Abstract
The Ndt80p transcription factor modulates azole tolerance in Candida albicans by controlling the expression of the gene for the drug efflux pump Cdr1p. To date, the contribution of this transcriptional modulator to drug tolerance is not yet well understood. Here, we investigate the role of Ndt80p in mediating fluconazole tolerance by determining its genome-wide occupancy using chromatin immunoprecipitation coupled to high-density tiling arrays. Ndt80p was found to bind a large number of gene promoters with diverse biological functions. Gene ontology analysis of these Ndt80p targets revealed a significant enrichment in gene products related to the cell wall, carbohydrate metabolism, stress responses, hyphal development, multidrug transport, and the cell cycle. Ndt80p was found on the promoters of ergosterol biosynthesis genes, including on the azole target Erg11p. Additionally, expression profiling was used to identify fluconazole-responsive genes that require Ndt80p for their proper expression. We found that Ndt80p is crucial for the expression of numerous fluconazole-responsive genes, especially genes involved in ergosterol metabolism. Therefore, by combining genome-wide location and transcriptional profiling, we have characterized the Ndt80p fluconazole-dependent regulon and demonstrated the key role of this global transcriptional regulator in modulating sterol metabolism and drug resistance in C. albicans.
Members of the Candida genus are the principal etiological agents of nosocomial fungal infections, with Candida albicans being the most commonly encountered species (15, 19, 38). C. albicans is a major cause of morbidity and mortality in bloodstream infections, particularly for immunosuppressed patients. This pathogen can also colonize various biomaterials and readily forms dense biofilms that are resistant to most known antifungal agents. The overall mortality for patients with candidemia is greater than 40% (12, 30), marking this opportunistic fungus as a serious public health menace.
The arsenal of clinically active antifungal compounds is limited. The eukaryotic nature of C. albicans makes it similar to its human host, thus reducing the number of potential drug targets (11). Most Candida infections can be treated with inhibitors that target either the biosynthesis of ergosterol, the main sterol of fungal membranes, or the biosynthesis of the key component of the fungal cell wall, (1,3)-β-d-glucan. However, the emergence of drug resistance in this pathogen sometimes reduces the effectiveness of antifungal drugs. C. albicans has evolved different resistance mechanisms to bypass the inhibitory effects of azole drugs (11), mainly through the overexpression of the ATP-binding cassette (ABC) transporter Cdr1p or Cdr2p or the major facilitator superfamily (MFS) transporter Mdr1p (36). Recently, Morschhauser et al. (32) have elucidated a mechanism that results in the overexpression of MDR1 in clinical isolates. Drug resistance was caused by a gain-of-function mutation in a zinc cluster transcription factor (TF) called Mrr1p. Inactivation of MRR1 abolishes the resistance of Mdr1p-overexpressing strains. Gain-of-function mutations in the zinc cluster TF Tac1p have also been shown to result in Cdr1p or Cdr2p overexpression (9, 10). Genome-wide location analysis using chromatin immunoprecipitation coupled to microarrays (ChIP-chip) showed that, in addition to Cdr1p and Cdr2p, Tac1p also targets the promoters of other resistance genes, such as the integral membrane flippase Rta3p and stress-related genes (29). Furthermore, Dunkel et al. (14) showed that azole resistance is linked to a gain-of-function mutation in the sterol metabolism regulator Upc2p, leading to constitutive expression of ergosterol biosynthesis genes. Genome-wide occupancy of Upc2p confirmed the direct binding of this TF to ergosterol genes, including the azole target Erg11p (45).
The TF Ndt80p has been identified as a positive regulator of CDR1 by screening a C. albicans expression library in a Saccharomyces cerevisiae strain carrying a CDR1-lacZ reporter construct (4). Furthermore, it has been shown that NDT80 is crucial for azole drug tolerance and activation of CDR1 expression in response to miconazole (4). The homologue of Ndt80p in S. cerevisiae regulates gene expression of middle sporulation genes and is required for exit from pachytene and for full meiotic recombination (23, 35, 37, 44). Thus far, the accurate role of this TF in regulating drug resistance in C. albicans remains to be assessed. To gain a more complete understanding of the role of Ndt80p in mediating azole resistance in C. albicans, we set out to investigate its genomic occupancy using genome-wide location analysis (ChIP-chip). By combining genome-wide location and expression analyses, we have characterized the Ndt80p fluconazole-dependent regulon for the first time and elucidated the key role of this TF as an activator of ergosterol metabolism genes.
MATERIALS AND METHODS
C. albicans strain construction, plasmids, and media.
The strains used in this study are listed in Table 1. Using the following general propagation and maintenance conditions, the strains were cultured at 30°C in yeast-peptone-dextrose (YPD) medium supplemented with uridine (2% Bacto peptone, 1% yeast extract, 2% dextrose, and 50 μg/ml uridine, with the addition of 2% agar for solid medium). For transformation, the one-step lithium acetate protocol (5) was used, with the modification that cells were incubated overnight at 30°C with the transforming DNA before plating.
TABLE 1.
Candida albicans strains used in this study
| Strain | Parental strain | Genotype or description | Reference |
|---|---|---|---|
| SC5314 | Clinical isolate | Gillum et al. (20a) | |
| BWP17 | SC5314 | ura3::λimm434arg4::hisG his1::hisG | Wilson et al. (43a) |
| ura3::λimm434arg4::hisG his1::hisG | |||
| AS30 | BWP17 | ura3::λimm434arg4::hisG his1::hisG ndt80::HIS1 | This study |
| ura3::λimm434arg4::hisG his1::hisG NDT80 | |||
| AS31 | AS30 | ura3::λimm434arg4::hisG his1::hisG ndt80::HIS1 | This study |
| ura3::λimm434arg4::hisG his1::hisG ndt80::URA3 | |||
| AS32 | AS31 | ura3::λimm434arg4::hisG his1::hisG ndt80::HIS1 | This study |
| ura3::λimm434arg4::hisG his1::hisG ndt80::ura3 | |||
| AS33 | AS32 | ura3::λimm434arg4::hisG his1::hisG::pURA3-NDT80 ndt80::HIS1 | This study |
| ura3::λimm434arg4::hisG his1::hisG ndt80 | |||
| AS34 | BWP17 | ura3::λimm434arg4::hisG his1::hisG NDT80-TAP-URA3 | This study |
| ura3::λimm434arg4::hisG his1::hisG NDT80 |
NDT80 was TAP tagged in vivo with a TAP-URA3 PCR product, as described by Lavoie et al. (26). Transformants were selected on YPD-uridine plates, and correct integration of the TAP tag was checked by PCR and sequencing. The NDT80-TAP construct was fully functional, based on complementation of the fluconazole sensitivity phenotype. Deletion of the nontagged allele in NDT80-TAP strain AS34 revealed that this strain has sensitivity comparable to both the single knockout and parental strains (data not shown).
Gene disruption was accomplished as follows. Plasmid pFA-HIS1 was used as the template to prepare the HIS1-PCR cassette, and plasmid pFA-URA3 was used to prepare the URA3 PCR cassette, as previously described by Gola et al. (21). Ndt80 was deleted in two steps. In the first step, one allele was replaced by homologous recombination with a PCR cassette containing the HIS1 gene. The BWP17 strain was transformed with the PCR fragments, and the cells were plated onto YPD plates minus histidine. Genomic DNA of the positive colonies was analyzed by PCR for the proper integration site of the cassette. The strain produced by deleting one allele of Ndt80 was named AS30. The second allele disruption was accomplished as done for the first allele using the URA3 PCR cassette, and the cells were plated onto YPD plates minus uridine. The colonies were again analyzed by PCR to identify the proper integration site. The complete deletion of candidate genes was confirmed by PCR, using primers internal to the recombination sites amplifying open reading frame (ORF) regions. The strain produced by deleting both alleles from Ndt80 (named AS31) was treated with 5-fluoroorotic acid to recycle the URA3 marker, and the resulting uridine-negative strain was named AS32.
The absence of aneuploidy was confirmed for mutant and revertant strains using comparative genomic hybridization. C. albicans genomic DNA was isolated from a saturated overnight culture with the Qiagen genomic DNA extraction kit and labeled with either Cy3 or Cy5 dye with the BioPrime CGH labeling kit (Invitrogen). Unincorporated nucleotides were removed with Qiagen PCR columns, and the labeled probes were then hybridized to DNA microarrays, as described by Nantel et al. (34).
For reintegration experiments, the NDT80 gene was reintegrated into the null mutant ndt80 strain. The wild-type NDT80 gene was amplified from genomic DNA using oligonucleotides REVF1 and REVR1 (Table 2) and Expand high-fidelity polymerase (Roche). The PCR fragment was digested with restriction enzymes ClaI and XhoI and cloned in the same sites of the CIp10 vector (33). The plasmid was sequenced to confirm the integrity of the NDT80 gene. Plasmid CIp10-NDT80 was digested with the StuI restriction enzyme and used to transform the ndt80 mutant strain (AS32). The uridine-positive colonies were analyzed by PCR, and the obtained wild-type fragment confirmed the reintegration of the NDT80 gene.
TABLE 2.
Primers used in this study
| Primer name | Primer sequence | Description | Purpose |
|---|---|---|---|
| NDT80 F1 | GCAGATCTCCTTCAAGCTACCACAAAGATAGAACTTCTGGGTATAGGGCTACAAACACCCCAACCCCTACTCCTCCACAGGGTCGACGGATCCCCGGGTT | 5′ NDT80 PCR cassette | C-terminal TAP tagging |
| NDT80 R1 | CTAAAAATTTTTTTGGTGCGGGTGATTGGTACACGACACCTGGCTCTGATATTTTGTGGGGATGGGGCAGTTACGGCTATAAATAAGATTACTATTACATCGATGAATTCGAGCTCGTT | 3′ NDT80 PCR cassette | C-terminal TAP tagging |
| NDT80 ExF1 | TCCTTCAGAGTTGCCTGACC | NDT80, forward—external | Validation of 5′ cassette insertion |
| NDT80 ExR1 | AGCCACCAGCTAGACCTGAC | NDT80, reverse—external | Validation of 3′ cassette insertion |
| ASNdt80 KOF1 | ACAACAACAACAGCAACACCACCACCAGCTTCCTCACCATCCTATTCACTATCATGGTATTGCTCAGCAACAACAGTCCCAGCTTCCTCATTTCGCTATGGAAGCTTCGTACGCTGCAGGTC | 5′ NDT80 PCR cassette | Gene replacement |
| ASNdt80 KOR1 | TGTGTAATTATAATACAAAATTTTTTTATTTACTTTAAACTTTAAAATCAACCTTTCTTCGTCATCATCAAAAAAAAAAAAAAATCTATAGTTTTGCTTATCTGATATCATCGATGAATTCGAG | 3′ NDT80 PCR cassette | Gene replacement |
| NDT80 ExF2 | TGCCCAACGAAGATCCTAAC | NDT80, forward—external | Validation of 5′ NDT80 replacement |
| NDT80 ExR2 | CACGACACCTGGCTCTGATA | NDT80, reverse—external | Validation of 3′ NDT80 replacement |
| Ndt80 InS1 | AGACCAAGCTGACGCTCAAT | NDT80, forward—internal | Validation of 5′ NDT80 replacement |
| Ndt80 InR1 | TTGACAGTCTCGTGGTCAGGC | NDT80, reverse—internal | Validation of 5′ NDT80 replacement |
| U1 | TTGAAGGATTAAAACAGGGAGC | URA3, forward | Validation of 3′ cassette insertion |
| U2 | ATACCTTTTACCTTCAATATCTGG | URA3, reverse | Validation of 5′ cassette insertion |
| H1 | TTTAGTCAATCATTTACCAGACCG | HIS1, forward | Validation of 3′ cassette insertion |
| H2 | TCTATGGCCTTTAACCCAGCTG | HIS1, reverse | Validation of 5′ cassette insertion |
| NDT80-RevF1 | CCTTTCCCATCTCCATATTACCATCTTC | NDT80, forward | Revertant construct |
| NDT80-RevR1 | TACTGTGGAGGAGTAGGGGTTG | NDT80, reverse | Revertant construct |
Tiling array design.
Starting with sequences from the C. albicans Genome Assembly 21 (41) and the mating type-like alpha locus (24), we extracted a continuous series of 242,860 60-bp oligonucleotides, each overlapping by 1 bp. We then eliminated 2,062 probes containing stretches of 13 or more A or T nucleotides. The remaining 240,798 sequences were then used to produce antisense whole-genome tiling arrays using the Agilent Technologies eArray service (https://earray.chem.agilent.com/earray/).
Whole-genome location profiling by ChIP-chip and data analysis.
Cells were grown to an optical density at 600 nm (OD600) of 2 in 40 ml of YPD. The subsequent steps of DNA cross-linking, DNA shearing, ChIP, and DNA labeling with Cy dyes were conducted exactly as described by Lavoie et al. (26). Tiling arrays were cohybridized with tagged immunoprecipitated (Cy5-labeled) and mock immunoprecipitated (untagged BWP17 strain; Cy3-labeled) DNA samples. Microarray hybridizing, washing, and scanning were performed, as described by Nantel et al. (34). Prehybridization and hybridization solutions consisted of DIG Easy Hyb solution (Roche Diagnostics, Mannheim, Germany), with 0.45% salmon sperm DNA and 0.45% yeast tRNA. The hybridization was carried out at 42°C for 20 h in a SlideBooster Hyb chamber SB800 (Advalytix, Brunnthal, Germany), with regular microagitation of the sample. Slides were washed once in 1.0% SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), once in 0.2% sodium dodecyl sulfate at 42°C for 5 min, twice in 0.1% SSC, twice in 0.2% sodium dodecyl sulfate at 42°C for 5 min, and once in 0.1% SSC at 24°C for 5 min, followed by four rinses in 0.1% SSC. Chips were air dried before being scanned using a ScanArray Lite microarray scanner (PerkinElmer). Fluorescence intensities were quantified using ImaGene software (BioDiscovery, Inc.), background corrected, and normalized for signal intensity (using Lowess normalization). The significance cutoff was determined using the distribution of log ratios for each factor. It was set at two standard deviations from the mean of log-transformed enrichments. Values shown are an average of two biological replicates derived from independently isolated transformants of tagged and mock constructs. Peak detection was performed using Gaussian edge detection applied to the smoothed probe signal curve, as described by Tuch et al. (39).
Motif discovery.
For de novo identification of the consensus-binding site, the sequences were analyzed with the Multiple EM for Motif Elucidation (MEME) program (http://meme.sdsc.edu/meme/website/meme.html). For sequences of 150 bp upstream and downstream, the detected signal peak was used. The motifs were allowed to have any sequence and length (between 6 and 50 bp) and could be present anywhere in the sequence.
Gene expression profiling.
Overnight cultures of designated C. albicans strains were diluted to a starting OD600 of 0.1 in 100 ml fresh YPD-uridine media. The cultures were incubated with shaking at 30°C to an OD600 of 0.8 and split into two 50-ml cultures. Fluconazole was added to the experimental culture to a final concentration of 100 μg per ml, while an equal volume of dimethyl sulfoxide was added to the control culture. The cultures were incubated for 60 min. Cells were harvested by centrifugation and stored at −80°C.
To extract RNA from cells, samples stored at −80°C were placed on ice, and RNeasy buffer RLT was added to pellets at a ratio of 10:1 (vol/vol) buffer/pellet. The pellet was allowed to thaw in the buffer while being vortexed briefly at high speed. The resuspended pellet was placed back on ice and divided into 1-ml aliquots in 2-ml screw-cap microcentrifuge tubes containing 0.6 ml of 3-mm-diameter acid-washed glass beads. Samples were homogenized five times, for 1 min each, at 4,200 rpm using BeadBeater. Samples were placed on ice for 1 min after each homogenization step. After homogenization, the Qiagen RNeasy protocol was followed, as recommended. Total RNA samples were eluted in RNase-free H2O. RNA quality and integrity were assessed using an Agilent 2100 bioanalyzer.
cDNA labeling and microarray production were performed, as previously described (34). Briefly, 20 μg of total RNA was reverse transcribed using oligo(dT)21 in the presence of Cy3 or Cy5-dCTP (Invitrogen) and SuperScript III reverse transcriptase (Invitrogen). Thereafter, template RNA was degraded by adding 2.5 units RNase H (USB) and 1 μg RNase A (Pharmacia), followed by incubation for 15 min at 37°C. The labeled cDNAs were purified with the QIAquick PCR purification kit (Qiagen). Prior to hybridization, Cy3/Cy5-labeled cDNA was quantified using a NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop) to confirm dye incorporation. Microarray hybridization, washing, and scanning were performed as described for ChIP-chip experiments. Genes with statistically significant changes in transcript abundance in each experiment were identified using a 1.5 cutoff point and Welch's t test with a false discovery rate of less than 5%.
RESULTS
Ndt80p binds to the promoter regions of 23% of C. albicans genes.
Ndt80p was shown to control azole drug tolerance in C. albicans through its regulation of expression of the ABC transporter CDR1 (4). To gain a more complete understanding of the role of Ndt80p in mediating drug resistance, we set out to investigate its genomic occupancy using genome-wide location analysis (ChIP-chip). DNA regions bound by Ndt80p were identified in duplicate ChIP-chip experiments using a high-density tiling array that contains 240,798 unique 60-mer probes, covering most of the C. albicans genome and overlapping by one nucleotide. Reproducible signal peaks were detected, as described in Materials and Methods. Using a twofold enrichment cutoff, Ndt80p was found to be associated with 1,446 genomic regions (see Table S1 in the supplemental material). A detailed analysis of Ndt80p data set occupancy of genomic regions can be found in Fig. S1 in the supplemental material. To assess the reliability of the ChIP-chip results, the immunoprecipitated DNA from two additional independent ChIP experiments was quantified using a single-spot microarray containing 5,423 intergenic and 6,394 intragenic 70-mer oligonucleotide probes, as described in Lavoie et al. (26). A total of 100 probes close to signal peaks detected with the tiling array were randomly selected. We confirmed that 100% (100/100) of these were real Ndt80p ChIP enrichment events, thus suggesting that this data set is of high quality (see Table S2 in the supplemental material).
The vast majority of Ndt80p-bound sites were found at gene promoter regions. Intragenic occupancy was exclusively detected in ORFs that are considered spurious or dubious (2). Among the 90 ORFs bound by Ndt80p, 25 are annotated as spurious, 49 as dubious, and 16 as uncharacterized ORFs not experimentally validated. Dubious and spurious ORFs have no orthologs in other eukaryotes and had expression profiles that were not significantly correlated with those of other genes in the genome. Consequently, those regions are more likely to be intergenic rather than intragenic sequences. We thus conclude that Ndt80p binding occurs specifically in promoter regions.
Ndt80p targets are enriched in genes related to metabolism, stress, development, and drug resistance.
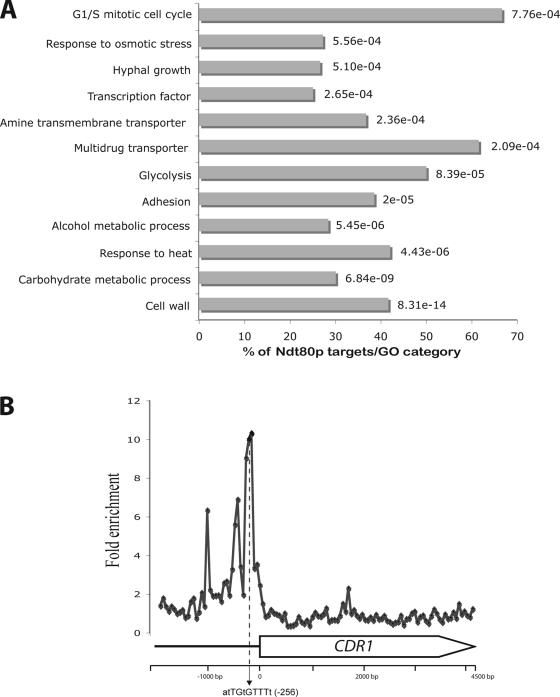
To reveal general functional features of the biological processes modulated by Ndt80p, we conducted a gene ontology (GO) analysis by analyzing the 1,446 genes whose promoters are associated with Ndt80p. For these analyses, all GO categories of genes having an enrichment P value of <0.0001 were selected. As shown in Fig. 1A, we found that Ndt80p binds to a wide range of genes with diverse biological functions. This analysis revealed a significant enrichment in genes related to the cell wall (P = 8.31e−14), carbohydrate metabolism (P = 6.84e−09), response to heat (P = 4.43e−06) and osmotic stress (P = 5.56e−04), alcohol metabolism (P = 5.45e−06), cell adhesion (P = 2e−05), glycolysis (P = 8.39e−05), the G1/S cell cycle (P = 7.76e−04), hyphal growth (P = 5.1e−04), and amine transmembrane transport (P = 2.36e−04). Examination of Ndt80p target genes revealed that they were remarkably enriched in promoter genes coding for transcription factors and transcriptional regulators (P = 2.65e−04) associated with each of these different functions. Transcriptional regulators that were found among Ndt80p targets include regulators of hyphal growth (Efg1p, Nrg1p, Ume6p, Tec1p, Cph2p, Flo8p, Czf1p, Ssn6p, Rfg1p), carbohydrate metabolism (Rgt1p, Tye7p, Gal4p, Mig1p), the cell cycle (Swi4p, Ash1p), lipid metabolism (Ino2p, Opi1p, Ctf1p), translation and amino acid metabolism (Cbf1p, Gln3p, Gcn4p), stress (Cat8p, Hac1p, Cas5p), and general transcriptional regulators (Sua71p, Tbp1p, Stp1p, Stp2p, Stp3p, Stp4p).
FIG. 1.
(A) GO biological process annotation of Ndt80p-bound promoters. The P value was calculated using hypergeometric distribution, as described on the GO Term Finder website (www.candidagenome.org/cgi-bin/GO/goTermFinder). (B) CDR1 promoter occupancy by Ndt80p.
Notably, promoters of multidrug transporter genes were significantly enriched in our data set (P = 2.09e−04). As expected, Ndt80p was found in the promoter region of the ABC transporter CDR1 (Fig. 1B), together with other ABC transporter genes, namely CDR2, CDR4, and orf19.4531. Furthermore, Ndt80p bound to promoter regions of MFS drug transporters, such as MDR1, FLU1, NAG3, and NAG4 as well as the two flippases RTA3 and RTA2. Ndt80p target genes also included other C. albicans drug resistance determinants, such as PDR16, ERG3, and the target of azole antifungal compounds ERG11.
De novo motif analysis of Ndt80p-bound promoters.
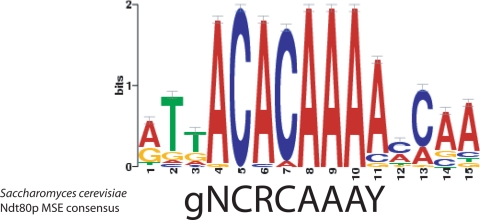
Structural studies demonstrated that the Ndt80p homologue of S. cerevisiae binds to the middle sporulation element (MSE) (18, 25, 31, 37). Based on both statistical and mutational analyses, the MSE has been well defined and corresponds to the 9-bp consensus, as follows: 5′-gNCRCAAAY-3′ (where lowercase letters indicate semiconserved residues, R indicates a purine, N indicates any nucleotide, and Y indicates either a thymine or a cytosine) (25, 37, 43). Using 100 experimentally determined Ndt80-bound loci ranking at the top of highly enriched peaks, we sought to characterize properties of Ndt80p-DNA interaction by assessing motif enrichment to detect de novo consensus of Ndt80p. As shown in Fig. 2, our result illustrated that the top-scoring minimotifs correspond to the consensus, as follows: 5′-NaCacAAAa-3′ (P = 10e−27). This motif is highly similar to that of the S. cerevisiae MSE, suggesting a conservation of the Ndt80p binding site in C. albicans. To assess the occurrence of the core MSE consensus, 5′-ACACAAA-3′, in all Ndt80p targets, we scanned the 1,446 promoters and found that 1,326 of these contain the Ndt80p binding consensus.
FIG. 2.
Logo of the top-scoring motif discovered by MEME using 100 experimentally determined Ndt80-bound loci, ranking at the top of the highly enriched peaks. Saccharomyces cerevisiae MSE consensus is shown on the bottom of the logo.
Identification of target genes whose expression is regulated by Ndt80p in response to fluconazole.
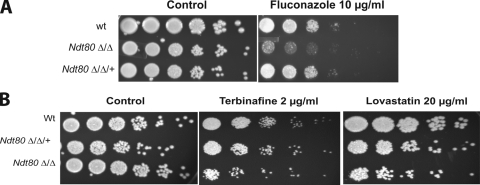
To gain insight into the role of Ndt80p in mediating drug tolerance in C. albicans, we performed transcriptional profiling experiments. We produced our own ndt80 mutant and confirmed the resulting fluconazole hypersensitivity that was first observed by Chen et al. (4) (Fig. 3A). Sensitivity of the ndt80 mutant toward two ergosterol inhibitors, terbinafine and lovastatin, which target Erg1p and Hmg1p, respectively, was also tested. As shown in Fig. 3B, the growth of the ndt80 strain was moderately inhibited in the presence of terbinafine and lovastatin compared to the growth of the revertant and parental strains.
FIG. 3.
The absence of Ndt80p causes sensitivity to ergosterol biosynthesis inhibitors. Serial dilutions (10-fold) of wild-type (wt), revertant, and ndt80 mutant strains were grown on YPD supplemented with fluconazole at 10 μg/ml (A) or terbinafine at 2 μg/ml and lovastatin at 20 μg/ml (B) and grown at 30°C for 48 h.
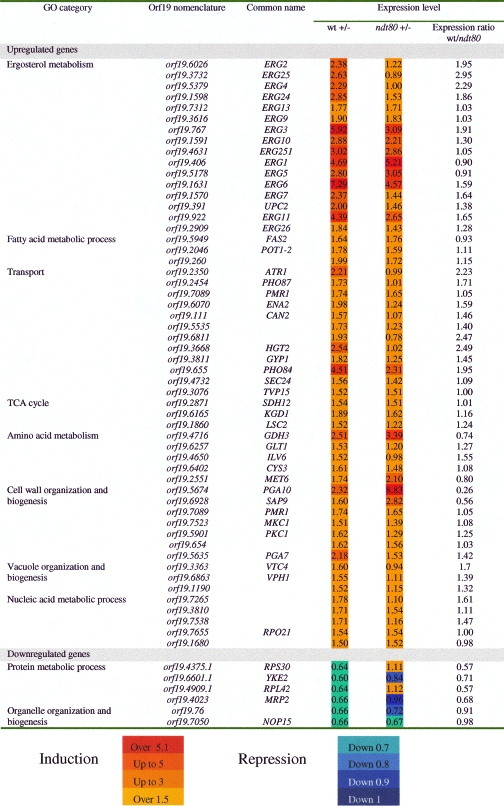
Fluconazole-responsive genes were identified by comparing the transcriptional profiles of wild-type cells exposed to fluconazole for 30 min to the transcriptional profiles of nontreated wild-type cells. Using a statistical significance analysis with an estimated false discovery rate of 5%, in addition to a stringent cutoff of 1.5-fold, we identified 128 fluconazole-responsive genes, including 95 upregulated genes and 33 downregulated genes (Table 3). GO enrichment analysis of upregulated genes revealed ergosterol metabolism as the largest significantly enriched category (P = 1.08e−20). The genes found to be responsive in this study include ERG1, ERG2, ERG3, ERG4, ERG5, ERG6, ERG7, ERG9, ERG10, ERG11, ERG13, ERG24, ERG25, ERG251, and ERG26 (Table 3). This expression signature is similar to those previously reported for transcriptional azole responses in S. cerevisiae (1), C. albicans (8, 13, 28), Mycosphaerella graminicola (7), and Aspergillus fumigatus (12). Interestingly, the transcript level of the regulator of sterol metabolism and drug resistance Upc2p was induced by fluconazole. The category of genes with the next largest number of responses to fluconazole was transport, including MFS transporters Hgt2p and orf19.2350.
TABLE 3.
Effect of fluconazole on gene expression of the C. albicans wild-type strain and the ndt80 mutanta
Only genes significantly belonging to a functional GO category are presented.
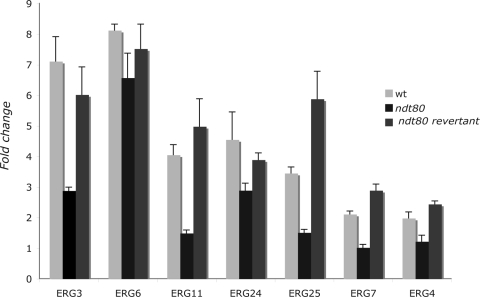
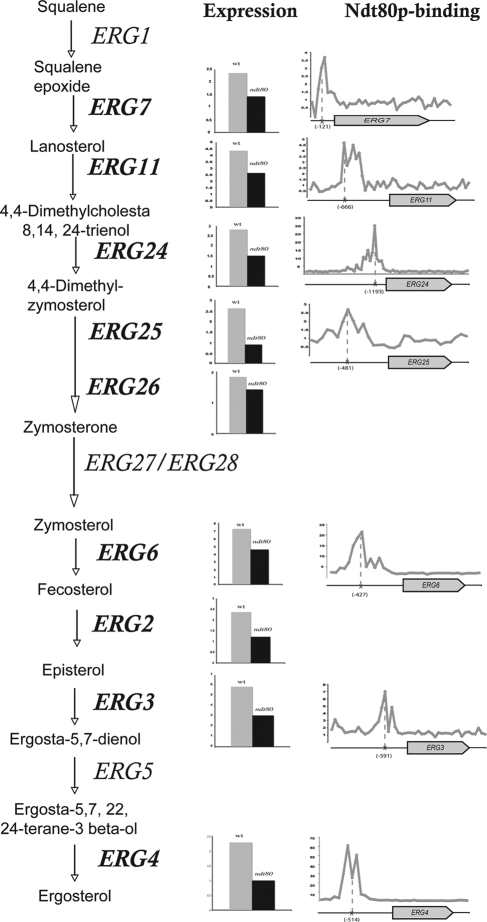
Since the ndt80 mutant was hypersensitive to fluconazole, we focused our investigation on fluconazole-responsive genes. Thus, gene expression of fluconazole-responsive genes in the ndt80 mutant was assessed by comparing transcriptional profiles of ndt80 cells exposed to fluconazole and nontreated ndt80 cells. Our results show that transcript level of fluconazole-responsive genes in ndt80 mutant was obviously altered (Table 3). Notably, there was a defect in transcriptional induction of 10 ERG genes, namely ERG3, ERG6, ERG11, ERG10, ERG24, ERG25, ERG2, ERG7, ERG4, and ERG26. Their expression was significantly decreased but not completely abolished in ndt80. Real-time quantitative PCR was performed to confirm microarray data, and the obtained result showed that part of the azole inducibility of ERG genes (ERG3, ERG4, ERG6, ERG7, ERG11, ERG24, ERG25) is indeed dependent on Ndt80p (Fig. 4). Among the 10 ERG gene promoters that are transcriptionally dependent on Ndt80p, 7 are bound by Ndt80p (Fig. 5). In addition, the transcript level of UPC2 was reduced in ndt80 compared to that of the wild type. Taking into account the role of this TF as a regulator of sterol metabolism and drug resistance, this is consistent with the reduction of ERG gene expression and the hypersensitivity of the ndt80 mutant to fluconazole.
FIG. 4.
Real-time quantitative PCR validation of microarray data. Expression levels of ERG3, ERG4, ERG6, ERG7, ERG11, ERG24, and ERG25 were determined in the wild type (wt), the ndt80 mutant, and the revertant strain, using ACT1 as a reference. The reported values are the means ± standard deviations from two independent experiments.
FIG. 5.
Effect of fluconazole on expression of C. albicans post-squalene ergosterol biosynthesis genes carried by the wild-type strain and ndt80 mutant. Ndt80p promoter binding of ergosterol genes is also shown. The positions of the MSE motifs are shown.
DISCUSSION
Exposure to antifungal drugs is perceived as an environmental stress which triggers a mechanism of tolerance that ultimately leads, in the long term, to the selection of strains that constitutively respond to stress (3). Thus, studying the mechanisms that govern the stress and tolerance responses is of major importance for understanding multidrug resistance in C. albicans. However, despite the considerable accumulated knowledge in the yeast model S. cerevisiae, efforts to characterize these pathways must be expended in C. albicans itself, because these two species exhibit extensive rewiring of their transcriptional regulatory network and cell signaling pathway (16, 17, 20). In S. cerevisiae, Ndt80p is a meiosis-specific TF essential for crossing-over, disassembly of the synaptonemal complex, and general cell cycle progression past prophase I (6, 44). In this study, by using genome-wide methods, we show that the functions of Ndt80p in C. albicans are broader and more distinct than those in S. cerevisiae. In fact, this report brings new insight into the role of Ndt80p in the control of C. albicans drug tolerance and demonstrates an unexpected role in the transcriptional regulation of ergosterol biosynthesis genes.
In this study, we used ChIP-chip technology to investigate the roles of Ndt80p in C. albicans. Several lines of evidence, summarized below, demonstrate the usefulness of this strategy. Indeed, out of 100 promoters that were randomly selected for the confirmation of the ChIP tiling array results, none were shown to be false positive. Furthermore, our results confirmed the observation that Ndt80p binds to the promoter region of CDR1, as suggested by Chen et al. (4), who demonstrated that this TF alleviates the LacZ activity of a CDR1-lacZ construct. Additionally, de novo motif analysis revealed a high enrichment of the MSE motif in Ndt80p promoter targets, thus demonstrating a high degree of conservation for this binding site in C. albicans. This was expected since the Ndt80p DNA-binding domain is conserved between C. albicans and S. cerevisiae. Thus, our results demonstrate that the ChIP-chip technique used here is a compelling tool for genome-wide occupancy analysis and for devising the function of transcription factors.
The finding that Ndt80p occupies the promoter regions of 23% of C. albicans genes suggests that this TF plays a global role in transcriptional regulation. This hypothesis is buttressed by the fact that the list of Ndt80p targets shows remarkable enrichment for genes whose products are transcriptional regulators acting in different biological process. This suggests that Ndt80p is a high-level hierarchical regulator acting in the transcriptional cascade of diverse biological process. Intriguingly, Ndt80p was found to bind promoters of hyphal growth regulators, such as the TF Cph2p, Efg1p, Ume6p and Flo8p, which activate filamentation-specific genes when C. albicans undergoes morphological switching. Based on this observation, Ndt80p might play a critical role in regulating morphogenesis in this fungal pathogen. Indeed, the ndt80 mutant is not able to form hyphae under different filamentation-promoting conditions (A. Sellam and A. Nantel, unpublished data), and the precise mechanisms involving Ndt80p in hyphal growth are under investigation.
To help with the identification of genes that require Ndt80p for fluconazole-dependent expression, we first produced our own list of fluconazole-responsive genes carried by the wild-type strain. Only 10 of our fluconazole-responsive genes were shown to be activated during the exposure to the same drug by Lepak et al. (27), with 8 of these common genes being involved in ergosterol biosynthesis (ERG1, ERG3, ERG4, ERG5, ERG6, ERG11, ERG24, ERG26). Copping et al. (8) have also investigated the transcriptional response of C. albicans to fluconazole. Likewise, 10 upregulated genes were common with those of our study, including 8 ERG genes (ERG1, ERG3, ERG4, ERG5, ERG6, ERG7, ERG11, ERG251) and also the fatty acyl coenzyme A synthase FAS2. These findings suggests that, even when using different strains and growth and treatment conditions on different microarray platforms, C. albicans responds to fluconazole by activating mainly the ergosterol pathway, as was indeed demonstrated by several fungi following the exposure to other azole drugs (1, 12, 7, 27, 28). In the wild-type strain, upregulation of ERG genes is considered to be a compensatory response used to overcome the inhibition of the lanosterol demethylase Erg11p by fluconazole. Based on our transcriptional profiling data, ndt80 was not able to activate as many ERG genes as in the wild-type strain, suggesting that these compensatory mechanisms are altered in this mutant. Thus, our study suggests that Ndt80p affects drug resistance by controlling the expression of genes involved in the biosynthesis of ergosterol. Direct analysis using ChIP-chip revealed that many promoters of ERG genes, namely, ERG3, ERG4, ERG6, ERG7, ERG11, ERG13, ERG24, ERG25, and ERG251, were bound by Ndt80p in vivo, thus suggesting that this TF directly regulates their expression.
Ndt80p is a transcriptional activator that plays a key role in the progression of the meiotic divisions in the yeast S. cerevisiae (23). In C. albicans, Ndt80p has been identified as a positive regulator of CDR1p (4). C. albicans Ndt80p appears to have functionally diverged from the role of its S. cerevisiae homologue, since ScNdt80p fails to activate the expression of the CDR1p-lacZ heterologous reporter gene (42). In our study, we demonstrate that Ndt80p binds the promoter of ERG biosynthesis genes and is required for their transcriptional activation in response to fluconazole. Such function has not been demonstrated in S. cerevisiae. Indeed, genome-wide mapping of Ndt80p in S. cerevisiae was performed; however, no significant binding was observed in the promoters of ergosterol biosynthesis genes (22). Furthermore, even with the absence of a complete sexual cycle in C. albicans, meiosis genes are quite conserved (40). However, no significant enrichment for genes involved in meiosis was obtained in this study. Taken together, these findings are a further confirmation that the Ndt80p regulon has been rewired in C. albicans.
Supplementary Material
Acknowledgments
We are grateful to Hervé Hogues for bioinformatics assistance.
This work was supported by grant MOP-42516 from the Canadian Institutes of Health Research (CIHR) to A.N.
This is NRC manuscript 50651.
Footnotes
Published ahead of print on 19 June 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Agarwal, A. K., P. D. Rogers, S. R. Baerson, M. R. Jacob, K. S. Barker, J. D. Cleary, L. A. Walker, D. G. Nagle, and A. M. Clark. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 27834998-35015. [DOI] [PubMed] [Google Scholar]
- 2.Braun, B. R., M. van Het Hoog, C. d'Enfert, M. Martchenko, J. Dungan, A. Kuo, D. O. Inglis, M. A. Uhl, H. Hogues, M. Berriman, M. Lorenz, A. Levitin, U. Oberholzer, C. Bachewich, D. Harcus, A. Marcil, D. Dignard, T. Iouk, R. Zito, L. Frangeul, F. Tekaia, K. Rutherford, E. Wang, C. A. Munro, S. Bates, N. A. Gow, L. L. Hoyer, G. Kohler, J. Morschhauser, G. Newport, S. Znaidi, M. Raymond, B. Turcotte, G. Sherlock, M. Costanzo, J. Ihmels, J. Berman, D. Sanglard, N. Agabian, A. P. Mitchell, A. D. Johnson, M. Whiteway, and A. Nantel. 2005. A human-curated annotation of the Candida albicans genome. PLoS Genet. 136-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannon, R. D., E. Lamping, A. R. Holmes, K. Niimi, K. Tanabe, M. Niimi, and B. C. Monk. 2007. Candida albicans drug resistance another way to cope with stress. Microbiology 1533211-3217. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. G., Y. L. Yang, H. I. Shih, C. L. Su, and H. J. Lo. 2004. CaNdt80 is involved in drug resistance in Candida albicans by regulating CDR1. Antimicrob. Agents Chemother. 484505-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 2183-84. [DOI] [PubMed] [Google Scholar]
- 6.Chu, S., and I. Herskowitz. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1685-696. [DOI] [PubMed] [Google Scholar]
- 7.Cools, H. J., B. A. Fraaije, T. P. Bean, J. Antoniw, and J. A. Lucas. 2007. Transcriptome profiling of the response of Mycosphaerella graminicola isolates to an azole fungicide using cDNA microarrays. Mol. Plant Pathol. 8639-651. [DOI] [PubMed] [Google Scholar]
- 8.Copping, V. M., C. J. Barelle, B. Hube, N. A. Gow, A. J. Brown, and F. C. Odds. 2005. Exposure of Candida albicans to antifungal agents affects expression of SAP2 and SAP9 secreted proteinase genes. J. Antimicrob. Chemother. 55645-654. [DOI] [PubMed] [Google Scholar]
- 9.Coste, A., V. Turner, F. Ischer, J. Morschhauser, A. Forche, A. Selmecki, J. Berman, J. Bille, and D. Sanglard. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 1722139-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coste, A. T., M. Karababa, F. Ischer, J. Bille, and D. Sanglard. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 31639-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowen, L. E., and W. J. Steinbach. 2008. Stress, drugs, and evolution: the role of cellular signaling in fungal drug resistance. Eukaryot. Cell 7747-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.da Silva Ferreira, M. E., I. Malavazi, M. Savoldi, A. A. Brakhage, M. H. Goldman, H. S. Kim, W. C. Nierman, and G. H. Goldman. 2006. Transcriptome analysis of Aspergillus fumigatus exposed to voriconazole. Curr. Genet. 5032-44. [DOI] [PubMed] [Google Scholar]
- 13.De Backer, M. D., T. Ilyina, X. J. Ma, S. Vandoninck, W. H. Luyten, and H. Vanden Bossche. 2001. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob. Agents Chemother. 451660-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunkel, N., T. T. Liu, K. S. Barker, R. Homayouni, J. Morschhauser, and P. D. Rogers. 2008. A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot. Cell 71180-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eggimann, P., J. Garbino, and D. Pittet. 2003. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect. Dis. 3685-702. [DOI] [PubMed] [Google Scholar]
- 16.Enjalbert, B., A. Nantel, and M. Whiteway. 2003. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 141460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enjalbert, B., D. A. Smith, M. J. Cornell, I. Alam, S. Nicholls, A. J. Brown, and J. Quinn. 2006. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 171018-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fingerman, I. M., K. Sutphen, S. P. Montano, M. M. Georgiadis, and A. K. Vershon. 2004. Characterization of critical interactions between Ndt80 and MSE DNA defining a novel family of Ig-fold transcription factors. Nucleic Acids Res. 322947-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fridkin, S. K., and W. R. Jarvis. 1996. Epidemiology of nosocomial fungal infections. Clin. Microbiol. Rev. 9499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasch, A. P., A. M. Moses, D. Y. Chiang, H. B. Fraser, M. Berardini, and M. B. Eisen. 2004. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2e398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198179-182. [DOI] [PubMed] [Google Scholar]
- 21.Gola, S., R. Martin, A. Walther, A. Dunkler, and J. Wendland. 2003. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 201339-1347. [DOI] [PubMed] [Google Scholar]
- 22.Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac, T. W. Danford, N. M. Hannett, J. B. Tagne, D. B. Reynolds, J. Yoo, E. G. Jennings, J. Zeitlinger, D. K. Pokholok, M. Kellis, P. A. Rolfe, K. T. Takusagawa, E. S. Lander, D. K. Gifford, E. Fraenkel, and R. A. Young. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 43199-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hepworth, S. R., H. Friesen, and J. Segall. 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 185750-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hull, C. M., and A. D. Johnson. 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 2851271-1275. [DOI] [PubMed] [Google Scholar]
- 25.Lamoureux, J. S., D. Stuart, R. Tsang, C. Wu, and J. N. Glover. 2002. Structure of the sporulation-specific transcription factor Ndt80 bound to DNA. EMBO J. 215721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavoie, H., A. Sellam, C. Askew, A. Nantel, and M. Whiteway. 2008. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics 9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lepak, A., J. Nett, L. Lincoln, K. Marchillo, and D. Andes. 2006. Time course of microbiologic outcome and gene expression in Candida albicans during and following in vitro and in vivo exposure to fluconazole. Antimicrob. Agents Chemother. 501311-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, T. T., R. E. Lee, K. S. Barker, L. Wei, R. Homayouni, and P. D. Rogers. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 492226-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, T. T., S. Znaidi, K. S. Barker, L. Xu, R. Homayouni, S. Saidane, J. Morschhauser, A. Nantel, M. Raymond, and P. D. Rogers. 2007. Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryot. Cell 62122-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macphail, G. L., G. D. Taylor, M. Buchanan-Chell, C. Ross, S. Wilson, and A. Kureishi. 2002. Epidemiology, treatment and outcome of candidemia: a five-year review at three Canadian hospitals. Mycoses 45141-145. [DOI] [PubMed] [Google Scholar]
- 31.Montano, S. P., M. L. Cote, I. Fingerman, M. Pierce, A. K. Vershon, and M. M. Georgiadis. 2002. Crystal structure of the DNA-binding domain from Ndt80, a transcriptional activator required for meiosis in yeast. Proc. Natl. Acad. Sci. USA 9914041-14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morschhauser, J., K. S. Barker, T. T. Liu, B. W. J. Bla, R. Homayouni, and P. D. Rogers. 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 3e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murad, A. M., P. R. Lee, I. D. Broadbent, C. J. Barelle, and A. J. Brown. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16325-327. [DOI] [PubMed] [Google Scholar]
- 34.Nantel, A., T. Rigby, H. Hogues, and M. Whiteway. 2006. Microarrays for studying pathogenicity in Candida albicans, p. 181-209. In K. Kavanagh (ed.), Medical mycology: cellular and molecular techniques. John Wiley & Sons, Chichester, England.
- 35.Pak, J., and J. Segall. 2002. Role of Ndt80, Sum1, and Swe1 as targets of the meiotic recombination checkpoint that control exit from pachytene and spore formation in Saccharomyces cerevisiae. Mol. Cell. Biol. 226430-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 452676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierce, M., K. R. Benjamin, S. P. Montano, M. M. Georgiadis, E. Winter, and A. K. Vershon. 2003. Sum1 and Ndt80 proteins compete for binding to middle sporulation element sequences that control meiotic gene expression. Mol. Cell. Biol. 234814-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan, L., X. Sun, X. Zhu, Z. Zhang, J. Li, and Q. Shu. 2004. Epidemiology of nosocomial pneumonia in infants after cardiac surgery. Chest 125410-417. [DOI] [PubMed] [Google Scholar]
- 39.Tuch, B. B., D. J. Galgoczy, A. D. Hernday, H. Li, and A. D. Johnson. 2008. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tzung, K. W., R. M. Williams, S. Scherer, N. Federspiel, T. Jones, N. Hansen, V. Bivolarevic, L. Huizar, C. Komp, R. Surzycki, R. Tamse, R. W. Davis, and N. Agabian. 2001. Genomic evidence for a complete sexual cycle in Candida albicans. Proc. Natl. Acad. Sci. USA 983249-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van het Hoog, M., T. J. Rast, M. Martchenko, S. Grindle, D. Dignard, H. Hogues, C. Cuomo, M. Berriman, S. Scherer, B. B. Magee, M. Whiteway, H. Chibana, A. Nantel, and P. T. Magee. 2007. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 8R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, J. S., Y. L. Yang, C. J. Wu, K. J. Ouyang, K. Y. Tseng, C. G. Chen, H. Wang, and H. J. Lo. 2006. The DNA-binding domain of CaNdt80p is required to activate CDR1 involved in drug resistance in Candida albicans. J. Med. Microbiol. 551403-1411. [DOI] [PubMed] [Google Scholar]
- 43.Wang, W., J. M. Cherry, Y. Nochomovitz, E. Jolly, D. Botstein, and H. Li. 2005. Inference of combinatorial regulation in yeast transcriptional networks: a case study of sporulation. Proc. Natl. Acad. Sci. USA 1021998-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1811868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu, L., M. Ajimura, R. Padmore, C. Klein, and N. Kleckner. 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 156572-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Znaidi, S., S. Weber, O. Z. Al-Abdin, P. Bomme, S. Saidane, S. Drouin, S. Lemieux, X. De Deken, F. Robert, and M. Raymond. 2008. Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot. Cell 7836-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.