The mutant pGEX-SSO Habc fusions were generated in a manner similar to that used for the wild-type GST-Habc fusions by performing PCR using pUN50-SSO1R45A R63D and pUN40-SSO2A49R D67R as templates and the appropriate primers.
In vivo BODIPY-PC analysis.
Cells were sporulated in liquid medium as described previously (37), and 2-decanoyl-1{O-[11-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionyl)amino]undecyl}-sn-glycero-3-phosphocholine (BODIPY-PC) was added directly to the cultures at a final concentration of 4 μM after 2 h at 30°C. Cells were harvested 3 h later. All assays were performed in triplicate, and BODIPY-PC cellular internalization was verified by examination of live cultures with fluorescence microscopy. Total lipid content of cultures was harvested and analyzed as described previously (47). The percent conversion of BODIPY-PC to BODIPY-PA was obtained by densitometry of pixel intensities using AlphaEaseFC, version 4.0.0 software (Alpha Innotech).
Expression and purification of GST fusion proteins.
All glutathione S-transferase (GST) fusion constructs were transformed into protease-deficient Escherichia coli strain BL21; expression and purification of GST fusions were performed using ProMega MagneGST beads according to the manufacturer's protocol (ProMega, Madison, WI).
Liposome binding assays.
All phospholipids were acquired from Avanti Polar Lipids (Alabaster, AL). Sucrose-laden liposomes were generated using appropriate phospholipid mixtures as described previously (6, 39). The resulting liposomes were incubated with 5 μg of bovine serum albumin and incubated with molar equivalents (∼70 pmol) of experimental protein for 30 min on ice. Vesicles were centrifuged as previously, and supernatant fractions were treated with trichloroacetic acid for protein precipitation. Both supernatant and pelleted fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Imperial Protein Stain (Pierce, Thermo Fisher Scientific, IL) staining. Gel images were acquired using a Hewlett Packard ScanJet G4010 scanner, and densitometry of pixel intensities was performed on the images using AlphaEaseFC, version 4.0.0, software (Alpha Innotech). All assays were conducted in triplicate except where noted. All assays were performed with lipids in molar excess to protein (data not shown).
RESULTS
Sso1p functions in parallel or downstream of Spo14p in PSM formation.
To determine if Sso1p is required for efficient Spo14p PLD activity during sporulation, we examined the conversion of the fluorophore-conjugated lipid BODIPY-PC to BODIPY-PA, which we previously found accurately reflects in vivo PLD activity (7, 47), in wild-type, sso1Δ, spo14Δ, and sso1Δ spo14Δ cells. As expected from previous studies (7, 47), very little BODIPY-PC was converted to BODIPY-PA in spo14Δ cells (Table 4). In contrast, similar levels of BODIPY-PA were generated in sso1Δ and wild-type cells (Table 4). Analysis of the sso1Δ spo14Δ double mutant indicated that spo14Δ is epistatic to sso1Δ for PLD activity (Table 4). Thus, Sso1p acts either in parallel or downstream of Spo14p PLD activity in PSM precursor vesicle fusion.
TABLE 4.
In vivo hydrolysis of internalized BODIPY-PC
| Strain | Relevant genotype | % BODIPY-PAa |
|---|---|---|
| AN120 | SPO14SSO1 | 2.5 ± 0.7 |
| Y4451 | spo14Δ | 0.1 ± 0.1 |
| Y4454 | sso1Δ | 3.2 ± 1.1 |
| Y4459 | spo14Δ sso1Δ | 0.1 ± 0.0 |
The percent conversion of internalized BODIPY-PC to BODIPY-PA was determined in cells induced to sporulate at 30°C, as described in Materials and Methods. Values are means ± SEM for three independent experiments.
Sso1p and Sso2p bind PA in vitro.
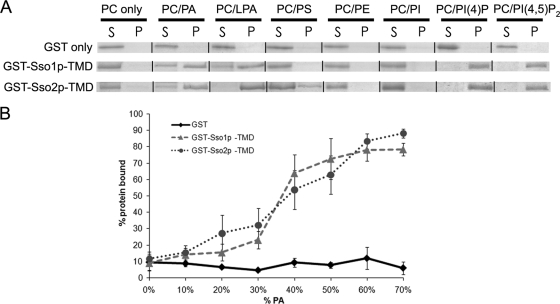
Given the importance of lipid interactions in SNARE-mediated fusion and the finding that Sso1p does not affect Spo14p PLD activity, we examined whether Sso1p is a target of PA, as has been shown for the sporulation-specific homolog of SNAP-25, Spo20p (40). To that end, we performed in vitro liposome binding assays (6) with a GST fusion to Sso1p lacking the TMD, termed GST-Sso1p-TMD, and GST alone as a negative control. As shown in Fig. 1A and Table 5, GST-Sso1p-TMD bound PA-containing liposomes, while GST alone did not exhibit any binding. As Sso1p and Sso2p are functionally redundant at the PM during vegetative growth (2), we also examined the ability of GST-Sso2p-TMD to bind PA. Similar to GST-Sso1p-TMD, GST-Sso2p-TMD bound PA-containing liposomes (Fig. 1A, Table 5).
FIG. 1.
Sso proteins have similar in vitro binding profiles to different lipids. (A) Representative images of binding assays. Liposomes were prepared in a 1:1 molar ratio with 34:1 PC and the lipid species listed above corresponding assays. Supernatant (S) and pellet (P) represent the components of a single assay. (B) Graph of percent protein bound for liposomes containing 0 to 70 mol% PA. Quantification was performed as described in Materials and Methods. Each data point represents a minimum of three independent assays; error bars represent standard error. PE, phosphatidylethanolamine; PS, phosphatidylserine; PI, phosphatidylinositol.
TABLE 5.
Percent GST fusion proteins bound in liposome assays
| GST protein | % Protein bound (SEM)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| PC | PA | LPA | PS | PE | PI | PI(4)P | PI(4,5)P2 | |
| GST only | 7.24 (3.63) | 7.84 (1.88) | 8.28 (0.83) | 10.03 (2.03) | 7.38 (1.74) | 15.65 (2.71) | 3.40 | 8.31 (1.05) |
| GST-Sso1p-TMD | 9.06 (4.70) | 72.61 (12.21) | 77.39 (10.04) | 13.35 (1.24) | 14.85 (4.21) | 17.40 (2.14) | 62.11 (20.79) | 85.52 (9.43) |
| GST-Sso2p-TMD | 11.71 (4.10) | 62.92 (11.78) | 88.62 (0.64) | 27.18 (2.09) | 12.45 (2.69) | 11.55 (3.05) | 93.95 (4.60) | 92.57 (0.16) |
Liposomes are composed of a 1:1 molar ratio of PC and the listed phospholipids. Densitometry was performed on scanned images of SDS-PAGE gels. Values are the percent protein bound to the pellet fraction and are the average from a minimum of three separate binding assays for each data point, with the exception of the PI(4)P value for the GST-only protein, which is from one experiment. PE, phosphatidylethanolamine; PS, phosphatidylserine; PI, phosphatidylinositol.
To better understand the nature of this interaction, we performed assays with liposomes comprising different lipid compositions. Both GST-Sso proteins bound liposomes containing LPA, a molecule possessing only one acyl chain but the same negatively charged polar headgroup as PA. In addition, both GST-Sso proteins bound PI(4)P and PI(4,5)P2 similarly (Fig. 1A and Table 5). GST alone did not exhibit binding to any of these liposome preparations (Fig. 1A and Table 5). These results suggest that Sso proteins preferably bind lipids whose headgroups possess one or more phosphate moieties.
We also examined the binding of GST-Sso fusions to liposomes containing 0 to 70 mol% PA. No significant difference was observed between GST-Sso1p-TMD and GST-Sso2p-TMD in binding abilities to the liposomes over all mole percentages of PA (Fig. 1B). GST alone did not exhibit binding to liposomes regardless of PA composition (Fig. 1B). Thus, both Sso proteins can bind similarly to PA-containing liposomes in addition to liposomes containing other negatively charged lipids.
The SNARE domains of both Sso1p and Sso2p bind PA.
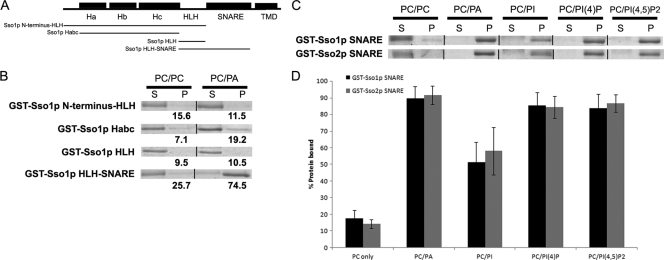
To determine the PA-binding region(s) of Sso1p and Sso2p, we generated a series of GST fusions to different regions of Sso1p for use in liposome binding assays (Fig. 2A). Sso1p, like other syntaxin homologs, possesses a region proximal to the amino terminus known as the Habc domain in addition to the HLH region, the conserved H3 SNARE motif, and TMD, located adjacent to the carboxy terminus of the protein (Fig. 2A) (4, 12, 57). A GST fusion consisting of Sso1p from its amino terminus through the HLH region (GST-Sso1p-start-HLH) did not bind PA-containing liposomes to a greater extent than PC-only liposomes; all GST fusions to portions of Sso1p within this region also did not show binding to liposomes containing PA (Fig. 2B). However, GST-Sso1p-HLH-SNARE showed binding to PA-containing liposomes (Fig. 2B). As GST fused to the HLH region alone showed no significant binding, the SNARE domain of Sso1p appears to mediate binding to PA.
FIG. 2.
The SNARE (H3) domains of both Sso proteins mediate binding to PA and other phosphoinositides in vitro. (A) Schematic of the structural organization of Sso1, with regions fused to GST used in liposome binding assays indicated below. (B) Images of binding assays with GST fusions on SDS-PAGE gels stained with Coomassie. Liposomes were prepared in a 1:1 molar ratio with 34:1 PC and the lipid species listed above corresponding assays. Supernatant (S) and pellet (P) represent the components of a single assay. Values below images of pellet fractions are percent protein bound as determined by densitometry from individual assays (see Materials and Methods). (C) Representative images of binding assays with GST-Sso SNARE proteins; liposomes were prepared as described for panel B. (D) Graph of percent protein bound for all liposome species tested with GST-Ssop SNARE fusions. All assays were performed in triplicate; error bars represent SEM.
As GST-Sso2p-TMD showed a similar binding profile to GST-Sso1p-TMD (Fig. 1A, Table 5), we tested GST fusions to the H3 SNARE domain of both Sso proteins. Both SNARE domains bound similarly to liposomes of various compositions, showing significant binding to PA and phosphoinositide-containing liposomes (Fig. 2C and D). These data demonstrate that the SNARE domains of both Sso proteins are sufficient to mediate PA binding and can also interact with phosphoinositides. Thus, both Sso proteins bind PA, indicating that PA binding is not the basis of the sporulation-specific requirement for Sso1p.
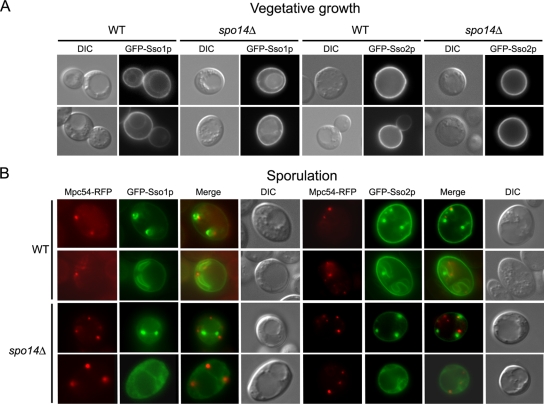
The Sso proteins localize to the PSM during sporulation.
One explanation for the specialized function of Sso1p in sporulation is that Sso1p, but not Sso2p, localizes to PSM precursor vesicles to mediate fusion. To test this hypothesis, we examined GFP fused to either Sso1p or Sso2p during vegetative growth and in cells induced in sporulation medium. As expected, both GFP-Sso fusions localized to the PM during vegetative growth in both wild-type and spo14Δ cells; GFP-Sso1p was also observed at the vacuolar membrane (Fig. 3A). In sporulating wild-type cells, both GFP-Sso proteins were at the developing PSM at an early stage of PSM fusion in addition to a later stage of PSM membrane growth and expansion (Fig. 3B). The signal intensity of GFP-Sso2p at the PM during PSM formation appeared to be greater than that of GFP-Sso1p (Fig. 3B). We examined GFP-Sso protein localization in spo14Δ cells to determine if Spo14p is required for in vivo localization. Since this mutant fails to form PSMs (40, 44, 46), we used strains expressing Mpc54p-RFP, a sporulation-specific component of the modified SPB, the initiation site of PSM formation (25), to mark the presumptive sites of PSM formation. The localization of both GFP-Sso proteins appeared somewhat variable in spo14Δ cells. A subpopulation of cells showed GFP-Sso protein signal in a diffuse cytoplasmic pattern while in other cells distinct green puncta were observed (Fig. 3B). In either case, GFP signal is found adjacent to Mpc54-RFP puncta, indicating that both GFP-Sso proteins are present at the SPB in the absence of Spo14p. Together, these data indicate that both Sso proteins are at the developing PSM during sporulation and that Spo14p is not required for their localization.
FIG. 3.
GFP-Ssop fusions localize similarly in both wild-type and spo14Δ cells. (A) Wild-type (WT; Y7834 and Y8026) and spo14Δ (Y7852 and Y8034) cells expressing either GFP-Sso1p or GFP-Sso2p driven by the vegetative promoter TEF2 were examined by fluorescence and differential interference contrast (DIC) microscopy. (B) Wild-type (Y7836 and Y7945) and spo14Δ (Y7854 and Y7953) cells expressing either GFP-Sso1p or GFP-Sso2p driven by the sporulation-specific promoter SPO20 and the modified SPB marker Mpc54p-RFP were induced in sporulation medium and examined by fluorescence and differential interference contrast microscopy.
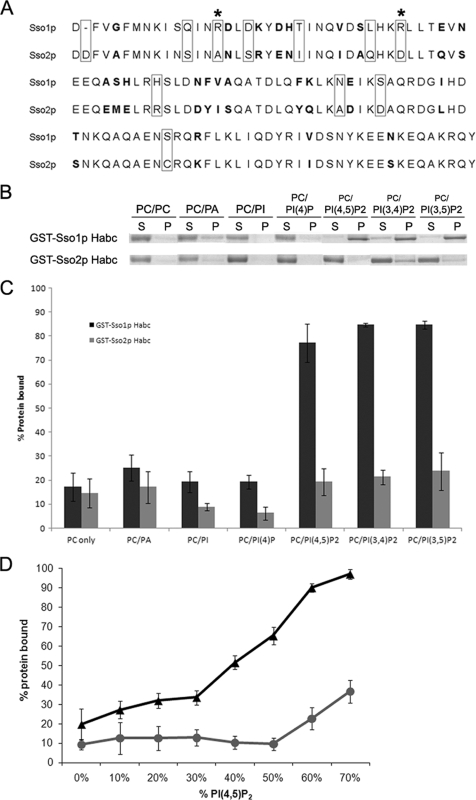
The Sso1p Habc domain binds phosphorylated phosphoinositides.
A previous study found that the Sso1p amino-terminal Habc region plays a role in sporulation function (43); the reason for this is unknown. Sequence alignment of the Sso proteins' Habc regions reveals a higher level of divergence than in other regions. These regions comprise ∼120 residues and have different residues at 35 positions and ∼70% identity (Fig. 4A). We performed liposome binding assays to explore the differential function of the Habc domains of the Sso proteins. While no significant differences were observed for binding to PC only, PC-PA, PC-PI, or PC-PI(4)P liposomes, GST-Sso1p Habc had a significantly greater ability to bind PI(4,5)P2-containing liposomes than GST-Sso2p Habc (Student's paired t test, P < 0.01) (Fig. 4B and C). We also tested the ability of GST-Sso1/2p Habc fusions to bind PI(3,4)P2- and PI(3,5)P2-containing liposomes and found that binding paralleled that observed for PI(4,5)P2-containing liposomes (P < 0.02) (Fig. 4B and C).
FIG. 4.
The Habc domain of Sso1p binds phosphorylated phosphoinositides. (A) Sequence alignment of equivalent regions of the Habc domains of Sso1p and Sso2p used in liposome binding assays. Bolded residues are conservative substitutions; boxed residues are nonconservative substitutions; asterisks indicate sites of mutation for each Habc domain. (B) Representative images of binding assays with GST-Ssop Habc fusion proteins. Liposomes were prepared in a 1:1 molar ratio with 34:1 PC and the lipid species listed above corresponding assays. Supernatant (S) and pellet (P) represent the components of a single assay. (C) Graph of percent protein bound for all liposome species tested with GST-Ssop Habc fusions. Quantifications were performed as described in Materials and Methods. All assays were performed in triplicate; error bars represent SEM. (D) Graph of percent protein bound for liposomes containing 0 to 70 mol % PI(4,5)P2. Quantification was performed as described in Materials and Methods. Each data point represents a minimum of three independent assays; error bars represent SEM.
To further determine the lipid specificity of the Habc region, we examined binding of both GST-Habc fusions to a range of 0 to 70 mol% PI(4,5)P2 liposomes. More GST-Sso1p Habc than GST-Sso2p Habc was bound to PI(4,5)P2 liposomes at all concentrations of PI(4,5)P2 tested; at 70 mol% PI(4,5)P2, ∼95% of Sso1p-Habc bound compared to ∼35% for Sso2p-Habc (Fig. 4D). These data demonstrate that the Habc domains of Sso1p and Sso2p show differential binding to phosphorylated phosphatidylinositides.
Overexpression of Sso2p and/or Mss4p stimulates PSM formation in sso1Δ cells.
SSO2 expressed from a multicopy plasmid, but not a single-copy plasmid, results in the formation of a very small number of tetrads (<1%) in sso1Δ cells (21, 43). These studies were performed using strains derived from the W303 background; we examined single and multicopy SSO2 plasmids for their ability to rescue sso1Δ cells in the SK-1 background; wild-type SK-1 cells are capable of robust sporulation. We found that while the single-copy plasmid failed to support sporulation, the multicopy-plasmid-bearing strain had a sporulation efficiency of ∼8 to 9% (Table 6). This result suggests that high levels of Sso2p can partially alleviate the sporulation-specific requirement for Sso1p.
TABLE 6.
Sporulation in wild-type and sso1Δ cells
| Strain | Relevant genotypea | % Spore formation (mean ± SD)b |
|---|---|---|
| Y8230 | SSO1 (SSO1 CEN) | 81.0 ± 9.7 |
| Y8232 | SSO1(SSO2 CEN) | 93.4 ± 1.8 |
| Y8234 | SSO1(CEN) | 84.2 ± 2.5 |
| Y8262 | sso1Δ (SSO1 CEN) | 61.8 ± 3.6 |
| Y8238 | sso1Δ (SSO2 CEN) | <0.1 |
| Y8240 | sso1Δ (CEN) | <0.1 |
| Y8121 | SSO1(SSO1 2μ) | 82.6 ± 9.2 |
| Y8124 | SSO1(SSO2 2μ) | 67.6 ± 4.0 |
| Y8127 | SSO1(2μ) | 90.0 ± 0.7 |
| Y8265 | sso1Δ (SSO1 2μ) | 72.4 ± 4.6 |
| Y8116 | sso1Δ (SSO2 2μ) | 8.0 ± 5.2 |
| Y8119 | sso1Δ (2μ) | <0.1 |
| Y8208 | SSO1(MSS4 2μ) | 79.0 ± 6.1 |
| Y8211 | SSO1(2μ) | 82.8 ± 3.6 |
| Y8214 | sso1Δ (MSS4 2μ) | 0.3 ± 0.1 |
| Y8217 | sso1Δ (2μ) | <0.1 |
| Y8220 | SSO1(MSS4 2μ SPO14 2μ) | 74.0 ± 7.5 |
| Y8222 | SSO1 (SPO14 2μ) | 67.8 ± 4.5 |
| Y8224 | sso1Δ (MSS4 2μ SPO14 2μ) | 0.2 ± 0.2 |
| Y8226 | sso1Δ (SPO14 2μ) | <0.1 |
| Y8159 | sso1Δ (MSS4 2μ SSO2CEN) | 0.2 ± 0.2 |
| Y8298 | sso1Δ (MSS4 2μ SSO2 2μ) | 21.3 ± 6.1 |
| Y8319 | sso1Δ (sso1R45A R63DCEN) | 73.4 ± 6.9* |
| Y8294 | sso1Δ (sso2A49R D67RCEN) | <0.1* |
| Y8347 | sso1Δ (sso2A49R D67R 2μ) | 21.1 ± 8.3* |
Genes in parentheses are carried by the plasmids.
A total of 300 and 1,000 cells were counted from three independent experiments for SSO1 and sso1Δ strains, respectively. Asterisks indicate strains in which 500 cells were counted per experiment. Sporulation was induced on solid medium. All cells with 1 to 4 spores visible by differential interference contrast optics were included.
As GST-Sso2p Habc could bind liposomes with high levels of PI(4,5)P2, we examined the effects of overexpressing the PI(4)P 5-kinase Mss4p in both wild-type and sso1Δ cells induced to sporulate. The sporulation efficiencies of all wild-type strains, either with Mss4p-expressing multicopy plasmid or empty vector, were not significantly different (Table 6). As expected, cells with a deletion of SSO1 and supplied with a single-copy plasmid bearing SSO1 were able to sporulate (Table 6). Interestingly, cells expressing Mss4p from the multicopy vector and either single-copy SSO2 or empty vector had sporulation efficiencies slightly greater than 0%; corresponding cells lacking Mss4p overexpression failed to form any asci (Table 6).
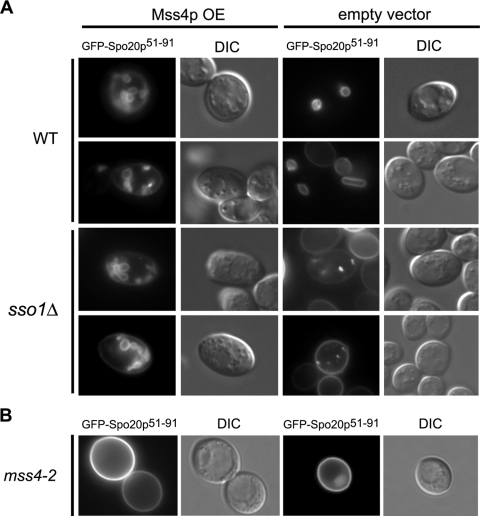
To determine if the small increase in sporulation in sso1Δ cells overexpressing Mss4p alone was due to increased PSM vesicle fusion, we examined if PSM formation can initiate in these cells by observing the localization of GFP fused to Spo20p carrying residues 51 to 91 (GFP-Spo20p51-91), which localizes to developing PSMs in wild-type cells or the modified SPB in sso1Δ cells (40). As seen in Fig. 5A, sso1Δ cells coexpressing Mss4p and GFP-Spo20p51-91 show small internal membrane compartments, which are presumably developing PSMs, while cells expressing GFP-Spo20p51-91 display only a punctate pattern of localization, consistent with SPB locations with no fusion. Signal quantification was performed on sso1Δ strains expressing the GFP fusion constructs; in Mss4p-overexpressing cells, approximately 50% of all cells displayed GFP signal, and of those cells, ∼37% showed PSMs; in strains lacking Mss4p overexpression, ∼46% displayed signal with ∼11% punctate localization and no PSMs. Of the cells not displaying localization to the PSM or SPB, GFP-Spo20p51-91 was localized to the PM, consistent with previous observations of its localization upon initial expression (40). These results reveal that PSM formation is indeed induced in cells lacking Sso1p upon PI(4)P 5-kinase overexpression.
FIG. 5.
Mss4p overexpression stimulates PSM formation in sso1Δ cells and depletion of Mss4p blocks PSM generation. (A) Wild-type (WT) or sso1Δ cells overexpressing Mss4p or empty vector and the PSM marker GFP-Spo20p51-91. (B) mss4-ts cells expressing GFP-Spo20p51-91 at restrictive temperature. All cells were induced in sporulation medium and examined with fluorescence and differential interference contrast (DIC) microscopy.
As either Sso2p or Mss4p overexpression alone leads to partial rescue of the sporulation defect in sso1Δ cells, we examined whether overexpression of both these proteins simultaneously would lead to increased sporulation efficiency. Intriguingly, Mss4p and Sso2p overexpression in sso1Δ cells increases sporulation efficiency to 21.3%, more than twofold higher than cells overexpressing Sso2p alone (Table 6). These data suggest that increased levels of PI(4,5)P2 and Sso2p together during sporulation can better rescue the sporulation defect of sso1Δ cells.
Previously, it was shown that Mss4p depletion results in a severe sporulation defect (48). To see if this defect is due to a block in PSM formation, we examined mss4-ts cells bearing the reporter GFP-Spo20p51-91 shifted to restrictive temperature after initiation of sporulation. Most of these cells display GFP signal at the PM, indicating that PSM formation is largely disrupted in this mutant (Fig. 5B). These cells were also examined with the DNA marker DAPI (4′,6′-diamidino-2-phenylindole) to monitor meiotic progression; the cells appeared to complete the meiotic divisions normally (data not shown). Taken together, these results suggest that PI(4,5)P2 and SNAREs interact to facilitate PSM formation during sporulation.
Overexpression of Mss4p does not increase PLD activity in vivo.
As PI(4,5)P2 stimulates the PLD activity of Spo14p in vivo (48), one possibility for the effect of Mss4p on PSM formation is that the resultant increase in PI(4,5)P2 levels leads to higher levels of Spo14p-generated PA, and it is this increase that stimulates PSM formation in sso1Δ cells. To examine PA levels under these conditions, we monitored the conversion of BODIPY-PC to BODIPY-PA in sporulating wild-type and sso1Δ cells possessing either the Mss4p overexpression plasmid or empty vector and observed no significant difference in PLD activity (Table 7). We also examined both wild-type and sso1Δ cells supplied with a high-copy-number plasmid expressing SPO14 in addition to the MSS4 overexpression plasmid or empty vector and again observed no difference in activity (Table 7). Furthermore, the conversion of BODIPY-PC to BODIPY-PA in the Spo14p-overexpressing cells was not significantly different from that with empty vector alone. These results suggest that the stimulation of PSM formation by Mss4p overexpression in sso1Δ cells is not due to an increase in PLD activity. Thus, it appears that PI(4,5)P2-Sso1p Habc domain interaction is important for PSM vesicle fusion events.
TABLE 7.
BODIPY-PC hydrolysis and percent spore formation
| Strain | Relevant genotypea | % BODIPY-PAb |
|---|---|---|
| Y8208 | SSO1(MSS4 2μ) | 1.3 ± 0.2 |
| Y8211 | SSO1(2μ) | 1.6 ± 0.3 |
| Y8214 | sso1Δ (MSS4 2μ) | 1.8 ± 0.3 |
| Y8217 | sso1Δ (2μ) | 1.9 ± 0.2 |
| Y8220 | SSO1(MSS4 2μ SPO14 2μ) | 1.8 ± 0.3 |
| Y8222 | SSO1 (SPO14 2μ) | 1.1 ± 0.1 |
| Y8224 | sso1Δ (MSS4 2μ SPO14 2μ) | 2.6 ± 0.3 |
| Y8226 | sso1Δ (SPO14 2μ) | 1.8 ± 0.1 |
Genes in parentheses are carried by the plasmids.
The conversion percentage of internalized BODIPY-PC to BODIPY-PA was determined in sporulated cells at 30°C as described in Materials and Methods. Values are means ± SEM for three independent experiments. Paired t tests for SSO1 and sso1Δ with and without Mss4p overexpression give P values of 0.13 and 0.20, respectively.
Introducing positively charged residues in Sso2p Habc promotes sporulation in sso1Δ and increases PI(4,5)P2 binding in vitro.
A comparison of Sso1p and Sso2p Habc domains reveals that there are two positively charged arginine residues present in the Habc domain of Sso1p not present in Sso2p (Fig. 4A). To determine if these residues are important for PI(4,5)P2 binding and function, we generated a single-copy SSO1 plasmid bearing mutations at these sites for expression of the equivalent Sso2p residues (Sso1pR45A R63D). We also generated SSO2 single-copy and multicopy plasmids bearing reciprocal mutations for expression of Sso1p residues (Sso2pA49R D67R).
We analyzed cells with a deletion of SSO1 and bearing these plasmids for their ability to rescue sporulation. The Sso1pR45A R63D plasmid rescued the sporulation defect as did wild-type Sso1p, indicating that these mutations have no effect on Sso1p function in vivo (Table 6). However, the multicopy Sso2pA49R D67R plasmid showed a greater than twofold increase in sporulation efficiency compared to the wild-type SSO2 plasmid (paired t test, P value of <0.02) (Table 6). The single-copy Sso2p mutant plasmid was unable to rescue sporulation (Table 6). These results indicate that introduction of positively charged residues into Sso2p Habc can increase its ability to rescue the sso1Δ sporulation defect but that these residues alone are not sufficient to mediate Sso1p Habc domain sporulation-specific function.
To examine whether mutations of these residues in the Habc domain affect binding to PI(4,5)P2 liposomes, we also performed lipid-binding assays with GST fused to the Sso1p Habc domain with the R45A R63D mutation (GST-Sso1p HabcR45A R63D) and GST-Sso2p HabcA49R D67R. Consistent with the in vivo sporulation efficiency data, GST-Sso1p HabcR45A R63D was able to bind PI(4,5)P2 liposomes at a level similar to that of GST-Sso1p Habc (95.2% ± 0.75% standard error of the mean [SEM]) However, GST-Sso2p HabcA49R D67R displayed an increased ability to bind PI(4,5)P2 liposomes compared to GST-Sso2p Habc (27.5% ± 1.2% SEM versus 12.6% ± 3.2% SEM). This binding difference was statistically significant; an unpaired t test gave a P value of <0.02. These results indicate that introduction of positively charged arginine residues in Sso2p Habc increases PI(4,5)P2 binding although replacing these residues in Sso1p indicates that the PI(4,5)P2 binding region has yet to be defined. Furthermore, these results support the hypothesis that SNARE-PI(4,5)P2 interactions are important for PSM fusion events.
DISCUSSION
SNARE-mediated fusion for PSM formation.
Although SSO1 and SSO2 are related genes arising from an evolutionarily recent duplication event (1, 2) and are functionally redundant during vegetative growth for exocytic fusion, Sso1p is specifically required for PSM synthesis during sporulation (2, 21). Similarly, two SNAP-25 homologs, Sec9p and Spo20p, are present in yeast; the former is required for exocytic function, and the latter is involved in PSM formation (5, 41). The presence of two syntaxin and two SNAP-25 homologs in yeast is consistent with the SNARE hypothesis, which postulates that a combination of different SNARE family members interacting in discrete SNAREpins may contribute to compartmental fusion specificity (53).
In vitro fusion assays also suggest interaction specificity between the different SNAREs. The Ssop/Sec9p binary complex forms a more efficient SNAREpin than the Ssop/Spo20p complex, with the Sso2p/Spo20p complex the least fusogenic of the four possible t-SNARE complexes (30). Mutation of a glutamine within the ionic layer of the H3 motif of Sso1p impaired its ability to interact with Spo20p and resulted in a decrease in sporulation efficiency, suggesting that small changes in related SNARE helices can affect interactions between cognate SNAREs and hence effect intracellular fusion specificity (59). However, the glutamine residue within the H3 of Sso1p is also present in Sso2p, suggesting that this interaction alone cannot explain the specificity of Sso1p for PSM formation.
Lipid-SNARE interactions are essential for vesicle fusion.
The activities of multiple lipid-modifying enzymes are required during sporulation, in particular, PLD and phosphoinositide kinases (9). The finding that both PA and PI(4,5)P2 interact with Sso1p to promote vesicle fusion events during PSM formation suggests that these lipids are important for SNARE function and specificity. The juxtamembrane region of the Sso1p H3 domain is necessary for PA binding, and inclusion of PA into Sso1p/Sec9p proteoliposomes increases the rate of their fusion to Snc1p proteoliposomes in vitro (30). We demonstrate that the H3 domains of both Sso1p and Sso2p are sufficient for binding liposomes including PA or phosphorylated phosphoinositides. The mammalian Sso1p homolog syntaxin 1A also has PA and phosphoinositide-binding ability (28). These data suggest that fusogenic lipid binding by the H3 domain is a conserved feature of syntaxins and that this interplay gives rise to efficient fusion events.
The Habc motif of syntaxin forms a pseudo-SNAREpin with the H3 domain, maintaining the syntaxin in a closed conformation, which prevents its cognate SNAREs from interacting with the syntaxin (10, 14, 42). Analyses of Sso1/2p chimeras indicated that the Sso1p Habc region is important for sporulation (43). Here, we demonstrate greater in vitro PI(4,5)P2-binding ability for the Habc motif of Sso1p than for the equivalent region in Sso2p. While Sso1p Habc bound three PIP2 species similarly, PI(3,5)P2 is associated with vacuolar membranes and endosomal trafficking, and PI(3,4)P2 has not been detected in S. cerevisiae (8, 13, 50, 54). It is therefore likely that the PIP2 species with which the Sso1p Habc domain interacts in vivo during PSM formation is PI(4,5)P2. This phosphoinositide has previously been shown to localize to developing PSMs and activate Spo14 PLD activity (45, 48).
No significant difference in binding is observed between Sso1p-TMD and Sso2p-TMD for PI(4,5)P2, suggesting that the Habc domain does not contribute additional binding capacity to Sso1p-TMD in vitro. Previous studies have demonstrated that in solution isolated syntaxins lacking their carboxy TMDs form organized coiled-coil regions, suggesting that the Habc domain is interacting with the H3 domain in a closed conformation (10, 14, 42). It is therefore likely that the GST-Sso1/2p-TMD binds to PA and phosphoinositide-containing liposomes while in this conformation, whereas both GST-Ssop Habc fusion proteins are likely unstructured in solution, enabling access of the Sso1p Habc domain to PIP2 in the binding assays.
Analyses of Sso1p and Sso2p Habc mutants in vivo and in vitro indicate that mutation of two positively charged arginine residues in Sso1p Habc to the corresponding Sso2p residues has no effect on either sporulation efficiency or in vitro PI(4,5)P2 binding. However, the reciprocal mutations in Sso2p Habc effected an increase in sporulation efficiency when the Sso2p mutant was overexpressed; additionally, in vitro PI(4,5)P2 binding was improved. These findings indicate that while the arginine residues in Sso1p Habc are not necessary to promote either sporulation or PI(4,5)P2 binding, introduction of these positively charged residues into Sso2p Habc improves both sporulation and PI(4,5)P2 binding over that of wild-type Sso2p. Although the PI(4,5)P2 binding site(s) of Sso1p Habc have yet to be defined, these data support the correlation between in vivo sporulation and in vitro PI(4,5)P2 binding.
Neither the wild-type single-copy SSO2 nor the mutant single-copy plasmid rescued sporulation in sso1Δ cells, indicating that additional region(s) of Sso1p are required for sporulation-specific function. A previous study of Sso1p/Sso2p chimeras indicated that both the Habc region and the 3′ untranslated region of Sso1p are required for sporulation (43). Curiously, this is not a consequence of expression levels (43). Thus, it appears that Sso1p has evolved more than one mechanism to specify PSM fusion events.
The relationship between PI(4,5)P2 and PLD-generated PA.
Cells depleted of the PI(4)P 5-kinase Mss4p display sporulation defects, including a reduction in PLD activity (48). Here, we show that PSM formation is largely blocked in the mss4-ts mutant at restrictive temperature, suggesting a role for PI(4,5)P2 in PSM generation. Interestingly, overexpression of Mss4p in sso1Δ cells induced in sporulation medium results in PSM formation. Overexpression of Sso2p in sso1Δ results in a partial rescue of sporulation, which is improved more than twofold when Mss4p is also overexpressed. Additionally, the Sso2p Habc domain can bind liposomes with increasing mole percentages of PI(4,5)P2 in vitro. These data suggest that high levels of PI(4,5)P2 can promote Sso2p activity in PSM fusion events in vivo in sso1Δ cells. The PSMs forming in both wild-type and sso1Δ cells overexpressing Mss4p alone appear small and misshapen compared to those developing in wild-type cells, suggesting that increased levels of PI(4,5)P2 in vivo cause delayed PSM formation initiation and/or affects PSM expansion regulation.
It is unlikely that increased levels of PI(4,5)P2 are stimulating Spo14p activity as in vivo measurements of the PLD activity of sso1Δ cells showed no significant difference in the presence or absence of Mss4p overexpression. A caveat to this result is that Mss4p is expressed from a high-copy-number plasmid, which is lost in some portion of the cells due to lack of selection during cell cycle synchronization and induction in sporulation medium; however, it remains likely that elevated PI(4,5)P2 levels are present throughout the course of these assays. These data suggest that the partial rescue by PI(4,5)P2 in vivo is not mediated through Spo14p-generated PA and that this phosphoinositide has an additional regulatory role in PSM formation.
We propose that PI(4,5)P2 is involved in mediating the change from a closed to open conformation of Sso1p in wild-type cells to allow the formation of a functional SNAREpin comprising Sso1p, Spo20p, and Snc1/2p at the modified SPB. Sso2p, while at the SPB during sporulation, may largely remain in the closed conformation due to weaker interaction between PI(4,5)P2 and the Sso2p Habc region and therefore does not normally participate in fusions at the PSM (Fig. 6). As Sso1p recruitment to the developing PSM is not dependent on Spo14p PLD activity, this work also suggests that Spo14p-generated PA serves as a fusogenic lipid during PSM precursor vesicle fusions. Taken together, our data support the idea that both proteins and lipids are active participants in membrane fusion events, with both classes of molecules acting upon each other to promote fusion.
FIG. 6.
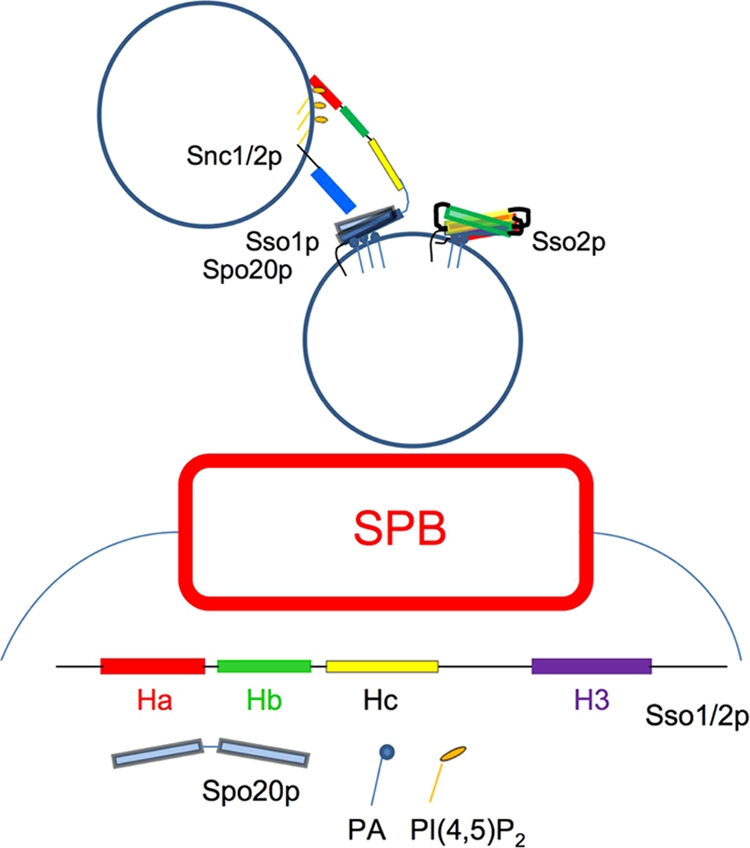
Schematic of vesicle fusion during PSM formation. The Habc domain of Sso1p on a PSM precursor vesicle interacts with PI(4,5)P2 from an incoming vesicle, enabling Spo20p and Snc1/2p access to its H3 motif for formation of the fusogenic SNAREpin while Sso2p remains in an conformationally inactive state.
Acknowledgments
We thank A. Neiman (State University of New York, Stony Brook, NY) for the generous gifts of SK-1-derived strains and plasmids, J. Jäntti (Institute of Biotechnology, University of Helsinki, Helsinki, Finland) for pGEX-SSO plasmids, the Nunnari laboratory (University of California, Davis) for help with the liposome binding assays, and J. Trimmer for useful advice, helpful discussions, and support.
Footnotes
Published ahead of print on 5 June 2009.
REFERENCES
- 1.Aalto, M. K., S. Keranen, and H. Ronne. 1992. A family of proteins involved in intracellular transport. Cell 68181-182. [DOI] [PubMed] [Google Scholar]
- 2.Aalto, M. K., H. Ronne, and S. Keranen. 1993. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 124095-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonin, W., D. Fasshauer, S. Becker, R. Jahn, and T. R. Schneider. 2002. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol. 9107-111. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, M. K., N. Calakos, and R. H. Scheller. 1992. Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science 257255-259. [DOI] [PubMed] [Google Scholar]
- 5.Brennwald, P., B. Kearns, K. Champion, S. Keranen, V. Bankaitis, and P. Novick. 1994. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell 79245-258. [DOI] [PubMed] [Google Scholar]
- 6.Buser, C. A., and Stuart McLaughlin (ed.). 1998. Ultracentrifugation technique for measuring the binding of peptides and proteins to sucrose-loaded phospholipid vesicles. Humana Press Inc., Totowa, NJ. [DOI] [PubMed]
- 7.Connolly, J. E., and J. Engebrecht. 2006. The Arf-GTPase-activating protein Gcs1p is essential for sporulation and regulates the phospholipase D Spo14p. Eukaryot. Cell 5112-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooke, F. T., S. K. Dove, R. K. McEwen, G. Painter, A. B. Holmes, M. N. Hall, R. H. Michell, and P. J. Parker. 1998. The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr. Biol. 81219-1222. [DOI] [PubMed] [Google Scholar]
- 9.Deng, L., J. Nagasawa, Y. Ono, Y. Ishikawa, T. Kakihara, R. Fukuda, and A. Ohta. 2008. Manipulation of major membrane lipid synthesis and its effects on sporulation in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 722362-2368. [DOI] [PubMed] [Google Scholar]
- 10.Dulubova, I., S. Sugita, S. Hill, M. Hosaka, I. Fernandez, T. C. Sudhof, and J. Rizo. 1999. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 184372-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elledge, S. J., and R. W. Davis. 1998. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene 70303-312. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez, I., J. Ubach, I. Dulubova, X. Zhang, T. C. Sudhof, and J. Rizo. 1998. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell 94841-849. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Borja, M., R. Wubbolts, J. Calafat, H. Janssen, N. Divecha, S. Dusseljee, and J. Neefjes. 1999. Multivesicular body morphogenesis requires phosphatidyl-inositol 3-kinase activity. Curr. Biol. 955-58. [DOI] [PubMed] [Google Scholar]
- 14.Fiebig, K. M., L. M. Rice, E. Pollock, and A. T. Brunger. 1999. Folding intermediates of SNARE complex assembly. Nat. Struct. Biol. 6117-123. [DOI] [PubMed] [Google Scholar]
- 15.Fratti, R. A., and W. Wickner. 2007. Distinct targeting and fusion functions of the PX and SNARE domains of yeast vacuolar Vam7p. J. Biol. Chem. 28213133-13138. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda, R., J. A. McNew, T. Weber, F. Parlati, T. Engel, W. Nickel, J. E. Rothman, and T. H. Sollner. 2000. Functional architecture of an intracellular membrane t-SNARE. Nature 407198-202. [DOI] [PubMed] [Google Scholar]
- 17.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2163-167. [DOI] [PubMed] [Google Scholar]
- 18.Hong, W. 2005. SNAREs and traffic. Biochim. Biophys. Acta 1744493-517. [PubMed] [Google Scholar]
- 19.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James, D. J., C. Khodthong, J. A. Kowalchyk, and T. F. Martin. 2008. Phosphatidylinositol 4,5-bisphosphate regulates SNARE-dependent membrane fusion. J. Cell Biol. 182355-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jantti, J., M. K. Aalto, M. Oyen, L. Sundqvist, S. Keranen, and H. Ronne. 2002. Characterization of temperature-sensitive mutations in the yeast syntaxin 1 homologues Sso1p and Sso2p, and evidence of a distinct function for Sso1p in sporulation. J. Cell Sci. 115409-420. [DOI] [PubMed] [Google Scholar]
- 22.Jenkins, G. M., and M. A. Frohman. 2005. Phospholipase D: a lipid centric review. Cell Mol. Life Sci. 622305-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jun, Y., and W. Wickner. 2007. Assays of vacuole fusion resolve the stages of docking, lipid mixing, and content mixing. Proc. Natl. Acad. Sci. USA 10413010-13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jun, Y., H. Xu, N. Thorngren, and W. Wickner. 2007. Sec18p and Vam7p remodel trans-SNARE complexes to permit a lipid-anchored R-SNARE to support yeast vacuole fusion. EMBO J. 264935-4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knop, M., and K. Strasser. 2000. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 193657-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kooijman, E. E., V. Chupin, B. de Kruijff, and K. N. Burger. 2003. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic 4162-174. [DOI] [PubMed] [Google Scholar]
- 27.Kooijman, E. E., V. Chupin, N. L. Fuller, M. M. Kozlov, B. de Kruijff, K. N. Burger, and P. R. Rand. 2005. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry 442097-2102. [DOI] [PubMed] [Google Scholar]
- 28.Lam, A. D., P. Tryoen-Toth, B. Tsai, N. Vitale, and E. L. Stuenkel. 2008. SNARE-catalyzed fusion events are regulated by Syntaxin1A-lipid interactions. Mol. Biol. Cell 19485-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, F., F. Pincet, E. Perez, W. S. Eng, T. J. Melia, J. E. Rothman, and D. Tareste. 2007. Energetics and dynamics of SNAREpin folding across lipid bilayers. Nat. Struct. Mol. Biol. 14890-896. [DOI] [PubMed] [Google Scholar]
- 30.Liu, S., K. A. Wilson, T. Rice-Stitt, A. M. Neiman, and J. A. McNew. 2007. In vitro fusion catalyzed by the sporulation-specific t-SNARE light-chain Spo20p is stimulated by phosphatidic acid. Traffic 81630-1643. [DOI] [PubMed] [Google Scholar]
- 31.Malsam, J., S. Kreye, and T. H. Sollner. 2008. Membrane fusion: SNAREs and regulation. Cell Mol. Life Sci. 652814-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMahon, H. T., and J. L. Gallop. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438590-596. [DOI] [PubMed] [Google Scholar]
- 33.McMaster, C. R. 2001. Lipid metabolism and vesicle trafficking: more than just greasing the transport machinery. Biochem. Cell Biol. 79681-692. [DOI] [PubMed] [Google Scholar]
- 34.McNew, J. A. 2008. Regulation of SNARE-mediated membrane fusion during exocytosis. Chem. Rev. 1081669-1686. [DOI] [PubMed] [Google Scholar]
- 35.Melia, T. J., D. You, D. C. Tareste, and J. E. Rothman. 2006. Lipidic antagonists to SNARE-mediated fusion. J. Biol. Chem. 28129597-29605. [DOI] [PubMed] [Google Scholar]
- 36.Mima, J., C. M. Hickey, H. Xu, Y. Jun, and W. Wickner. 2008. Reconstituted membrane fusion requires regulatory lipids, SNAREs and synergistic SNARE chaperones. EMBO J. 272031-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morishita, M., and J. Engebrecht. 2005. End3p-mediated endocytosis is required for spore wall formation in Saccharomyces cerevisiae. Genetics 1701561-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morishita, M., R. Mendonsa, J. Wright, and J. Engebrecht. 2007. Snc1p v-SNARE transport to the prospore membrane during yeast sporulation is dependent on endosomal retrieval pathways. Traffic 81231-1245. [DOI] [PubMed] [Google Scholar]
- 39.Nakanishi, H., P. de los Santos, and A. M. Neiman. 2004. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell 151802-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakanishi, H., M. Morishita, C. L. Schwartz, A. Coluccio, J. Engebrecht, and A. M. Neiman. 2006. Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J. Cell Sci. 1191406-1415. [DOI] [PubMed] [Google Scholar]
- 41.Neiman, A. M. 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 14029-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholson, K. L., M. Munson, R. B. Miller, T. J. Filip, R. Fairman, and F. M. Hughson. 1998. Regulation of SNARE complex assembly by an N-terminal domain of the t-SNARE Sso1p. Nat. Struct. Biol. 5793-802. [DOI] [PubMed] [Google Scholar]
- 43.Oyen, M., J. Jantti, S. Keranen, and H. Ronne. 2004. Mapping of sporulation-specific functions in the yeast syntaxin gene SSO1. Curr. Genet. 4576-82. [DOI] [PubMed] [Google Scholar]
- 44.Riedel, C. G., M. Mazza, P. Maier, R. Korner, and M. Knop. 2005. Differential requirement for phospholipase D/Spo14 and its novel interactor SmaI for regulation of exocytotic vesicle fusion in yeast meiosis. J. Biol. Chem. 28037846-37852. [DOI] [PubMed] [Google Scholar]
- 45.Rose, K., S. A. Rudge, M. A. Frohman, A. J. Morris, and J. Engebrecht. 1995. Phospholipase D signaling is essential for meiosis. Proc. Natl. Acad. Sci. USA 9212151-12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudge, S. A., A. J. Morris, and J. Engebrecht. 1998. Relocalization of phospholipase D activity mediates membrane formation during meiosis. J. Cell Biol. 14081-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudge, S. A., T. R. Pettitt, C. Zhou, M. J. Wakelam, and J. A. Engebrecht. 2001. SPO14 separation-of-function mutations define unique roles for phospholipase D in secretion and cellular differentiation in Saccharomyces cerevisiae. Genetics 1581431-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rudge, S. A., V. A. Sciorra, M. Iwamoto, C. Zhou, T. Strahl, A. J. Morris, J. Thorner, and J. Engebrecht. 2004. Roles of phosphoinositides and of Spo14p (phospholipase D)-generated phosphatidic acid during yeast sporulation. Mol. Biol. Cell 15207-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sciorra, V. A., S. A. Rudge, J. Wang, S. McLaughlin, J. Engebrecht, and A. J. Morris. 2002. Dual role for phosphoinositides in regulation of yeast and mammalian phospholipase D enzymes. J. Cell Biol. 1591039-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaw, J. D., H. Hama, F. Sohrabi, D. B. DeWald, and B. Wendland. 2003. PtdIns(3,5)P2 is required for delivery of endocytic cargo into the multivesicular body. Traffic 4479-490. [DOI] [PubMed] [Google Scholar]
- 51.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 6731-40. [DOI] [PubMed] [Google Scholar]
- 53.Sollner, T., S. W. Whiteheart, M. Brunner, H. Erdjument-Bromage, S. Geromanos, P. Tempst, and J. E. Rothman. 1993. SNAP receptors implicated in vesicle targeting and fusion. Nature 362318-324. [DOI] [PubMed] [Google Scholar]
- 54.Strahl, T., and J. Thorner. 2007. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta 1771353-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sutton, R. B., D. Fasshauer, R. Jahn, and A. T. Brunger. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature 395347-353. [DOI] [PubMed] [Google Scholar]
- 56.Weber, T., B. V. Zemelman, J. A. McNew, B. Westermann, M. Gmachl, F. Parlati, T. H. Sollner, and J. E. Rothman. 1998. SNAREpins: minimal machinery for membrane fusion. Cell 92759-772. [DOI] [PubMed] [Google Scholar]
- 57.Weimbs, T., S. H. Low, S. J. Chapin, K. E. Mostov, P. Bucher, and K. Hofmann. 1997. A conserved domain is present in different families of vesicular fusion proteins: a new superfamily. Proc. Natl. Acad. Sci. USA 943046-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, Y., F. Zhang, Z. Su, J. A. McNew, and Y. K. Shin. 2005. Hemifusion in SNARE-mediated membrane fusion. Nat. Struct. Mol. Biol. 12417-422. [DOI] [PubMed] [Google Scholar]
- 59.Yang, H. J., H. Nakanishi, S. Liu, J. A. McNew, and A. M. Neiman. 2008. Binding interactions control SNARE specificity in vivo. J. Cell Biol. 1831089-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]