Abstract
Many apicomplexan parasites, including Plasmodium falciparum, harbor a so-called apicoplast, a complex plastid of red algal origin which was gained by a secondary endosymbiotic event. The exact molecular mechanisms directing the transport of nuclear-encoded proteins to the apicoplast of P. falciparum are not well understood. Recently, in silico analyses revealed a second copy of proteins homologous to components of the endoplasmic reticulum (ER)-associated protein degradation (ERAD) system in organisms with secondary plastids, including the malaria parasite P. falciparum. These proteins are predicted to be endowed with an apicoplast targeting signal and are suggested to play a role in the transport of nuclear-encoded proteins to the apicoplast. Here, we have studied components of this ERAD-derived putative preprotein translocon complex in malaria parasites. Using transfection technology coupled with fluorescence imaging techniques we can demonstrate that the N terminus of several ERAD-derived components targets green fluorescent protein to the apicoplast. Furthermore, we confirm that full-length PfsDer1-1 and PfsUba1 (homologues of yeast ERAD components) localize to the apicoplast, where PfsDer1-1 tightly associates with membranes. Conversely, PfhDer1-1 (a host-specific copy of the Der1-1 protein) localizes to the ER. Our data suggest that ERAD components have been “rewired” to provide a conduit for protein transport to the apicoplast. Our results are discussed in relation to the nature of the apicoplast protein transport machinery.
The apicomplexan parasite Plasmodium falciparum is the etiological agent of malaria tropica, the most severe form of human malaria, responsible for over 250 million infections and 1 million deaths annually (61). Many apicomplexan parasites, including P. falciparum, harbor a so-called apicoplast, a complex plastid of red algal origin which was gained by a secondary endosymbiotic event (27, 58). Although during the course of evolution this plastid organelle has lost the ability to carry out photosynthesis, it is still the site of several important biochemical pathways, including isoprenoid and heme biosynthesis, and as such is essential for parasite survival (60). As in other plastids, the vast majority of genes originally encoded on the plastid genome have been transferred to the nucleus of the host. As a result, their gene products (predicted to constitute up to 10% of all nucleus-encoded proteins) must be imported back into the apicoplast (12). The apicoplast is surrounded by four membranes (55), and this protein import process thus represents a major cell biological challenge and has attracted much research interest, not least due to the importance of P. falciparum as a human pathogen (16, 50).
The signals directing transport of nucleus-encoded proteins to complex plastids, including the apicomplexan apicoplast, have been studied in great detail in recent years, and reveal that such proteins are endowed with specific N-terminal targeting sequences, referred to as a bipartite topogenic signals (BTS), that direct their transport to this compartment (50). BTS are composed of an N-terminal endoplasmic reticulum (ER)-type signal sequence, which initially allows proteins to enter the secretory system via the Sec61 complex (59). Following this, proteins are carried via a Golgi complex-independent transport step to the second outermost membrane, from where they are then translocated across the remaining three apicoplast membranes, directed by the second part of the BTS, the transit peptide (51). Based on evolutionary considerations, it has long been suggested that transport across the inner two apicoplast membranes occurs via a Toc/Tic-like (where Toc and Tic are translocons of the outer and inner chloroplast envelopes, respectively) protein translocase machinery, and this is supported by a recent publication that provides evidence for an essential role of a Toxoplasma gondii Tic20 homologue in this transport process (50, 57). Despite this progress, it is still unclear how proteins travel across the second and third outer apicoplast membranes. Several models have been discussed to account for this transport step, including vesicular shuttle and translocon-based mechanisms (recently reviewed in reference 19), but until recently no actual molecular equipment had been found which could account for these membrane translocation events. To address this question, Sommer et al. screened the nucleomorph genome of the chromalveolate cryptophyte Guillardia theta (which, similar to P. falciparum, contains a four-membrane-bound plastid organelle) for genes encoding potential translocon-related proteins (49). Surprisingly, the authors identified genes encoding proteins usually involved in the ER-associated protein degradation pathway (ERAD), which recognizes incorrectly folded protein substrates and retrotranslocates them to the cell cytosol for degradation by the ubiquitin (Ub)-proteasome system (35, 44). As such, the ERAD system functions as a translocation complex, capable of transporting proteins across a biological membrane. Further characterization of one of these proteins (G. theta Der1-1, a homologue of yeast Der1p, a component of the ERAD system) provided strong evidence for a plastid localization. These data suggested an attractive solution to the mechanistic problem of transport across the second and third outermost membrane of complex plastids by hypothesizing a role for an ERAD-derived protein translocon complex. Intriguingly, this study also identified several members of this ERAD-derived translocon complex (apicoplast ERAD [apERAD]) in the nuclear genome of P. falciparum endowed with an N-terminal BTS (49). The BTS derived from one of these proteins, P. falciparum sDer1-1 [PfsDer1-1], was sufficient to direct transport of green fluorescent protein (GFP) to the apicoplast of P. falciparum, suggesting that this ERAD-like machinery is ubiquitous among chromalveolates with four membrane-bound plastids (49). In this current report we extend our study of the P. falciparum apERAD complex.
MATERIALS AND METHODS
Bioinformatics.
Homologues of the ERAD pathway in apicomplexan parasites were identified by a BLAST (2) search implemented in the Eukaryotic Pathogens Database Resources (http://eupathdb.org/eupathdb/) and PlasmoDB (version 5.1) (3). Preliminary Babesia bovis sequence data was obtained from Washington State University/USDA ARS website (http://www.vetmed.wsu.edu/research_vmp/program-in-genomics). Sequences were analyzed by SignalP, version 3.0 (5), PlasmoAP (12), and PATS (62) for identification of N-terminal bipartite signals. Sequence alignments were carried out using Clustal (31) (standard settings are available at http://www.ebi.ac.uk/Tools/clustalw2/index.html).
For analysis of amino acids at the +1 position following signal peptide cleavage, predicted apicoplast and secreted nonapicoplast (signal peptide containing) data sets were retrieved from PlasmoDB and subjected to analysis by SignalP, version 3.0. Protein sequences and SignalP predictions were then fed into a custom-designed Matlab script (available upon request from J. Hiss) which performed in silico signal peptide cleavage and sorting of the proteins depending on the +1 amino acid (aromatic or nonaromatic). Alignments of the 20 amino acid sequences (FASTA format) (see List SA1 in the supplemental material) were then prepared using Weblogo (10).
Transmembrane (TM) domain prediction of all Plasmodium sp. PfDer1-1 sequences was carried out using the programs PHOBIUS, TMHMM, and MINNOU (9). Amino acid sequences corresponding to predicted TM domains were analyzed for both length and hydrophobicity (using the Woods [23] and Doolittle [29] scales) and statistically analyzed by the Kolmogorov-Smirnov (KS) statistic (40). The KS statistics regards the TMD lengths (predicted using the tools above) as a distribution in the host and the parasite, respectively. If they differ on a 5% niveau of the KS test, this means that the null hypothesis that both distributions were drawn from the same underlying distribution must be rejected. A KS test was used because a standard distribution of the values could not be assumed.
Expression constructs.
All primers used in generation of constructs are listed in Table SA2 in the supplemental material. Regions encoding the BTS of PfsUba1 (PF13_0182; bases 1 to 390), PfsCdc48 (PF07_0047; bases 1 to 420), PfsUb (PF08_0067; bases 1 to 300), and PfDer1-2 (PFC0590c; bases 1 to 405) were PCR amplified from genomic P. falciparum DNA using specific oligonucleotides (xBTS_for and xBTS_rev) introducing a 5′ XhoI and a 3′ AvrII restriction site. The amplicons were then digested with XhoI and AvrII and cloned into similarly digested pARL-GFP-DHFR (where DHFR is dihydrofolate reductase) (13) in front of the GFP coding sequence. For integration of the GFP coding sequence into the 3′ region of PF14_0498 (PfsDer1-1), 758 bp from the 3′ end of the gene (leaving out the stop codon) were amplified from P. falciparum 3D7 genomic DNA using primers int_for/int_rev, introducing 5′ NotI and 3′ KpnI restriction sites, and ligated into similarly restricted pARL-GFP-DHFR (removing the chloroquine resistance transporter promoter region). For integration of the GFP coding sequence into the 3′ end of PF13_0182 (PfsUba1), 968 bp from the 3′ end of the gene (leaving out the stop codon) were amplified from P. falciparum 3D7 genomic DNA using primers int_for/int_rev, introducing a 5′ NotI and a 3′ KpnI restriction site, and ligated into similarly restricted pARL-GFP-DHFR as above. For localization of PfhDer1-1, the entire cDNA sequence (lacking the stop codon) was amplified from 3D7 RNA using a Superscript II 1-Step RT-PCR kit (Invitrogen) according to the manufacturer's protocol, using primers PfhDer1-1_F and PfhDer1-1_R. The resulting PCR product was restricted with XhoI and KpnI and cloned into similarly restricted pARL-GFP-DHFR. All constructs were verified by automated sequencing. Plasmid DNA was isolated via a Qiagen Maxiprep protocol. Regions encoding acyl carrier protein (ACP)-DsRed (ACP accession number, PFB0385w) and HSP-DsRed (Hsp60 accession number, PFL1545c) were amplified from pSSPF2/PfACP-DsRed (kindly provided by S. Sato) (47) using primers ACP-F/HSP-F and DsRed-R and cloned into the pARL-BSD vector, which contains a blasticidin-S-deaminase (BSD)-selectable marker.
Cell culture, transfection, and generation of GFP-integrant lines.
The P. falciparum 3D7 line was cultured in human O+ erythrocytes according to standard protocols (53), except that cultures were incubated in gassed flasks. Transfection was carried out by electroporation of infected human O+ erythrocytes as previously described (11). GFP transfectants were selected with 5 nM WR99210 (kindly supplied by D. Jacobus) for human DHFR-based vectors or 8.7 nM blasticidin for BSD-based constructs. Integrant parasites were selected by repeated drug cycling (3 weeks on and 3 weeks off), and integration was checked via PCR. Positive parasite populations were then cloned by limiting dilution. Integration was confirmed in each clone by integration-specific PCR (see text for details), followed by sequencing to determine the exact integration site.
IFAs and live-cell imaging.
Immunofluorescence assays (IFAs) were carried out following fixation using 4% paraformaldehyde-0.00075% glutaraldehyde as previously described (52) except that fixation was carried out at 37 °C for 30 min, and quenching was performed with 125 mM glycine-phosphate-buffered saline (PBS). Primary antibodies used were the following: rabbit anti-ACP (1:500; kindly provided by GI McFadden), rabbit anti-BiP (1:2,200; kindly provided by T. Gilberger), chicken anti-GFP (1/1,000; Abcam), and anti-chicken-Cy2 and anti-rabbit-Cy3 (both 1/2,000; Dako). Antibodies were diluted in 3% bovine serum albumin-PBS. Hoechst 33258 (Molecular probes) was used at a concentration of 50 ng/ml for fixed parasites or 10 μg/ml for live parasites. MitoTrackerOrange (Molecular Probes) was used at 20 nM. All images were acquired at either 37 °C (live cells) or room temperature (fixed cells) on a Zeiss Cell Observer using appropriate filter sets. Individual images were imported into Image J64 (version 1.39u; available at http://rsb.info.nih.gov/ij), converted to 8-bit grayscale, subjected to background subtraction, and overlaid. Colocalization analysis was carried out using the ImageJ Plugin written by Pierre Bourdoncle (available at http://rsb.info.nih.gov/ij/plugins/colocalization.html). To create figures, TIF files were imported into PowerPoint (Microsoft) and assembled, and slides were exported as TIF files. No gamma adjustments were applied to any images, and all data are presented in accordance with the recommendations of Rossner and Yamada (45).
Protein biochemistry and immunoblotting.
For membrane fractionation parasites were lysed on 10 mM Tris-2 mM EDTA. The pellet was resuspended in 0.1 M sodium carbonate buffer-1 mM EDTA, pH 11, and incubated on ice for 30 min. The insoluble fraction was obtained by centrifugation at 36,000 × g at 4 °C for 30 min. Urea extraction was carried out as previously described (36). High-salt treatment was carried out by Tris lysis (as above), followed by incubation of membrane fractions in high-salt buffer (50 mM HEPES, pH 7.5, 0.6 M KCl, 5 mM dithiothreitol, 3 mM MgCl2) on ice for 30 min, after which the pellet was separated from the supernatant and subjected to Western blot analysis. For Triton X-100 solubilization, Tris-lysed parasites were resuspended in different concentrations of Triton X-100-PBS. The solution was incubated at 4 °C overnight and centrifuged at 36,000 × g for 30 min. Protein samples were added to 1 volume of 2× Laemmli sample buffer and boiled for 10 min. For Western blot analyses, protein samples were prepared from saponin-isolated parasites. A total of 2 × 107 (4 × 107 for integrant lines) parasite cell equivalents were separated by 10 to 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane. Primary monoclonal mouse anti-GFP antibodies (Roche) were used in a concentration of 1/2,000 for BTS fusion proteins and 1/1,000 for the integration line. Rabbit anti-Exp1 (1:500) and monoclonal mouse anti-PfHsp70 (1/1,000; a gift of Thierry Blisnick) have previously been described (17). Anti-mouse horseradish peroxidase and anti-rabbit horseradish peroxidase (Dako, Hamburg) were used at 1/2,000. Immunoblots were developed via chemiluminescence using an enhanced chemiluminescence system.
Time course.
Isolated parasites were collected from highly synchronized cultures every 6 h over a period of 48 h via saponin lysis; proteins were separated via 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane for subsequent Western blot analysis, as detailed above.
RESULTS
Identification of apERAD proteins in apicomplexa.
A previous bioinformatic analysis identified duplicated P. falciparum homologues of ERAD components: the putative channel protein Der1p, the AAA-type ATPase Cdc48, the Cdc48 cofactor Ufd1, and the E1 Ub-activating enzymes Uba1 (49). In addition to these components, we are now also able to identify further duplicated ERAD components: the E1 Ub-activating enzyme Uba2, an E2 Ub-conjugating enzyme Ubc, as well as Ub itself (Table 1). We hypothesized that if an ERAD-like transport system is essential for protein transport to the apicoplast, we should be able to identify duplicated ERAD components in other members of plastid-bearing apicomplexa. For this reason, we screened the genomic sequences of Plasmodium spp., T. gondii, B. bovis, and Theileria parva for homologues to ERAD components (4). We were able to identify duplicate ERAD components in all of these genomes (Table 1; see also Table SA3 in the supplemental material). Both ER (host-specific ERAD [hERAD]) and apERAD paralogues of Ubc4 and Ufd1 were identified in all organisms, while other predicted proteins were less conserved among species (Table 1; see also Table SA3 in the supplemental material). In Plasmodium spp. the genes appear to be spread across all chromosomes, with no particular linkage between the location of the ER and the respective apicoplast paralogues. All genes seem to be located in perfectly syntenic regions, away from places of large-scale chromosomal rearrangements or single-gene insertions or deletions (28). Based on regions of synteny, we are able to assign putative chromosome numbers to several of the “missing” components. Thus, a Plasmodium knowlesi hUfd1 and a Plasmodium chabaudi sDer1-1 protein are both predicted to be encoded on the respective chromosome 13 (Table 1). Ongoing assembly, gap closing, and analysis of these genomes is likely to reveal these genes.
TABLE 1.
apERAD homologues in Plasmodium spp. identified by bioinformatics approachesa
| Protein name |
P. falciparum protein in:
|
P. vivax protein in:
|
P. yoelii protein in:
|
P. knowlesi protein in:
|
P. berghei protein in:
|
P. chabaudi protein in:
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ER
|
Apicoplast
|
ER
|
Apicoplast
|
ER
|
Apicoplast
|
ER
|
Apicoplast
|
ER
|
Apicoplast
|
ER
|
Apicoplast
|
|||||||||||||
| Acc. no. | Ch. | Acc. no. | Ch. | Acc. no. | Ch. | Acc. no. | Ch. | Acc. no. | Ch. | Acc. no. | Ch. | Acc. no. | Ch. | Acc. no. | Ch. | Acc. no. | Ch. | Acc. no. | Ch. | Acc. no. | Ch. | Acc. no. | Ch. | |
| Cdc48 | PFF0940c/Mal8P1.92 | 6 | PF07_0047 | 7 | PVX_114095 | 11 | PVX_088085g | 1 | PY03639 | 11 | PY05787g,h | 12 | PKH_113000 | 11 | PKH_010920 | 1 | PB000171.02.0 | 11 | PB000404.03.0h | 12 | PC000445.00.0 | 11 | PC000038.00.0f,g,h | 12 |
| Der1-1 | PF14_0653e | 14 | PF14_0498 | 14 | PVX_117040 | 12 | PVX_117865 | 12 | PY02870f | 13 | PY06810 | 13 | PKH_123690 | 12 | PKH_125400 | 12 | PB000599.01.0f | 13 | PB000620.00.0f,g | 13 | PC301995.00.0f | 13 | ND | 13d |
| Der1-2 | PF10_0317 | 10 | PFC0590cf | 3 | PVX_111100 | 6 | PVX_119810f | 8 | PY03142 | 5 | PY02283g | 6 | PKH_061690 | 6 | PKH_082580f | 8 | PB001618.02.0 | 5 | PB301046.00.0f,g | 4 | PC000384.05.0 | 5 | PC300480.00.0f,g,h | 4 |
| Hrd1b | PF14_0215 | 14 | ND | PVX_085355 | 13 | ND | PY00025f,g | 10 | ND | PKH_132660 | 13 | ND | PB001204.02.0 | 10 | ND | PC000714.01.0f,g/PC000098.01.0.01 | 10 | ND | ||||||
| Hrd3c | PFC0550w/PF14_0462 | 3/14 | ND | PVX_118065/PVX_119750 | 12/8 | ND | PY04510/ PY051121 | 4/13 | ND | PKH_125790/PKH_082700 | 12/8 | ND | PB000257.00.01/PB000807.2.0 | 4/13 | ND | PC300752.00.0f/PC000360.2.0f | 4/13 | ND | ||||||
| Npl4 | PFE0380c | 5 | ND | PVX_097945 | 10 | ND | PY05126 | 11 | ND | PKH_102520 | 10 | ND | PB001631.02.0 | 11 | ND | PC000343.04.0 | 11 | ND | ||||||
| Ub | PFL0585w | 12 | PF08_0067 | 8 | PVX_084620 | 13 | PVX_089620h | 5 | PY03971f | 6 | PY00539g | 7 | PKH_131070 | 13 | PKH_051680 | 5 | PB000763.03.0 | 6 | PB000118.00.0f | 7 | PC000735.00.0 | 6 | PC300085.00.0 | 7 |
| Uba1b | PFL1245w | 12 | PF13_0182 | 13 | PVX_123920 | 14 | PVX_082590 | 12 | PY01879 | 14 | PY01851/PY06413f,g | 13 | PKH_144260 | 14 | PKH_121970 | 12 | PB000204.03.0f | 14 | PB000355.02.0f,g | 13 | PC001230.02.0f | 14 | PC000262.00.0 | 13 |
| Uba2 | PFL1790w | 12 | PF13_0344 | 13 | PVX_100800 | 14 | PVX_115230 | 11 | PY05539 | 14 | PY02846g,h | 11 | PKH_145560 | 14 | PKH_110530 | 11 | PB000980.01.0 | 14 | PB301007.00.0 | 11 | PC000817.00.0 | 14 | PC000463.02.0g,h | 11 |
| Ubc | PFL0190w | 12 | Mal13P1.227 | 13 | PVX_084235 | 13 | Pv0831752 | 12 | PY03025f | 4/6 | PY00590 | 13 | PKH_130250 | 13 | PKH_120890 | 12 | PB000336.03.0f | 6 | PB000746.02.0f,g | 13 | PC000554.00.0 | 6 | PC000271.02.0f,g | 13 |
| Ufd1b | PF14_0178 | 14 | PFI0810cf | 9 | PVX_085555 | 13 | PVX_099250 | 7 | PY01640f/ PY01641f/ PY01642f | 10 | PY04576f,g | 8 | ND | 13d | PKH_071380 | 7 | PB000251.00.0 | 10 | PB001013.03.0f,g,h | 8 | PC105816.00.0f/PC000154.03.0f | 10 | PC000751.03.0f,g,h | 8 |
Acc. no., PlasmoDB accession number; Ch, chromosomal location; ND, not detected.
Multiple PlasmoDB entries refer to only one actual gene.
Gene duplication.
Gene not detected, chromosomal location inferred from synteny.
Revised; GenBank accession no. FJ555561.
Gene model incomplete/lacking initiation methionine.
No signal peptide.
No transit peptide.
In addition to the duplicated ERAD components mentioned, we were also able to identify further components of the hERAD system (Hrd1, Hrd3, and Npl4) (Table 1; see also Table SA3 in the supplemental material). Npl4 is present in only a single copy and is thus unlikely to be involved in apERAD transport processes. Hrd3 was found to be encoded twice in the genome; however, neither copy is predicted to encode a BTS, also negating a role in the apERAD system (Table 1; see also Table SA3 in the supplemental material).
Many, but not all, of the apERAD homologues encode a BTS, suggesting that they are transported to the apicoplast. In several cases where a BTS appears to be missing, closer inspection of the sequences reveals that these protein sequences are incomplete and do not include the entire N-terminal part of the protein (no initiation methionine) (Table 1). In addition, several proteins are predicted to possess an N-terminal transit peptide but are missing an ER-type signal sequence, essential for entry into the secretory system (Table 1). Prediction of intron/exon boundaries is notoriously challenging in the P. falciparum system (33), and a plausible explanation for the lack of a signal peptide suggests that the missing N-terminal signal can be found on a 5′ exon which has not been included in the gene model. Indeed, based on cDNA sequences, we have previously reannotated the gene model for PFI0810c (encoding PfsUfd1) to include a signal peptide encoded on a previously “missed” 5′ exon (49). Coding of BTS on extra exons is common in many organisms and may reflect their evolutionary history (39). An additional reason for failing to identify a recognizable BTS in homologues from all organisms is that current software designed to predict apicoplast transit peptides has been trained on P. falciparum proteins and may therefore not be able to consistently predict transit peptides of B. bovis or T. parva proteins (62). Additionally, based on cDNA sequencing, we have now reannotated the gene PF14_0653, encoding PfhDer1-1 (GenBank accession no. FJ555561) (Table 1).
Earlier studies have highlighted the importance of a DnaK (Hsp70) binding site within P. falciparum transit peptides in high-fidelity protein traffic to the apicoplast (12), suggesting that binding of Hsp70 at the trans side of the target membrane plays a role in membrane translocation. Gould et al. and Sommer et al. additionally identified putative periplastid-resident Hsp70 proteins in Phaeodactylum tricornutum and G. theta, respectively (15). During the course of our bioinformatic studies, we also identified a member of the Hsp70 family predicted to have a BTS (encoded by Mal13P1.540). It appears unlikely that this protein is a bona fide apicoplast protein as the region predicted to be a transit peptide overlaps with the Hsp70 ATPase domain, and the protein contains a C-terminal KDEL ER retrieval sequence, features not contained in the P. tricornutum or G. theta proteins.
In further support of our analyses, we also screened the genome of Cryptosporidium parvum for ERAD components (1, 4). C. parvum, while phylogenetically belonging to the phylum Apicomplexa, no longer contains an apicoplast, which appears to have been lost during the course of evolution (24). Indeed, although we are able to identify many components of the hERAD system (see Table SA3 in the supplemental material), we cannot find duplications of these genes or of predicted proteins with N-terminal extensions containing a BTS, further supporting a role for apERAD components exclusively in apicoplast-related processes.
PfsDer1-1, encoded by PF14_0498, is a structural orthologue of yeast Der1p.
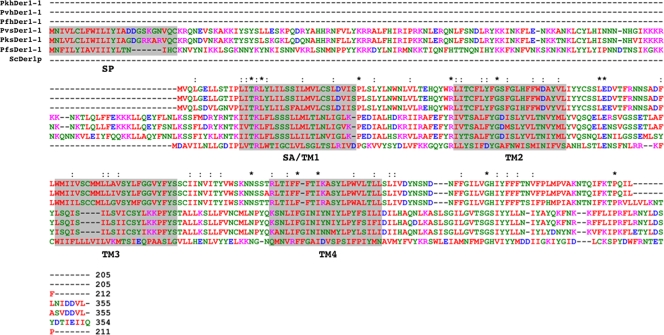
In yeast, Der1p is a central component of the ERAD system (26). This approximately 25-kDa protein contains four membrane-crossing segments, the first of which is predicted to act as a signal anchor, both recruiting the protein to the ER and initiating its integration into the membrane (21). Der1p integrates into membranes with both N and C termini exposed to the cytosol (21). PfsDer1-1 shows 16% identity and 42% similarity to yeast Der1p and is predicted to contain a Der1-like domain (PFAM code PF04511). Additionally, predicted membrane-crossing segments align well between both sequences (Fig. 1), and in all Plasmodium sDer1-1 proteins and hDer1-1 (Fig. 1). These data support the hypothesis that PF14_0498 encodes a structural orthologue of yeast Der1p.
FIG. 1.
Alignment of Plasmodium spp. ER (hDer1-1) and apicoplast (sDer1-1) protein sequences. SP, predicted signal peptide; SA/TM1, signal anchor or transmembrane domain 1; TM2/3/4, predicted TM domains. Asterisk, amino acid conservation at this point; colon, conservative amino acids change. Pk, Plasmodium knowlesi; Pv, Plasmodium vivax. The yeast Der1p sequence is included to allow comparison. Protein identity values are as follows: Saccharomyces cerevisiae Der1p [ScDer1p]/PfhDer1-1, 11%; ScDer1p/PfsDer1-1, 16%; PfhDer1-1/PfsDer1-1, 20%. The color scheme is as follows: red, small hydrophobic residues (A, V, F, P, M, I, L, and W; aromatics apart from Y included); blue, acidic (D and E); magenta, basic (R and K); green, hydroxyl, amine, and basic (S, T, Y, H, C, N, G, Q); gray, all others.
The N termini of apERAD homologues target GFP to the apicoplast.
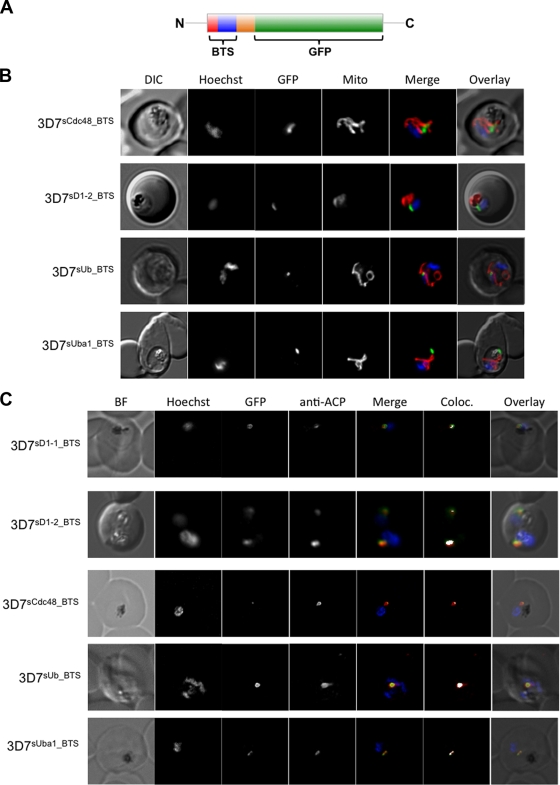
We have previously demonstrated that the N-terminal region of PfsDer1-1 efficiently targets GFP to an intraparasitic compartment suggested to represent the apicoplast (49). We were interested in investigating whether further predicted BTS derived from apERAD components are capable of directing reporter protein transport to this organelle. We therefore generated 3D7 transfectant parasite lines expressing GFP fused C-terminally to predicted BTS derived from PfsCdc48, PfsDer1-2, PfsUba1, and PfsUb (referred to as 3D7PfsD1-2-BTS, 3D7PfsCcd48-BTS, 3D7PfsUba1-BTS, and 3D7PfsUb-BTS lines, respectively) (Fig. 2A). Live-cell imaging reveals that BTS derived from all four proteins are sufficient to target GFP to a punctate structure within the parasite (Fig. 2B), generally in apposition to both the mitochondrion and nucleus, which is suggestive of an apicoplast localization (54). To verify this, we carried out IFAs using antibodies against the ACP, an apicoplast marker (a kind gift of Geoff McFadden, University of Melbourne). In all parasite lines, we observe colocalization between the GFP and ACP signals, confirming an apicoplast localization (Fig. 2C).
FIG. 2.
(A) Structure of the reporter proteins used in this study. Red, predicted signal peptide; blue, transit peptide; orange, remainder of protein sequence. The following numbers of N-terminal amino acids were included to ensure inclusion of a functional BTS: PfsUba1, 130; PfsCdc48, 140; PfsUb, 100; PfDer1-2, 135. (B) Live-cell imaging of BTS-GFP transfectant lines. Transfectant parasite lines were stained with MitoTracker to visualize mitochondrion and with Hoechst to visualize the nucleus. No colocalization can be observed between the GFP and MitoTracker signals. DIC, differential interference contrast; Mito, Mitotracker; Merge, GFP (green) with Hoechst (blue) and MitoTracker (red); Overlay, DIC plus GFP and MitoTracker. (C) IFA of BTS-GFP transfectant lines. Transfectant lines were analyzed by labeling with anti-ACP antiserum (a marker for the apicoplast) followed by Cy3-coupled secondary antibodies. In all lines studied there is significant overlap between ACP and GFP signals, verifying an apicoplast localization for the GFP reporter. DIC, differential interference contrast; BF, bright field; ACP, acyl carrier protein; Merge, GFP (green) with ACP (red) and Hoechst (blue); Overlay, DIC, GFP, Hoechst, and ACP; Coloc, colocalization.
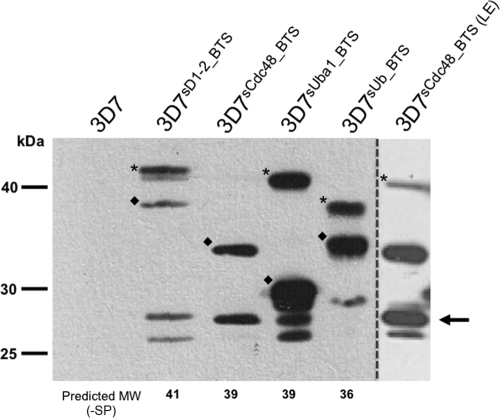
Upon import to the apicoplast, transit peptides are generally cleaved by a transit peptide peptidase (56). Western blot analysis of our transfectant lines using anti-GFP antibodies reveals multiple bands. These bands can, on the basis of their molecular masses, be assigned to either the full-length chimeric preprotein with an uncleaved transit peptide (Fig. 3), a lower-molecular-mass species probably representing the chimera after transit peptide cleavage (Fig. 3), and a previously described degradation product running at the size of GFP alone (Fig. 3) (59). The relative size shift between preprotein and mature protein varies between BTS-GFP reporters. Transit peptides are known to vary greatly in length (20 to 100 amino acids) (36), and this result indicates that pre-PfsCdc48 and pre-PfsUba1 contain longer transit peptides than the other apERAD components investigated here.
FIG. 3.
Western blot analysis of BTS-GFP transfectant lines. Predicted molecular masses of preproteins are presented. Based on these predictions, an asterisk indicates the unprocessed preprotein (after removal of signal peptide), a diamond indicates the processed mature protein, and an arrow indicates unspecific GFP degradation products. No immunoreactive bands are detected in protein samples prepared from the parental 3D7 line. LE, long exposure to reveal higher-molecular-mass bands in 3D7sCdc48-BTS samples. The values for the predicted molecular weight (MW) after removal of the signal peptide (−SP) are given in kDa.
Taken together, these results show that predicted BTS derived from PfsDer1-2, PfsCdc48, PfsUba1, and PfsUb are able to target GFP to the apicoplast and suggest that their function is to mediate delivery of the full-length proteins to this compartment.
Full-length PfsDer1-1 and PfsUba1 localize to the apicoplast, whereas PfhDer1-1 is an ER-resident protein.
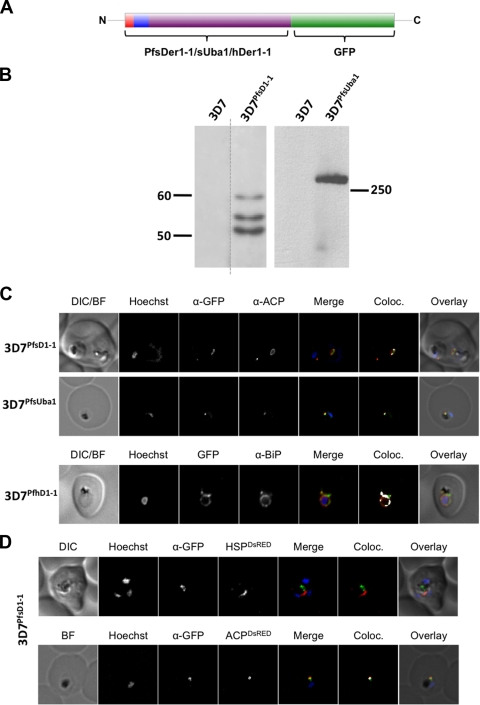
Our preliminary results, based on bioinformatic sequence analyses and BTS-GFP reporters, strongly suggest that apERAD components localize to the apicoplast; however, we wished to verify that full-length gene products are also present at the same location. Initially, we attempted to fuse the full-length gene encoding PfsDer1-1 to the GFP coding sequence and express the chimeric reporter from episomally maintained plasmids. Despite repeated attempts at transfection, we were unable to obtain a drug-resistant population, suggesting that expression of the reporter protein under these conditions is toxic to the parasite, possibly due to incorrect levels/timing of expression. For this reason, we decided to engineer the endogenous gene locus by single-crossover homologous recombination, incorporating the in-frame GFP coding sequence into the 3′ end of the respective gene (shown schematically in Fig. SA4 in the supplemental material). After transfection, selection of drug-resistant parasites, and drug cycling to select for integration, we were able to isolate clonal parasite populations that had integrated the transfection vector into their genomes. To verify integration into the correct gene loci, we carried out integration-specific PCR, followed by sequencing of the PCR products and Southern blotting (see Fig. SA4 in the supplemental material; also data not shown). In these parasites, expression of the PfsDer1-1-GFP or PfsUba1-GFP fusion protein is under the control of the endogenous promoter.
We analyzed our clonal integrant parasite lines 3D7PfsD1-1 and 3D7PfsUba1 by Western blotting. In the case of 3D7PfsD1-1, in addition to a band migrating at the size of the mature, processed PfsDer1-1-GFP chimera (50 kDa), several higher-molecular-mass bands could be detected, probably representing transit peptide cleavage intermediates (Fig. 4B). Analysis of 3D7PfsUba1 revealed a single >250-kDa band, which was absent in the parental 3D7 strain, in agreement with the predicted molecular mass of the GFP-tagged chimeric protein. Cleavage intermediates could not be visualized, most likely due to poor separation at this high molecular mass. The general expression levels of both proteins are low and required us to load approximately four times as many parasite cell equivalents (4 × 107 versus 1 × 107) as is usual in our laboratory in order to obtain signals of sufficient strength for analysis.
FIG. 4.
(A) Structure of the GFP-reporter constructs used in this study. Red, predicted signal peptide; blue, transit peptide; purple, remainder of protein sequence. (B) Western blot analysis of integrant parasite lines 3D7PfsD1-1 and 3D7PfsUba1 with anti-GFP antibodies. Several bands are recognized in the 3D7PfsD1-1 transgenic but not the 3D7 parental line. This multiple-band pattern is probably due to transit peptide processing. A single high-molecular-mass band is evident in the 3D7PfsUba1 transfectant line but not in the parental 3D7 line. Size markers are in kilodaltons. (C) IFA of GFP fusion transfectant lines. DIC, differential interference contrast; BF, bright field; Coloc, colocalization. In all transfectant parasite lines, the ACP signal is seen to overlap with the GFP signal, verifying an apicoplast localization of the GFP reporter. (D) Cotransfection and IFA of the 3D7PfsD1-1 transgenic parasite line. HSP, N terminus of mitochondrial PfHsp60; ACP, N terminus of apicoplast marker ACP. The GFP signal is seen to overlap with DsRed in the ACP-DsRed but not HSP-DsRed cotransfectant lines, excluding a mitochondrial localization and verifying an apicoplast localization of the GFP reporter. α, anti.
Epifluorescence microscopy of 3D7PfsD1-1 and 3D7PfsUba1 revealed only a weak GFP signal, consistent with the low protein abundance noted above. As subcellular localization of the GFP-chimera by live-cell imaging was limited by detection sensitivity, we carried out IFA using antibodies against GFP and ACP. In both 3D7PfsD1-1 and 3D7PfsUba1 lines, GFP and ACP signals colocalized (Fig. 4C), suggesting that both PfsDer1-1-GFP and PfsUba1-GFP are resident apicoplast proteins. As a control, we expressed PfhDer1-1 fused to GFP from episomally maintained plasmids, generating the 3D7PfsD1-1 line. Fluorescence microscopy of this parasite line revealed GFP fluorescence in a perinuclear ring structure, indicative of an ER localization. Immunofluorescent colocalization using antibodies recognizing the ER marker protein BiP (a kind gift of T. Gilberger) substantiated this, with good colocalization of GFP and BiP signals (Fig. 4C, bottom). As an additional control, we cotransfected 3D7PfsD1-1 with a construct encoding either the BTS of the apicoplast-resident protein ACP or the mitochondrial targeting sequence of mitochondrial PfHsp60 fused to the red fluorescent protein DsRed. In fixed cells colabeled with anti-GFP antibodies, ACP-DsRed and GFP signals colocalized while HSP-DsRed and GFP signal did not (Fig. 4D). These data further support our hypothesis that the full-length PfsDer1-1-GFP fusion protein is transported to the apicoplast, whereas PfhDer1-1 is a bona fide ER-resident protein.
PfsDer1-1 tightly associates with apicoplast membranes.
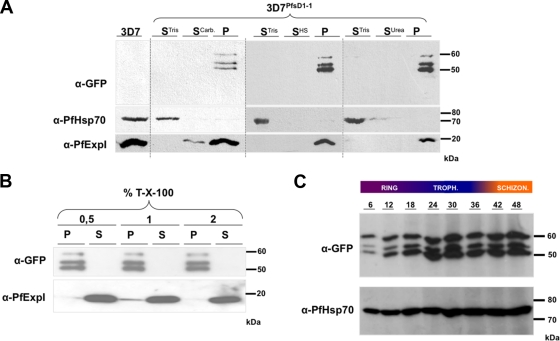
To function as part of a protein-conducting channel for the translocation of preproteins, PfsDer1-1 is expected to integrate into biological membranes. We therefore investigated the membrane association of our PfsDer1-1-GFP fusion construct. To this end, we carried out sequential membrane fractionation and extraction on 3D7PfsD1-1-infected erythrocytes, followed by Western blotting with anti-GFP antibodies. As a control for the fractionation and extraction protocol, we also analyzed the distribution of PfHsp70 (a soluble protein of the parasite cytosol) and PfExp1 (a single spanning TM protein of the parasitophorous vacuolar membrane) (17). As expected, PfHsp70 was found only in the soluble fraction following hypotonic lysis with 10 mM Tris (pH 7.4) (Fig. 5A), and PfExp1 was found largely in the carbonate-insoluble pellet fraction (Fig. 5A) with a weak signal in the supernatant following carbonate extraction, as previously described (38). GFP signals could be detected in only the final carbonate-insoluble pellet fraction, consistent with a strong association of PfsDer1-1-GFP with a lipid bilayer (Fig. 5A). Similarly, GFP signal could be detected only in the pellet fraction following membrane extraction with urea (Fig. 5A, P), with control proteins PfHsp70 and PfExp1 present in the Tris-soluble fraction (Fig. 5A) and final membrane pellet fraction (Fig. 5A), respectively, as expected and previously described (36). Upon treatment with increasing concentrations of the detergent Triton X-100, the control protein PfExp1 could be easily extracted from the membrane fraction (Fig. 5B), whereas PfsDer1-1-GFP was highly resistant to solubilization (Fig. 5B). Taken together, these data verify that PfsDer1-1 is tightly associated with apicoplast membranes and is probably an integral membrane protein of this organelle.
FIG. 5.
(A) Membrane extraction of 3D7PfsD1-1-infected erythrocytes. STris, supernatant after 50 mM Tris lysis; SCarb, SHS, and SUrea refer to supernatant after extraction with, respectively, carbonate, high salt, or urea buffer; P, final membrane pellet. In all cases PfsDer1-1-GFP can be detected exclusively in the final membrane pellet fraction, suggestive of a tight membrane association. Control proteins PfHsp70 (soluble protein) and PfExp1 (protein containing a TM domain) can be detected in the STris and P fractions, respectively, as expected. (B) Triton X-100 (T-X-100) extraction of 3D7PfsD1-1-infected erythrocytes. P, pellet after extraction; S, supernatant following extraction. PfsDer1-1-GFP can de detected in the pellet fraction only after treatment, whereas PfExp1 is solubilized already with 0.5% Triton X-100. (C) Developmental expression of PfsDer1-1-GFP. Protein extracts from highly synchronized 3D7PfsD1-1 parasites were prepared and analyzed by Western blotting with anti-GFP antibodies. As a control, we analyzed expression of PfHsp70 using anti-PfHsp70 antibodies. Expression of PfsDer1-1-GFP appears (in comparison to PfHsp70) to gradually increase during the 48-h intraerythrocytic life cycle of the parasite. Time scale refers to hours postinvasion. Troph, trophozoite; Schizon, schizont; α, anti.
PfsDer1-1 is expressed throughout the intraerythrocytic life cycle.
According to the study of Le Roch et al., mRNA abundance of genes encoding P. falciparum apERAD components, including PfsDer1-1, increases dramatically during the late trophozoite and early schizont stages of the parasite's intraerythrocytic cycle (32). To investigate whether these mRNA expression profiles correlated with protein abundance, we tightly synchronized 3D7PfsD1-1 parasite cultures using sorbitol (30) and removed samples for analysis every 6 h. Using anti-GFP antibodies, we could observe that the protein appeared to be present throughout the entire intraerythrocytic life cycle (Fig. 5B) but that protein abundance increased throughout the ∼48-h cycle, with the highest levels of protein being found in late trophozoites and early schizont parasites, consistent with the higher mRNA abundance during these stages and suggesting regulation at the transcriptional level. It appears that highest expression of PfsDer1-1 takes place late in the parasite's developmental cycle, correlating well with the time period in which both nuclear and apicoplast division is taking place (54).
Apicoplast targeting signals in P. falciparum proteins appear divergent from those in most other chromalveolates.
The study by Sommer et al. (49) leads us to predict that apERAD components are situated either in the second outermost membrane itself (membrane-bound components such as PfsDer1-1) or in the space between the second and third apicoplast membrane, referred to as the periplastidic compartment (PPC; soluble components such as PfsCdc48). This indicates that, in contrast to enzymes involved in biochemical pathways in the apicoplast stroma, apERAD components only need to be targeted across two of the four membranes surrounding the apicoplast. As the small size of the apicoplast and the diffraction limits associated with light microscopy do not allow us to verify this localization via light microscopy, we asked if bioinformatic analyses would provide support for a PPC localization. Previous studies on protein transport to complex plastids of red algal origin have shown that differential sorting to the plastid stroma (across four membranes) or the PPC (across only two membranes) is directed by the amino acid present in the +1 position following signal peptide cleavage. Proteins required to cross all four membranes usually possess an aromatic amino acid, or leucine, at this position, whereas those localizing to the PPC do not (14, 15, 19, 37). To investigate whether we could predict the localization of P. falciparum apERAD components based on these criteria, we analyzed the amino acid residue at the +1 position following in silico signal peptide cleavage. This analysis revealed that none of the proteins studied is predicted to expose an aromatic amino acid at the N terminus of the transit peptide, and only one protein (PfsUba) exposes a leucine residue at this point (see Fig. SA5 in the supplemental material). For comparison, we also analyzed the +1 amino acid of apERAD components encoded by P. tricornutum, a diatom algae that also contains a four-membrane bound plastid. None of the P. tricornutum sequences reveals an aromatic amino acid (or leucine) at the +1 position (see Fig. SA5 in the supplemental material). In itself, this result is suggestive of a predicted PPC localization for P. falciparum apERAD proteins; however, a larger bioinformatic analysis encompassing the predicted +1 amino acids of 194 predicted apicoplast proteins reveals that only 56 (29%) of these proteins do in actual fact obey the +1 rule (see Fig. SA5, upper panel, in the supplemental material). As a negative bioinformatic control, we analyzed the +1 amino acid of 277 P. falciparum proteins predicted to contain an N-terminal ER-type signal sequence but no transit peptide. Of these 277 proteins, 67 (24%) contain an aromatic amino acid, or leucine, at the +1 site (see Fig. SA5, lower panel, in the supplemental material). These data suggest that the +1 rule does not apply to P. falciparum apicoplast-targeted proteins and, therefore, cannot be used as a tool to predict a definitive localization of P. falciparum proteins to either the PPC or apicoplast stroma.
Predicted TM domains of sDer1-1 show unusual features.
To function as a protein-conducting channel, sDer1-1 must firstly cotranslationally enter the secretory pathway before being carried to, and inserting into, an internal apicoplast membrane. This is an unusual situation, as it implies that a highly hydrophobic TM protein must potentially cross several membranes before reaching its site of action. Although transport of proteins destined to posttranslationally insert into membranes is poorly understood, several studies have revealed unusual properties of predicted TM domains in proteins trafficked in this manner (34, 42, 43). For this reason, we performed a statistical comparison of the length distribution of the predicted membrane-spanning regions of all Plasmodium Der1-1 orthologues (three sDer1-1 and three hDer1-1 proteins). A KS statistic (40) reveals that the lengths of the first and the third TM domains differ significantly (5% level) between ER and apicoplast copies (for TM1 sDer1-1, 19.3 ± 0.7 amino acids; TM1 hDer1-1, 21.2 ± 2.2 amino acids; TM3 sDer1-1, 19.8 ± 5.4 amino acids; TM3 hDer1-1, 23.7 ± 5.4 amino acids) (see Table SA6 in the supplemental material).
DISCUSSION
Here, we report the identification and initial characterization of an ERAD-derived potential preprotein translocon complex within the apicoplast, referred to as apERAD, of the human malaria parasite P. falciparum. Utilizing a bioinformatic approach, we are able to show that all plastid-bearing apicomplexan parasites so far studied encode an apERAD. Furthermore, using transfection technology paired with protein biochemistry, we were able to demonstrate that the predicted BTS derived from several apERAD homologues are sufficient to target GFP to the apicoplast. Additionally, as proof of principle, we localized PfsDer1-1 and PfsUba1 (homologues of yeast ERAD components) to the apicoplast. Taken together, these data are consistent with a role for PfsDer1-1 in import of nucleus-encoded preproteins to the apicoplast.
Although several models have been suggested to account for the trafficking of nucleus-encoded preprotein to the complex plastid of chromalveolates containing four membrane-bound plastids until recently no molecular machinery had been identified that could actually carry out such transport processes (recently reviewed in reference 50). Numerous early studies of protein translocation across biological membranes revealed several criteria which must be fulfilled by PCCs and their associated factors, including the creation of a hydrophilic pore within the target membrane, the means to distinguish substrate proteins, and the necessity for a driving force to either pull or push polypeptide chains through the channel (6, 7, 48). The discovery of duplicated ERAD-derived systems in chromalveolates thus provided an attractive solution to the mechanistic problem as an ERAD-based protein translocon would be able to provide a hydrophilic pore across the membrane (based around sDer1-1, possibly together with sDer1-2) and the necessary pulling force for passage through the pore (provided by associated factors including sCdc48).
Integration of the GFP-coding sequence into the PfsDer1-1 and PfsUba1 gene loci allowed us to localize these proteins by fluorescence microscopy. Despite low endogenous expression levels of the reporter protein, we were able to show that these proteins colocalize with the apicoplast marker ACP. Due to the small size of the apicoplast and diffraction limits associated with light microscopy, we were not able to directly assign the proteins to a particular subcompartment of the apicoplast. Based on studies of an ERAD-derived translocon in chromalveolate algae (49) and the diatom alga P. tricornutum (20), we suggest that PfsDer1-1 inserts into the second outermost apicoplast membrane. Although we cannot formally discount the possibility that P. falciparum apERAD components are transported to the third outer, or indeed inner, apicoplast membrane, this would appear unlikely for several reasons. If PfsDer1-1 is involved in protein transport processes, insertion in the third outer membrane would result in a topology incongruous with transport into the apicoplast, requiring a reversal of the transport direction, a situation for which no precedent exists (49). Additionally, a recent study in T. gondii has convincingly demonstrated a role for T. gondii Tic20 in protein transport to the apicomplexan apicoplast, a situation which is likely to also hold true for P. falciparum, thus making a role for PfsDer1-1 in this process unlikely. Future studies will aim to experimentally address the exact localization of this protein, possibly by using novel cell biological tools such as self-assembling GFP (8).
PfsDer1-1 is extremely tightly associated with membranes, as evidenced by membrane extraction experiments. Unexpectedly, we were unable to solubilize PfsDer1-1 even under harsh (2% Triton X-100) conditions. Although insolubility under high Triton X-100 concentrations may seem indicative of an association with cytoskeletal components, there is no evidence for such structures in the apicoplast. Analysis of further apicoplast membrane proteins may shed light on this unusual solubility profile.
In yeast, Cdc48-mediated translocation through the ERAD translocon is driven by ubiquitinylation of substrate proteins upon emergence from the translocon (35, 41). In the course of this present study, we identified a Ub predicted to be endowed with a BTS for transport to the apicoplast (PfsUb). The BTS derived from this protein was able to target GFP to the apicoplast. PfsUb contains the essential K48 and K63 residues required for its involvement in both mono- and polyubiquitinylation of substrate proteins (see Fig. SA7 in the supplemental material). We were not able to categorically identify high-molecular-weight ubiquitinylated forms of our GFP reporter proteins, suggesting that if apicoplast targeted proteins are indeed ubiquitinylated during their trafficking, either Ub is subsequently cleaved from the proteins or the transit peptide (a possible site of ubiquitinylation) is cleaved together with the Ub moiety from the mature protein. The latter situation would seem unlikely, given that the transit peptide is required for further membrane passage events. While the exact molecular details remain to be dissected, our data support a role for PfsUb in ubiquitinylation of apicoplast-targeted preproteins. Possibly, ubiquitinylation of transit peptides emerging from the trans side of the translocon acts as a trigger for PfsCdc48-driven membrane translocation. It is noteworthy that we failed to identify apERAD versions of the Ub ligases PfHrd1 or PfHrd3. Ub ligase-independent monoubiquitinylation of substrate proteins has previously been described (22), and the possibility exists that the reduced apERAD system instead relies on Ub conjugating (E2) enzymes to transfer Ub to substrate proteins. Alternatively, it is feasible that one of the two independent copies of PfHrd3 we identified is transported to the apicoplast, albeit in a manner which does not require a recognizable transit peptide. Such a mechanism has been demonstrated for the delivery of FtsH to membranes of the T. gondii apicoplast (25).
One observation which is of particular interest is that transit peptides derived from apERAD components also appear to be cleaved upon organellar import (Fig. 3). Transit peptide cleavage has previously been observed in preproteins transported to the lumen of the apicoplast, and a putative stromal-processing peptidase has been identified (56). Processing of PPC-localized plastid preproteins has previously been observed in P. tricornutum and was taken as evidence to suggest the existence of a periplastid-processing peptidase (15). Our results further support a model in which preproteins destined for the PPC are processed by an as yet unidentified protease.
In the course of our study, we also investigated whether it is possible to distinguish between PPC and stromal apicoplast proteins, based on the physiochemical properties of the amino acid residue exposed after signal peptide cleavage. We find that, in contrast to several other systems studied, there is no significant difference at the +1 position between proteins trafficked across all four apicoplast membranes and those that must passage across only the outer two. Indeed, there is no significant difference between the +1 residue in apicoplast proteins and that of other secretory proteins. Bioinformatic studies suggest that the phenylalanine motif is of ancestral origin (shared in the common ancestor of the green and red lineages) and has subsequently been lost in members of the green line. Additionally, transit peptides derived from haptophyte algae (red lineage) also have a relaxed requirement for phenylalanine at this position (37). Our data suggest that, at least at the level of the +1 position, P. falciparum transit peptides appear to share some properties with those of haptophytes. As a logical consequence, this result also suggests that P. falciparum apicoplast proteins (which do not obey the +1 rule) are differentially sorted to either PPC or plastid stroma based on sequence information in the downstream protein sequence. Further studies will be required to verify this hypothesis experimentally as well as to elucidate its significance for the nature of the apicoplast protein import system.
In yeast, ERAD substrates are generally recognized on the basis of distinct N-glycan modifications (18). Previous studies have determined that P. falciparum has little, if any, capacity for N-glycosylation (46), suggesting that even for the hERAD system, the signal required for ERAD recognition could differ from that common in yeast. As a result, how proteins are recognized by the apERAD system must also remain a matter of speculation at this point. What does appear clear is that, as all proteins trafficked to the apicoplast must first enter the parasite's ER, recognition of apERAD substrates must take place via signals distinct from that for hERAD. Likewise, we failed in all organisms studied to identify apERAD homologues of Npl4p, a protein which, in the yeast system, usually interacts with Cdc48p and Ufd1p. This Cdc48p-Ufd1p-Nlp4p is involved in recognition of polyubiquitinylated ERAD substrate proteins. A recent study of sERAD in the diatom P. tricornutum suggests that sDer1 itself is able to distinguish substrates (20). Thus, PfsDer1 itself may play a role in substrate recognition. It is clear that further studies will be required to dissect the exact sequence and/or substrate recognition requirements for this translocation process.
To conclude, our data provide strong evidence for the presence in the apicoplast of the malaria parasite P. falciparum and of other apicomplexans of components of an ERAD-derived translocon complex. Specifically, we can show that PfsDer1-1 and PfsUba1 are localized to the apicoplast and that the BTS derived from further sERAD components are also capable of targeting reporters to the apicoplast, suggesting that these proteins also fulfill their biological function in this compartment. This translocon complex is likely to be required for the import of nucleus-encoded preproteins to the organelle. Exactly how these components function in a coordinated fashion to allow passage of proteins across the multiple membranes of the apicoplast remains to be studied in detail but will provide the basis for future research efforts. The unusual intracellular life cycle of the malaria parasite has already revealed several novel cell biological phenomena, and our study suggests that P. falciparum still has many tricks up its sleeve.
Supplementary Material
Acknowledgments
This work was supported by a Ph.D. fellowship of the Philipps University Marburg (S.S.), the International Max-Planck Research School (C.T.), DFG grants PR1099/2-1 and SFBTR1 (J.M.P.), a long-term fellowship of the European Molecular Biology Organization (T.W.A.K., ALTF-763-2006), and SFB593 (U.G.M).
We especially thank Thierry Blisnick, Lars Bullmann, Franziska Hempel, Ming Kalanon, Simone Kuelzer, Christof Taxis, Klaus Lingelbach, Geoff McFadden, Nina Gehde, and our coworkers at the Marburg University Hospital blood bank for essential reagents and fruitful discussions. We especially acknowledge the help and advice of the entire PlasmoDB team.
Footnotes
Published ahead of print on 5 June 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Abrahamsen, M. S., T. J. Templeton, S. Enomoto, J. E. Abrahante, G. Zhu, C. A. Lancto, M. Deng, C. Liu, G. Widmer, S. Tzipori, G. A. Buck, P. Xu, A. T. Bankier, P. H. Dear, B. A. Konfortov, H. F. Spriggs, L. Iyer, V. Anantharaman, L. Aravind, and V. Kapur. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304441-445. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 3.Aurrecoechea, C., J. Brestelli, B. P. Brunk, J. Dommer, S. Fischer, B. Gajria, X. Gao, A. Gingle, G. Grant, O. S. Harb, M. Heiges, F. Innamorato, J. Iodice, J. C. Kissinger, E. Kraemer, W. Li, J. A. Miller, V. Nayak, C. Pennington, D. F. Pinney, D. S. Roos, C. Ross, C. J. J. Stoeckert, C. Treatman, and H. Wang. 2009. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37D539-D543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aurrecoechea, C., M. Heiges, H. Wang, Z. Wang, S. Fischer, P. Rhodes, J. Miller, E. Kraemer, C. J. J. Stoeckert, D. S. Roos, and J. C. Kissinger. 2007. ApiDB: integrated resources for the apicomplexan bioinformatics resource center. Nucleic Acids Res. 35D427-D430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 6.Blobel, G., and B. Dobberstein. 1975. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 67835-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blobel, G., and B. Dobberstein. 1975. Transfer to proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J. Cell Biol. 67852-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabantous, S., and G. S. Waldo. 2006. In vivo and in vitro protein solubility assays using split GFP. Nat. Methods 3845-854. [DOI] [PubMed] [Google Scholar]
- 9.Cao, B., A. Porollo, R. Adamczak, M. Jarrell, and J. Meller. 2006. Enhanced recognition of protein transmembrane domains with prediction-based structural profiles. Bioinformatics 22303-309. [DOI] [PubMed] [Google Scholar]
- 10.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 141188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fidock, D. A., and T. E. Wellems. 1997. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. USA 9410931-10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foth, B. J., S. A. Ralph, C. J. Tonkin, N. S. Struck, M. Fraunholz, D. S. Roos, A. F. Cowman, and G. I. McFadden. 2003. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science 299705-708. [DOI] [PubMed] [Google Scholar]
- 13.Gehde, N., C. Hinrichs, I. Montilla, S. Charpian, K. Lingelbach, and J. M. Przyborski. 2009. Protein unfolding is an essential requirement for transport across the parasitophorous vacuolar membrane of Plasmodium falciparum. Mol. Microbiol. 71613-628. [DOI] [PubMed] [Google Scholar]
- 14.Gould, S. B., M. S. Sommer, K. Hadfi, S. Zauner, P. G. Kroth, and U. G. Maier. 2006. Protein targeting into the complex plastid of cryptophytes. J. Mol. Evol. 62674-681. [DOI] [PubMed] [Google Scholar]
- 15.Gould, S. B., M. S. Sommer, P. G. Kroth, G. H. Gile, P. J. Keeling, and U. G. Maier. 2006. Nucleus-to-nucleus gene transfer and protein retargeting into a remnant cytoplasm of cryptophytes and diatoms. Mol. Biol. Evol. 232413-2422. [DOI] [PubMed] [Google Scholar]
- 16.Grellier, P., D. Depoix, J. Schrevel, and I. Florent. 2008. Discovery of new targets for antimalarial chemotherapy. Parasite 15219-225. [DOI] [PubMed] [Google Scholar]
- 17.Gunther, K., M. Tummler, H. H. Arnold, R. Ridley, M. Goman, J. G. Scaife, and K. Lingelbach. 1991. An exported protein of Plasmodium falciparum is synthesized as an integral membrane protein. Mol. Biochem. Parasitol. 46149-157. [DOI] [PubMed] [Google Scholar]
- 18.Helenius, A., and M. Aebi. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 731019-1049. [DOI] [PubMed] [Google Scholar]
- 19.Hempel, F., A. Bozarth, M. S. Sommer, S. Zauner, J. M. Przyborski, and U. G. Maier. 2007. Transport of nuclear-encoded proteins into secondarily evolved plastids. Biol. Chem. 388899-906. [DOI] [PubMed] [Google Scholar]
- 20.Hempel, F., L. Bullmann, J. Lau, S. Zauner, and U. G. Maier. 17 April 2009, posting date. ERAD-derived pre-protein transport across the 2nd outermost plastid membrane of diatoms. Mol. Biol. Evol. doi: 10.1093/molbev/msp079. [DOI] [PubMed]
- 21.Hitt, R., and D. H. Wolf. 2004. Der1p, a protein required for degradation of malfolded soluble proteins of the endoplasmic reticulum: topology and Der1-like proteins. FEMS Yeast Res. 4721-729. [DOI] [PubMed] [Google Scholar]
- 22.Hoeller, D., C. M. Hecker, S. Wagner, V. Rogov, V. Dotsch, and I. Dikic. 2007. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol. Cell 26891-898. [DOI] [PubMed] [Google Scholar]
- 23.Hopp, T. P., and K. R. Woods. 1981. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. USA 783824-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, J., N. Mullapudi, C. A. Lancto, M. Scott, M. S. Abrahamsen, and J. C. Kissinger. 2004. Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol. 5R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karnataki, A., A. E. Derocher, I. Coppens, J. E. Feagin, and M. Parsons. 2007. A membrane protease is targeted to the relict plastid of toxoplasma via an internal signal sequence. Traffic 81543-1553. [DOI] [PubMed] [Google Scholar]
- 26.Knop, M., A. Finger, T. Braun, K. Hellmuth, and D. H. Wolf. 1996. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 15753-763. [PMC free article] [PubMed] [Google Scholar]
- 27.Kohler, S., C. F. Delwiche, P. W. Denny, L. G. Tilney, P. Webster, R. J. Wilson, J. D. Palmer, and D. S. Roos. 1997. A plastid of probable green algal origin in apicomplexan parasites. Science 2751485-1489. [DOI] [PubMed] [Google Scholar]
- 28.Kooij, T. W., J. M. Carlton, S. L. Bidwell, N. Hall, J. Ramesar, C. J. Janse, and A. P. Waters. 2005. A Plasmodium whole-genome synteny map: indels and synteny breakpoints as foci for species-specific genes. PLoS Pathog. 1e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157105-132. [DOI] [PubMed] [Google Scholar]
- 30.Lambros, C., and J. P. Vanderberg. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65418-420. [PubMed] [Google Scholar]
- 31.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 232947-2948. [DOI] [PubMed] [Google Scholar]
- 32.Le Roch, K. G., Y. Zhou, P. L. Blair, M. Grainger, J. K. Moch, J. D. Haynes, P. De La Vega, A. A. Holder, S. Batalov, D. J. Carucci, and E. A. Winzeler. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 3011503-1508. [DOI] [PubMed] [Google Scholar]
- 33.Lu, F., H. Jiang, J. Ding, J. Mu, J. G. Valenzuela, J. M. Ribeiro, and X. Z. Su. 2007. cDNA sequences reveal considerable gene prediction inaccuracy in the Plasmodium falciparum genome. BMC Genomics 8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meier, S., W. Neupert, and J. M. Herrmann. 2005. Proline residues of transmembrane domains determine the sorting of inner membrane proteins in mitochondria. J. Cell Biol. 170881-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meusser, B., C. Hirsch, E. Jarosch, and T. Sommer. 2005. ERAD: the long road to destruction. Nat. Cell Biol. 7766-772. [DOI] [PubMed] [Google Scholar]
- 36.Papakrivos, J., C. I. Newbold, and K. Lingelbach. 2005. A potential novel mechanism for the insertion of a membrane protein revealed by a biochemical analysis of the Plasmodium falciparum cytoadherence molecule PfEMP-1. Mol. Microbiol. 551272-1284. [DOI] [PubMed] [Google Scholar]
- 37.Patron, N. J., and R. F. Waller. 2007. Transit peptide diversity and divergence: a global analysis of plastid targeting signals. Bioessays 291048-1058. [DOI] [PubMed] [Google Scholar]
- 38.Przyborski, J. M., S. K. Miller, J. M. Pfahler, P. P. Henrich, P. Rohrbach, B. S. Crabb, and M. Lanzer. 2005. Trafficking of STEVOR to the Maurer's clefts in Plasmodium falciparum-infected erythrocytes. EMBO J. 242306-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ralph, S. A., B. J. Foth, N. Hall, and G. I. McFadden. 2004. Evolutionary pressures on apicoplast transit peptides. Mol. Biol. Evol. 212183-2194. [DOI] [PubMed] [Google Scholar]
- 40.Rassokhin, D. N., and D. K. Agrafiotis. 2000. Kolmogorov-Smirnov statistic and its application in library design. J. Mol. Graph. Model. 18368-382. [DOI] [PubMed] [Google Scholar]
- 41.Richly, H., M. Rape, S. Braun, S. Rumpf, C. Hoege, and S. Jentsch. 2005. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 12073-84. [DOI] [PubMed] [Google Scholar]
- 42.Rojo, E. E., B. Guiard, W. Neupert, and R. A. Stuart. 1998. Sorting of D-lactate dehydrogenase to the inner membrane of mitochondria. Analysis of topogenic signal and energetic requirements. J. Biol. Chem. 2738040-8047. [DOI] [PubMed] [Google Scholar]
- 43.Rojo, E. E., B. Guiard, W. Neupert, and R. A. Stuart. 1999. N-terminal tail export from the mitochondrial matrix. Adherence to the prokaryotic “positive-inside” rule of membrane protein topology. J. Biol. Chem. 27419617-19622. [DOI] [PubMed] [Google Scholar]
- 44.Romisch, K. 2005. Endoplasmic reticulum-associated degradation. Annu. Rev. Cell Dev. Biol. 21435-456. [DOI] [PubMed] [Google Scholar]
- 45.Rossner, M., and K. M. Yamada. 2004. What's in a picture? The temptation of image manipulation. J. Cell Biol. 16611-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samuelson, J., S. Banerjee, P. Magnelli, J. Cui, D. J. Kelleher, R. Gilmore, and P. W. Robbins. 2005. The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc. Natl. Acad. Sci. USA 1021548-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato, S., and R. J. Wilson. 2004. The use of DsRED in single- and dual-color fluorescence labeling of mitochondrial and plastid organelles in Plasmodium falciparum. Mol. Biochem. Parasitol. 134175-179. [DOI] [PubMed] [Google Scholar]
- 48.Schatz, G., and B. Dobberstein. 1996. Common principles of protein translocation across membranes. Science 2711519-1526. [DOI] [PubMed] [Google Scholar]
- 49.Sommer, M. S., S. B. Gould, P. Lehmann, A. Gruber, J. M. Przyborski, and U. G. Maier. 2007. Der1-mediated preprotein import into the periplastid compartment of chromalveolates? Mol. Biol. Evol. 24918-928. [DOI] [PubMed] [Google Scholar]
- 50.Tonkin, C. J., M. Kalanon, and G. I. McFadden. 2008. Protein targeting to the malaria parasite plastid. Traffic 9166-175. [DOI] [PubMed] [Google Scholar]
- 51.Tonkin, C. J., N. S. Struck, K. A. Mullin, L. M. Stimmler, and G. I. McFadden. 2006. Evidence for Golgi-independent transport from the early secretory pathway to the plastid in malaria parasites. Mol. Microbiol. 61614-630. [DOI] [PubMed] [Google Scholar]
- 52.Tonkin, C. J., G. G. van Dooren, T. P. Spurck, N. S. Struck, R. T. Good, E. Handman, A. F. Cowman, and G. I. McFadden. 2004. Localization of organellar proteins in Plasmodium falciparum using a novel set of transfection vectors and a new immunofluorescence fixation method. Mol. Biochem. Parasitol. 13713-21. [DOI] [PubMed] [Google Scholar]
- 53.Trager, W., and J. B. Jensen. 1976. Human malaria parasites in continuous culture. Science 193673-675. [DOI] [PubMed] [Google Scholar]
- 54.van Dooren, G. G., M. Marti, C. J. Tonkin, L. M. Stimmler, A. F. Cowman, and G. I. McFadden. 2005. Development of the endoplasmic reticulum, mitochondrion and apicoplast during the asexual life cycle of Plasmodium falciparum. Mol. Microbiol. 57405-419. [DOI] [PubMed] [Google Scholar]
- 55.van Dooren, G. G., S. D. Schwartzbach, T. Osafune, and G. I. McFadden. 2001. Translocation of proteins across the multiple membranes of complex plastids. Biochim. Biophys. Acta 154134-53. [DOI] [PubMed] [Google Scholar]
- 56.van Dooren, G. G., V. Su, M. C. D'Ombrain, and G. I. McFadden. 2002. Processing of an apicoplast leader sequence in Plasmodium falciparum and the identification of a putative leader cleavage enzyme. J. Biol. Chem. 27723612-23619. [DOI] [PubMed] [Google Scholar]
- 57.van Dooren, G. G., C. Tomova, S. Agrawal, B. M. Humbel, and B. Striepen. 2008. Toxoplasma gondii Tic20 is essential for apicoplast protein import. Proc. Natl. Acad. Sci. USA 10513574-13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waller, R. F., P. J. Keeling, G. G. van Dooren, and G. I. McFadden. 2003. Comment on “A green algal apicoplast ancestor.” Science 30149. [DOI] [PubMed] [Google Scholar]
- 59.Waller, R. F., M. B. Reed, A. F. Cowman, and G. I. McFadden. 2000. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 191794-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiesner, J., A. Reichenberg, S. Heinrich, M. Schlitzer, and H. Jomaa. 2008. The plastid-like organelle of apicomplexan parasites as drug target. Curr. Pharm. Des. 14855-871. [DOI] [PubMed] [Google Scholar]
- 61.World Health Organization. 2008. World malaria report 2008. World Health Organization, Geneva, Switzerland. http://apps.who.int/malaria/wmr2008/malaria2008.pdf.
- 62.Zuegge, J., S. Ralph, M. Schmuker, G. I. McFadden, and G. Schneider. 2001. Deciphering apicoplast targeting signals—feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene 28019-26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.