Abstract
The gene coding for a C-5(6) sterol desaturase in Tetrahymena thermophila, DES5A, has been identified by the knockout of the TTHERM_01194720 sequence. Macronucleus transformation was achieved by biolistic bombardment and gene replacement through phenotypic assortment, using paromomycin as the selective agent. A knockout cell line (KO270) showed a phenotype consistent with that of the DES5A deletion mutant. KO270 converted only 6% of the added sterol into the C-5 unsaturated derivative, while the wild type accumulated 10-fold larger amounts under similar conditions. The decreased desaturation activity is specific for the C-5(6) position of lathosterol and cholestanol; other desaturations, namely C-7(8) and C-22(23), were not affected. Analysis by reverse transcription-PCR reveals that DES5A is transcribed both in the presence and absence of cholestanol in wild-type cells, whereas the transcribed gene was not detected in KO270. The growth of KO270 was undistinguishable from that of the wild-type strain. Des5Ap resembles known C-5(6) sterol desaturases, displaying the three typical histidine motifs, four hydrophobic transmembrane regions, and two other highly conserved domains of unknown function. A phylogenetic analysis placed T. thermophila's enzyme and Paramecium orthologues in a cluster together with functionally characterized C-5 sterol desaturases from vertebrates, fungi, and plants, although in a different branch.
Tetrahymena thermophila is a fresh-water protozoan that has been used successfully as a model system in cell biology (8). The advanced molecular and genetic tools developed for Tetrahymena have facilitated fundamental discoveries, such as the first descriptions of ribozymes, telomeres, and telomerases, thereby maintaining this organism at the forefront of fundamental research (2, 11, 30).
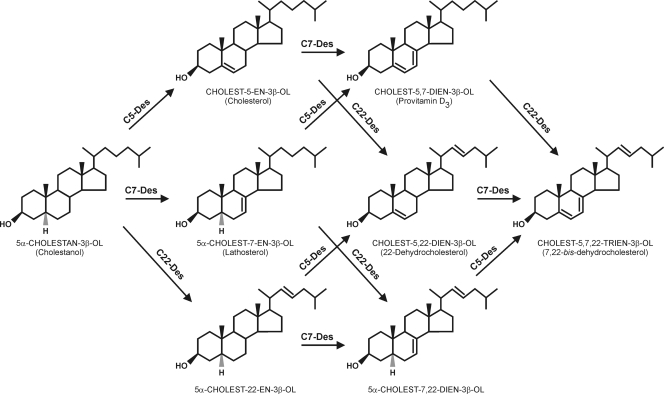
Conner et al. (5, 6) described the peculiar sterol metabolism in Tetrahymena that leads to the accumulation of provitamin D analogs due to the C-5(6), C-7(8), and C-22(23) sterol-desaturating activities present in the organism (Fig. 1). The transformation of cholesterol to the C-7 unsaturated derivative (provitamin D3 [cholest-5,7-dien-3β-ol]) particularly has attracted attention because of pharmaceutical and food-related applications (28, 29) to decrease the cholesterol content in foodstuffs and the coupled production of provitamin D3 in a single step (27). Despite the potential societal impact, progress on the isolation and purification of desaturases has been modest, mainly due to the loss of enzyme activity upon the dissociation of microsomal complexes (13).
FIG. 1.
Sterol desaturation in Tetrahymena; substrates and product formation. C5-Des, C-5(6) sterol desaturase; C7-Des, C-7(8) sterol desaturase; C22-Des, C-22(23) sterol desaturase.
The preliminary characterization of sterol-desaturating activities in T. thermophila indicated that the corresponding enzymes are located in the microsomal fraction and require cytochrome (Cyt) b5, Cyt b5 reductase, oxygen, and a reduced cofactor (NADH) (17). These biochemical requirements are characteristic of sterol C-5(6) desaturases and C-4 methyl oxidases (14). By using amino acid sequences of known C-5 desaturases as queries, eight putative desaturases/methyl oxidases were retrieved after a BLAST search of the T. thermophila genome. All of them have the three characteristic histidine boxes that represent the structural signature of this family of enzymes. The sequence with the highest score (TTHERM_01194720) was selected for further analysis.
As a first approach to unravel the pathway for sterol metabolism in T. thermophila, we report here the isolation and characterization by reverse genetics of the first C-5(6) sterol desaturase identified in a ciliate, as well as a detailed structural and phylogenetic analysis.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
T. thermophila strain CU428 (mpr1-1/mpr1-1; mp-s, VII), designated the wild type (WT) in this work, and plasmid pBS-MnB-3 were a gift from M. A. Gorovsky (University of Rochester, NY) (21). Cells were grown in 250-ml Erlenmeyer flasks containing 100 ml SPP (super proteose peptone) medium with the following composition (wt/vol): 1% proteose-peptone (Oxoid, United Kingdom), 0.1% yeast extract (Merck, Germany), 0.2% glucose (Merck, Germany), and 0.003% iron citrate (Sigma-Aldrich). In sterol desaturase activity assays, medium was supplemented with lathosterol (5α-cholest-7-en-3β-ol), cholestanol (5α-cholestan-3β-ol), or cholesterol (cholest-5-en-3β-ol) at a final concentration of 20 μg/ml, which was added from 1 mg/ml stock solutions in ethanol (17). When indicated, paromomycin (Sigma-Aldrich) was added from a 200 mg/ml stock solution in water, together with 1 μg/ml of CdCl2, which was prepared as a 100 μg/ml stock solution in water.
Cultures were inoculated daily with a 1:10 dilution of a 24-h culture. Cultivation was carried out in a rotary shaker (180 rpm) at 30°C.
Plasmid pBS-MnB-3, containing the neomycin resistance gene under a cadmium-inducible metallothionein (MTT) promoter (the neo 3 cassette) expressing paromomycin resistance, was used throughout this study (21).
Standard DNA and RNA manipulation procedures.
Genomic DNA of T. thermophila CU428 was prepared as previously described (10). The isolation of plasmid DNA was performed with a Wizard Plus SV Miniprep DNA Purification system kit (Promega). Total RNA was prepared from T. thermophila cultures grown for 6 and 24 h using TRIzol reagent (Invitrogen, Carlsbad, CA).
Nucleic acid fragments were amplified with PCR using Taq DNA polymerase (Go Taq; Promega). When high-fidelity PCR was required, the Triple Master PCR system (Eppendorf AG, Germany) plus Taq DNA polymerase was the choice. Amplifications involved an initial denaturation step at 94°C for 4 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 50 to 60°C for 1 min, and extension at 72°C for 2 min. The products were separated on 1% agarose gels, isolated, and recovered using a PCR Wizard Prep kit (Promega, Madison, WI). Sequencing reactions were performed in a Genius thermal cycler (Techne) using a Dye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq DNA polymerase (FS enzyme) (Applied Biosystems) by following the protocols supplied by the manufacturer, and they were analyzed in an ABI prism 377 sequencer (Applied Biosystems).
For RNA analysis, reverse transcription (RT) reactions were carried out using SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the manufacture's instructions. All RNA samples were treated with DNase I prior to amplification. The amplification was done for 35 cycles (94°C for 1 min, 55°C for 1 min, and 72°C for 1 min). cDNA synthesis was monitored by PCR with α-tubulin primers.
The primers used for all amplification reactions are listed in Table 1.
TABLE 1.
List of oligonucleotides used for PCR amplifications
| Description or purpose | Sequence IDa | Primer no. and sequence
|
|||
|---|---|---|---|---|---|
| No. | Forward | No. | Reverse | ||
| C270 fragment | UP | 1 | ATTAGCATTACTCCATAAGTTCC | 2 | GTGTATTTAAATTAAAGGAGTTATTCAGTATCTTTAATCCATTTAGCACG |
| DW | 3 | CCTCTTCACATACATGTTAGCTCTTTATTTTGTAAGCTTAATTATTCGC | 4 | GCTAGTGGAATAAGATTTAATGG | |
| neo 3 | 5 | TGAATAACTCCTTTAATTTAAATACAC | 6 | AGAGCTAACATGTATGTGAAGAGG | |
| KO270 mutant | WT allele | 7 | CTTACTGGGTTCCTGACAGG | 9 | GCTAGTGGAATAAGATTTAATGG |
| KO allele | 8 | TCCTCTTCACATACATGTTAGC | 9 | GCTAGTGGAATAAGATTTAATGG | |
| RT-PCR analysis | C5-Des exon | 10 | TTTGCCTGAATTTAAAGGAGATTT | 11 | GGAAGGTGTGGAGCCATCTA |
| α-Tub | 12 | TGTCGTCCCCAAGGAT | 13 | GTTCTCTTGGTCTTGATGGT | |
| DES5A gene | TTHERM_01194720 | 14 | ATGGTTTATTGGCTTATTGCTGAATAG | 15 | TCAATTCTTTTTTTGTTTAATTTATTTG |
ID, identity. neo 3 indicates the primers used for the neo 3 cassette in the plasmid pBS-MnB-3; WT allele and KO allele are the primers used to check the replacement of the endogenous gene in the KO270 mutant; C5-Des exon and α-Tub are the primers used for the amplification of the second exon of the TTHERM_01194720 gene and the α-tubulin gene used in the competitive RT-PCR analysis, respectively. DES5A gene TTHERM_01194720 is the primer used in the WT strain for the full amplification of the C5(6) sterol desaturase gene.
Construction of the transformation sequence C270 and the KO270 mutant.
For DES5A gene disruption in T. thermophila, we constructed the transformation sequence C270 for somatic knockout by overlapping PCR using two successive amplification rounds (23). In the first round, the flanking regions of the TTHERM_01194720 putative sequence (from the TIGR database) of 1.1 kb upstream (UP) and 0.9 kb downstream (DW) were amplified separately using T. thermophila genomic DNA as the template and primers 1 and 2 (for fragment UP) and 3 and 4 (for fragment DW). The neo 3 cassette (1.9 kb) from plasmid pBS-MnB-3, expressing paromomycin resistance under the control of a cadmium-inducible promoter (MTT) (21), was amplified separately with primers 5 and 6 (Table 1).
The three PCR products were purified from gels with the Wizard SV Gel and PCR Clean-Up system (Promega) and used as DNA templates for the second amplification round with primers 1 and 4. The entire 3.94-kb PCR product of the second-round amplification was purified and used for the transformation of Tetrahymena cells.
For the transformation of the recipient T. thermophila CU428, cells were grown in 50-ml cultures in SPP medium at 30°C to reach a density of 2 × 105 cells/ml. Cultures were starved overnight in 10 mM Tris buffer, pH 7.5, and transformed with 2.5 μg of purified C270 DNA fragment delivered with gold particles according to a biolistic gun protocol (4). Bombardment was performed in a Dupont Biolistic PDS-1000/He particle delivery system (Bio-Rad). Transformants were recovered in 50 ml SPP medium containing 1.0 μg/ml CdCl2. After 4 h, 80 μg/ml paromomycin was added, and the entire mixture was distributed in seven microtiter plates of 96 wells each.
Phenotypic assortment and gene replacement assays.
In our selection procedure, transformants first were grown in 96-well plates in SPP medium containing 1.0 μg/ml CdCl2 and 80 μg/ml paromomycin. Every 1 to 2 days, 10-μl aliquots of these cultures were transferred into fresh medium with an increasing paromomycin concentration of up to a maximum of 3 mg/ml. Cells resistant to this concentration of antibiotic were used to isolate clonal lines. With these procedures the mutant cell line KO270 was selected.
The level of gene segregation was checked by comparative PCR. Transcript levels were assayed by RT-PCR. In the first case, the WT gene and the corresponding fragment from the deletion mutant were amplified simultaneously, using primers 7 and 9 for the amplification of the WT copy of the DES5A gene and primers 8 and 9 for the deletion mutant.
For transcript-level assays, the WT and the KO270 mutant were grown in medium with or without cholestanol added. After RNA extraction and purification, cDNAs were obtained by PCR amplification with primers 10 and 11, corresponding to part of the second exon of the cDNA, and the products were separated on 1% agarose gels. The detection of α-tubulin transcript was used as a control using primers 12 and 13.
Growth curves.
Mid-log-phase cultures of WT T. thermophila and the KO270 mutant were used to inoculate 50 ml of fresh SPP medium at an initial cell density of 1 × 104 cells/ml. Cell numbers were determined after 0, 5, 10, 15, 20, and 36 h. Growth curves were plotted, and doubling times were calculated from the linear region of the growth curves plotted logarithmically.
Detection of C-5(6), C-7(8), and C-22(23) sterol desaturase activity.
Sterol desaturase activities were analyzed upon culture in medium with selected sterols added. For each specific activity, cells were grown for 24 h in SPP medium containing 20 μg/ml of lathosterol or cholesterol. After this period, 2-ml samples were withdrawn from the cultures, and cells were separated by centrifugation (3,000 × g, 5 min at 4°C), washed, resuspended in water, and submitted to lipid saponification by the addition of 1 volume of 2 M NaOH in methanol-water (1:1, vol/vol) at 60°C for 1 h (3). After being cooled and mixed, 5.6 ml of chloroform-methanol (3:2, vol/vol) was added. Sterols were extracted into the lower phase, concentrated under a nitrogen flow, and separated by high-performance liquid chromatography (HPLC) on a C18 Ultrasphere column, using methanol/water (98:2, vol/vol) as the mobile phase at 41°C. Sterol identification was performed by using standards and by mass spectrometry (MS) analysis (see further below).
Identification of sterols by gas chromatography-MS (GC-MS).
Cells from cultures with added sterol were collected by centrifugation at 3,000 × g for 5 min at 4°C and washed twice with 20 ml of distilled water, and the lipids were extracted according to Bligh and Dyer (3). The organic phase was evaporated under N2 and saponified. After twofold extraction with 2 ml hexane, the organic solvent was evaporated under an N2 stream, and the residue was resuspended in 50 μl of distilled pyridine. Fifty microliters of N-methyl-N-(trimethylsilyl) trifluoroacetamide was added, and the mixture was incubated for 40 min at 80°C. The composition of the steryl trimethylsilyl ether derivatives was analyzed by running samples through an SE-30 column (30-m tall, 0.22-mm inside diameter column; Scientific Glass Engineering, Ringwood, Australia) in a Perkin-Elmer AutoSystem XL gas chromatograph. The column was temperature programmed at 10°C/min from 100 to 310°C and subsequently held for 10 min at 310°C. MS was carried out using a Perkin-Elmer mass detector (model TurboMass) operated at an ionization voltage of 70 eV with a scan range of 20 to 500 Da. The retention time and mass spectrum of all new peaks obtained were compared to those of standards (Steraloids) and those available in the database NBS75K (National Bureau of Standards).
Phylogenetic analyses.
Available C-5(6) sterol desaturases, C-4 sterol methyl oxidases, and sphingolipid hydroxylase protein sequences were aligned using Clustal W (25). Phylogenetic analyses were carried out by the neighbor-joining method using the program MEGA4, version 4.0.2 (24), with 10,000 bootstrap samplings or by minimum evolution with 5,000 bootstrap replicates. Both methods gave very similar tree topologies.
Nucleotide sequence accession number.
The nucleotide sequence for gene DES5A has been deposited in GenBank under accession number FJ940725.
RESULTS
Identification and sequence analysis of genes in the Tetrahymena thermophila genome encoding putative sterol desaturases.
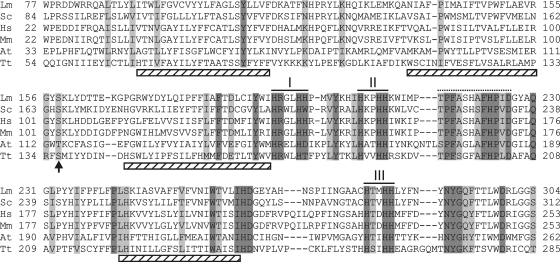
Amino acid sequence alignments of C-5(6) sterol desaturases from phylogenetically distantly related organisms show the remarkable feature that four hydrophobic segments and three histidine clusters are highly conserved. The histidine blocks (HX3H, HX2HH, and HX2HH), at a conserved mutual distance of 8 and 70 amino acids, respectively, presumably are involved as ligands of iron atom(s) complexed by the protein, a trait commonly displayed by all C-5(6) sterol desaturases and sterol C-4 methyl oxidases (14). Fatty acid desaturases share similar His motifs (HX(3/4)H, HX(2/3)HH, and H/QX(2/3)HH), but theirs are at a mutual distance of 31 and 134 amino acids (22), respectively.
The mechanism of sterol desaturation involves an electron transfer from NAD(P)H to the terminal oxidase (the desaturase itself) via Cyt b5 and Cyt b5 reductase, as has been documented for mammals (15), yeast (18), and plants (19).
Cyt b5 also is present in Tetrahymena and is required for the activity of fatty acid desaturases in microsomes (20), albeit with slightly different properties, as shown by the absorbance spectrum of oxidized conditions versus those of reduced conditions (9). We have reported previously that the presence of Cyt b5 and Cyt b5 reductase also were absolute requirements for C-7(8) and C-22(23) sterol desaturase activities in T. thermophila, presumably for the electron transfer from the reduced cofactor NAD(P)H (17). Based on these findings, the presence of the three highly conserved histidine blocks was investigated in the putative gene sequences from the TIGR database (http://www.tigr.org/) of the T. thermophila genome, and a BLAST search was performed using the complete protein sequence of C-5(6) sterol desaturases of Saccharomyces cerevisiae (accession number NP_013157) and Homo sapiens (NP_008849) as the query.
The search retrieved eight putative genes with significant similarity, as indicated in Table 2. Seven of these genes tentatively were assigned to be orthologs of C-5(6) sterol desaturases, C-4 sterol methyl oxidases, or, more distantly, C-4 sphingolipid hydroxylases according to a search in the OrthoMCL database (http://www.orthomcl.org/), while no orthologs were identified the for TTHERM_00438800 sequence. Based on the fact that it has the highest similarity to C-5(6) sterol desaturases, we selected the sequence TTHERM_01194720 as the putative gene, and it was named DES5A. The open reading frame has 1,324 bases, with a predicted structure comprising three exons from base 1 to 275 (exon I), 331 to 598 (exon II), and 864 to 1324 (exon III). The theoretical protein (Des5Ap) has 334 amino acids and a deduced molecular mass of 39,665 Da. Des5Ap showed the three conserved His motifs that are typical of all C-5(6) desaturases known so far at distances of 9 amino acids between the first and second motifs and 69 between the second and third. The conserved hydroxylated amino acid, described as being crucial for the enzymatic activity of the desaturases and located 32 to 34 amino acids N terminal of the first His motif (14), is present in the form of a serine (Fig. 2). It also was possible to assign a highly conserved domain [(T/S)PF(A/S)(S/G)(H/L/F)(A/S)FHP(V/I/L)DA] located 5 to 8 positions downstream of the second His cluster with yet-unknown function (14).
TABLE 2.
Putative sterol desaturase genes in the T. thermophila genome and amino acid sequence comparison to the most similar proteins
| Gene sequence no. (TIGR database) | Protein no. (NCBI database) | Amino acid sequence comparison to domain PFAM PF04116a
|
Ortholog(s)a | |
|---|---|---|---|---|
| Score | E value | |||
| TTHERM_01194720 | XP_001029976 | 101.9 | 2.2e-27 | C-5(6) desaturase |
| TTHERM_00446080 | XP_001023372 | 76.2 | 1.2e-19 | C-5(6) desaturase |
| TTHERM_00438800 | XP_001017777 | 51.5 | 3.2e-12 | Not foundb |
| TTHERM_00758950 | XP_001016978 | 78.5 | 2.4e-20 | C-4-sphingolipid hydroxylases |
| TTHERM_00487050 | XP_001032917 | 95.8 | 1.5e-25 | 4-Methyl-oxidase and serine/threonine protein kinases |
| TTHERM_00077800 | XP_001015720 | 102.0 | 2E-27 | 4-Methyl-oxidase |
| TTHERM_00876970 | XP_001016047 | 112.2 | 1.7e-30 | 4-Methyl-oxidase |
| TTHERM_00348230 | XP_001022978 | 106.2 | 1.1e-28 | 4-Methyl-oxidase |
Shown are sequence comparisons of putative T. thermophila sterol desaturases selected in the TIGR database (http://www.tigr.org/tdb/e2k1/ttg/) and the conserved domain PFAM PF04116 (http://www.sanger.ac.uk/) and orthologs found in the OrthoMCL database (http://www.orthomcl.org/).
No orthologs were found with an E value of less than 1e-5.
FIG. 2.
Multiple sequence alignment of all C-5(6) sterol desaturases using the Clustal X program with default parameters. The sequences displayed belong to Leptosphaeria maculans (Lm; accession number AAN27998), Saccharomyces cerevisiae (Sc; NP_013157), Homo sapiens (Hs; NP_008849), Mus musculus (Mm; NP_766357), Arabidopsis thaliana (At; NP_186907), and Tetrahymena thermophila (Tt; XP_001029976). Histidine clusters (I, II, and III) are indicated with black lines, and predicted transmembrane regions are indicated with cross-hatched rectangles. The conserved hydroxylated amino acid is marked with an arrow, and highly conserved domains are marked with a dotted line. Identical amino acid columns are shown in dark gray; conserved substitutions are shown in light gray.
The amino acid sequence alignment with several known C-5(6) sterol desaturases showed 43% similarity and 29% identity with those of Mus musculus (NP_766357), 39 and 24% with S. cerevisiae (NP_013157), 38 and 28% with Arabidopsis thaliana (NP_186907), and 43 and 25% with the sea urchin Strongylocentrotus purpuratus (XP_001188758), respectively.
The analysis of putative trans-membrane helices (12) indicated the presence of four motifs, with two of the conserved clusters of His residues located between trans-membrane helices 3 and 4 (Fig. 2). This same topology is shared by C-5(6) sterol desaturases isolated from A. thaliana, Candida glabrata, H. sapiens, and S. cerevisiae (14).
Knockout of T. thermophila DES5A gene.
To determine whether TTHERM_01194720 encodes a C-5(6) sterol desaturase, we targeted the DES5A gene for knockout mutagenesis. The transformation sequence C270 (see Materials and Methods), which provides paromomycin resistance, was introduced into the macronucleus (Fig. 3A), which is transcriptionally active and determines the phenotype of the cell, by somatic transformation by following a biolistic bombardment protocol (4). With this procedure, around 9 transformants per μg of DNA were obtained.
FIG. 3.
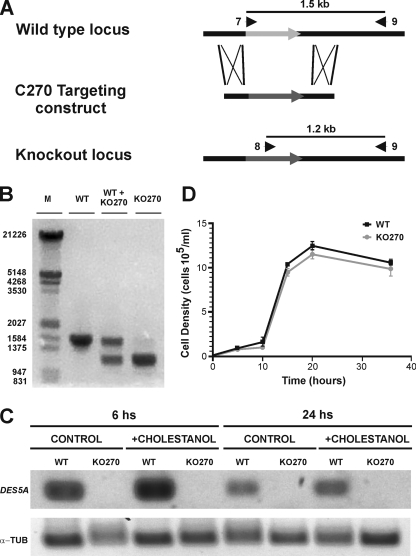
Schematic representation of gene replacement in the WT locus of the DES5A gene with the targeting construct (C270 fragment) by homologous recombination, generating the knockout locus in the KO270 mutant. (A) The numbered arrowheads indicate the primers used. (B) Shown are the PCR amplification products from genomic DNA of WT T. thermophila and the KO270 mutant using two allele-specific primers (7 and 8) and the locus specific primer (9) indicated above the gel (Table 1). In the WT, a 1.5-kb band was visible, while in KO270 a 1.2-kb fragment corresponding to the knockout locus was amplified. Sample WT + KO270 is a mixture of the two genomic DNAs, and it was used as a control. M, markers for fragment length. (C) Shown are RNA levels in the WT and KO270 grown in cholestanol-containing cultures, sampled at 6 and 24 h, and measured by RT-PCR. DES5A indicates the fragment amplified by using primers 10 and 11, designed on the second exon of DES5A. α-TUB indicates the fragment amplified from α-tubulin cDNA using primers 12 and 13 as a control. (D) Comparison of T. thermophila WT (•) and KO270 (▪) growth rates. Both strains showed similar growth rates and final biomass yields.
When integrative vectors are used in transformation experiments, only the partial replacement of the ∼45 endogenous copies of the genes present in the macronucleus initially occurs. The amitotic division of the macronucleus provides the basis for phenotypic assortment, in which an allele subsequently can be unequally segregated (26). From seven plates with 96 wells each, around 23 wells showed cell growth with 80 μg/ml paromomycin. Transformants were successively transferred to fresh SPP medium with increasing concentrations of paromomycin for at least 200 generations until no further growth could be obtained. The selected clones were those that grew at the higher paromomycin concentrations. One knockout mutant (KO270), which was resistant to 75 mg/ml paromomycin, showed extensive gene replacement in the locus, as indicated by comparative PCR. Figure 3B shows the results of the DNA amplification of specific fragments from WT and KO270 cells and a mixture of both using the allele-specific primers 7 and 8, respectively, and the locus-specific primer 9. In the WT, a predicted 1.5-kb fragment corresponding to the undisrupted sequence was amplified using primers 7 and 9, whereas in KO270, a main 1.2-kb fragment corresponding to the knockout locus and a faint 1.5-kb fragment were amplified under the same reaction conditions. These results confirm that the transforming fragment correctly targeted the DES5A locus, and that most, if not all, endogenous WT alleles in the macronucleus have been replaced after extensive phenotypic assortment. Although an incomplete replacement cannot be ruled out, the faint 1.5-kb band in KO270 most probably is due to the amplification of the micronucleus intact copy of the gene.
In addition, RNA expression from DES5A of the WT and knockout strains after 6 and 24 h of being cultured in medium with or without cholestanol was assayed. As shown in Fig. 3C, no transcript was detected in the KO270 deletion mutant, while expression both in the presence and absence of cholestanol was observed in the WT strain.
The complete gene then was amplified from CU428 genomic DNA, using primers 14 and 15 (Table 1), for sequence analysis. The gene isolated from CU428 (GenBank accession number FJ940725) was 99.8% identical to the one from SB210, the strain used by TIGR for the genome project (7).
C-5(6) sterol desaturase activity is strongly diminished in KO270 deletion mutant.
Tetrahymena has three sterol desaturase activities with similar properties: C-5(6), C-7(8), and C-22(23) desaturases (16). For the identification of the enzymes, the culture of the organism with specific sterols and the analysis of the conversion products is a straightforward possibility. Therefore, we cultured the WT and KO270 with lathosterol, cholesterol, and cholestanol for 24 h and compared their sterol composition. The list of expected products with the added sterols, both in the WT and mutant strains, are summarized in Table 3.
TABLE 3.
Sterol biotransformations expected in the T. thermophila WT and KO270 knockout mutanta
| Strain | Products in cultures supplemented with:
|
||
|---|---|---|---|
| Cholesterol | Cholestanol | Lathosterol | |
| WT | Cholest-5, 22-dien-3β-ol; cholest-5, 7-dien-3β-ol; cholest-5, 7, 22-trien-3β-ol (↑) | 5α-Cholest-22-en-3β-ol; lathosterol; cholesterol; cholest-5, 7-dien-3β-ol; cholest-5, 22-dien-3β-ol; 5α-cholest-7, 22-dien-3β-ol; cholest-5, 7, 22-trien-3β-ol (↑) | Cholest-5, 7-dien-3β-ol; cholest-5, 22- dien-3β-ol; cholest-5, 7, 22-trien-3β-ol (↑) |
| KO270 | Cholest-5, 22-dien-3β-ol; cholest-5, 7-dien-3β-ol; cholest-5, 7, 22-trien-3β-ol (↑) | 5α-Cholest-22-en-3β-ol; lathosterol; cholesterol (↓); cholest-5, 7-dien-3β-ol (↓); cholest-5, 22-dien-3β-ol (↓); 5α-cholest-7, 22-dien-3β-ol (↑); cholest-5, 7, 22-trien-3β-ol (↓) | Cholest-5, 7-dien-3β-ol (↓); 5α-cholest-7, 22-dien-3β-ol (↑); cholest-5, 7, 22-trien-3β-ol (↓) |
Arrows indicate whether an increase (↑) or decrease (↓) of the products is expected.
The analysis of conversion products formed from lathosterol, for instance, may help to identify C-5(6)- and C-22(23)-desaturating activities, while those formed from cholesterol help in the identification of C-7(8) or C-22(23) desaturases, and the ones formed with cholestanol are helpful for C-5(6), C-7(8), or C-22(23) desaturase identification. For example, a conversion of lathosterol into cholest-5,7,22-trien-3β-ol demonstrates C-5(6) and C-22(23) desaturation, while the conversion of cholestanol to cholest-5,7,22-trien-3β-ol demonstrates C-5(6), C-7(8), and C-22(23) desaturase activities. On the other hand, if either cholest-5,7-dien-3β-ol or cholest-5,7,22-trien-3β-ol is formed during growth with cholesterol, this may be an indication of C-7(8) desaturase and/or C-22(23) desaturase (Table 3).
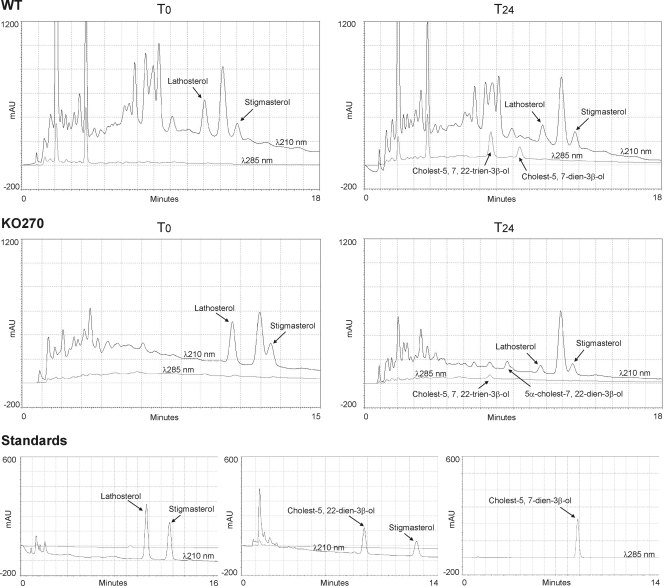
In our case, sterols formed by KO270 and the WT during growth in lathosterol showed significant differences. In particular, the formation of all C-5(6) unsaturated derivatives, such as cholest-5,7-dien-3β-ol and cholest-5,7,22-trien-3β-ol, were significantly impaired in the deletion mutant, showing an 87% decrease with respect to that of the WT. As displayed in Table 4, roughly 50% of the initial amount of lathosterol was recovered as C-5 unsaturated products in the WT, compared to only 6% in the KO270 mutant after 24 h of culture. All of the compounds recovered with their relative areas are displayed in Fig. 4 as HPLC graphs. It is worth noting that all C-5 sterol derivatives, such as cholest-5,7-dien-3β-ol and cholest-5,7,22-trien-3β-ol, can be measured at 285 nm due to the formation of a conjugated 5,7-diene, whereas (5α)-cholest-7,22-dien-3β-ol could be identified only by its retention time.
TABLE 4.
Recovery of sterols from WT T. thermophila and mutant KO270 cultured with cholesterol or lathosterola
| Sterol | Amount (in μg/ml) (%) of sterol at:
|
|||
|---|---|---|---|---|
| 0 h
|
24 h
|
|||
| WT | KO270 | WT | KO270 | |
| Cholesterol | 21.74 (100) | 18.82 (100) | 7.02 (32) | 8.32 (44) |
| Cholest-5, 7-dien-3β-ol | <0.05 (<1) | <0.05 (<1) | 0.84 (4) | 0.42 (2) |
| Cholest-5, 7, 22-trien-3β-ol | <0.05 (<1) | <0.05 (<1) | 13.46 (61) | 10.26 (55) |
| Lathosterol | 19.42 (100) | 22.54 (100) | 9.02 (46) | 12.03 (54) |
| Cholest-5, 7-dien-3β-ol | <0.05 (<1) | <0.05 (<1) | 2.8 (14) | <0.05 (<1) |
| Cholest-5, 7, 22-trien-3β-ol | <0.05 (<1) | <0.05 (<1) | 7.1 (36) | 1.46 (6) |
Cells were grown in 250-ml Erlenmeyer flasks containing 100 ml of SPP medium supplemented with cholesterol or lathosterol at 20 μg/ml (final concentration). Results shown are mean values from three independent experiments.
FIG. 4.
HPLC analysis of sterols extracted from WT T. thermophila and the KO270 mutant grown with lathosterol at zero (T0) and 24 (T24) h. For quantification, stigmasterol (cholest-5,22-dien-24β-ethyl-3β-ol) was added in all cases as an internal standard. Absorbance was recorded at 210 nm for all sterols and 285 nm for sterols displaying conjugated double bonds (5,7-diene derivatives).
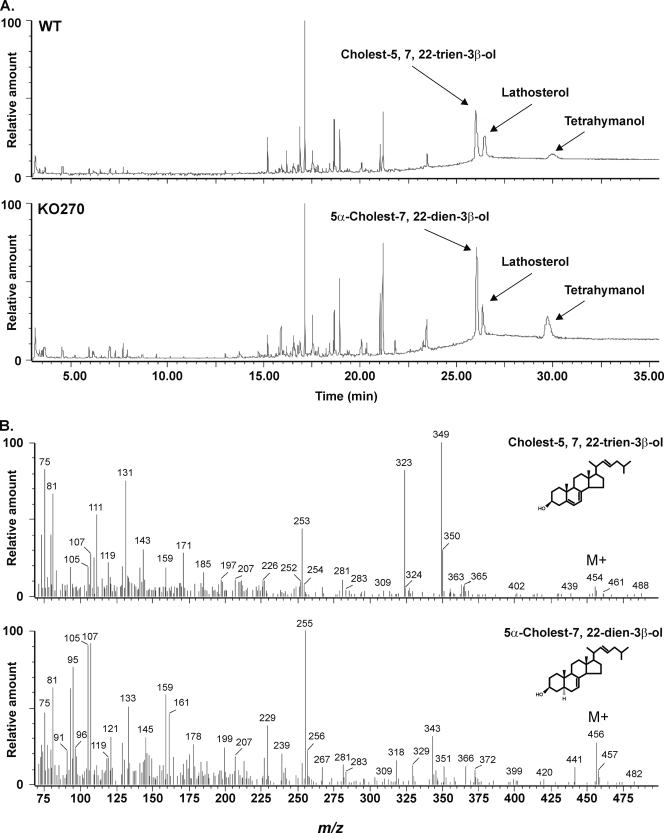
Further confirmation of the identity of the isolated sterols was obtained by GC-MS analysis. As shown in Fig. 5, the sterols recovered from cultures with lathosterol were, besides lathosterol itself, cholest-5,7,22-trien-3β-ol in the WT and 5α-cholest-7,22-dien-3β-ol in the KO270 mutant, confirming that there was no measurable C-5(6) desaturase activity in the latter.
FIG. 5.
GC-MS analysis of trimethylsilyl ether derivatives of total sterols isolated from WT T. thermophila and the KO270 mutant grown with lathosterol. (A) Sterols were recovered after 24 h of culturing. (B) Mass spectra of cholest-5,7,22-trien-3β-ol and (5α)-cholest-7,22-dien-3β-ol trimethylsilyl derivatives. The compounds were identified with the National Institute of Standards and Technology library.
The conversion of cholesterol, on the other hand, showed similar results between strains: 61 and 55% of cholest-5,7,22-trien-3β-ol was recovered in the WT and KO270 mutant, respectively, thus indicating that C-7(8) and C-22(23) desaturases were not impaired (Table 4). Taken together, these results confirmed that KO270 was indeed a DES5A mutant displaying the typical sterol profile expected in a C-5(6) sterol desaturase knockout (Table 3).
The disruption of the DES5A gene in the KO270 mutant had no other physiological consequences on the organism, as shown by its growth pattern and cellular behavior. As seen in Fig. 3D, the growth curves of the WT and mutant were very similar, with a duplication time of 2.63 and 2.83 h, respectively, with no significant differences in total biomass yield. Also, cellular morphology and movement were undistinguishable between the strains.
Phylogenetic analysis of T. thermophila C-5(6) sterol desaturase (Des5Ap).
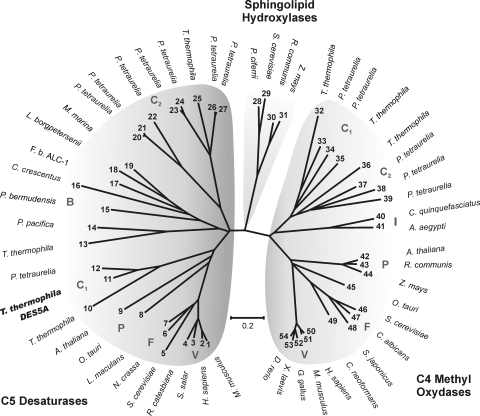
A consensus phylogenetic tree was constructed by the neighbor-joining and minimum evolution methods, with multiple alignments of 47 sequences of C-5(6) desaturases, C-4 methyl oxidases, and C-4 sphingolipid hydroxylases (all members of the fatty acid hydroxylase superfamily, which display the His boxes and use a similar electron transport system) and 7 T. thermophila sequences, which are listed in Table 2. Sequence TTHERM_00487050 was excluded from the analysis, as it has no apparent orthologs. It encodes a hypothetical 2,049-amino-acid protein, with a small C-terminal domain sharing 48% similarity with fungal C-4 methyl oxidases and an N-terminal domain that is similar to those of serine/threonine protein kinases.
The resulting tree (Fig. 6) is composed of three well-defined clusters, the first one grouping sterol C-5 desaturases, a second one represented by C-4 sphingolipid hydroxylases, and the third one grouping C-4 methyl oxidases. The second cluster does not contain ciliate orthologs and results in a good outgroup for the phylogenetic analysis. The first cluster is formed by vertebrate, fungal, plant, bacterial, and two ciliate (T. thermophila and Paramecium tetraurelia) branches; one ciliate branch (C1) contains Des5Ap, its paralog TTHERM_00446080, and a P. tetraurelia ortholog. The second ciliate branch (C2) is formed by TTHERM_00758950 and seven P. tetraurelia hypothetical proteins. Finally, TTHERM_00438800 is more related to the bacterial branch. The ciliate branch containing Des5Ap is very related to vertebrate, fungal, and plant C-5 desaturases, many of which have been characterized biochemically. This finding is in nice agreement with the results showed above, making the paralog protein encoded by TTHERM_00446080 a strong candidate to be the enzyme responsible for the remaining C-5-desaturating activity found in the DES5A mutant. The C-4 methyl oxidase cluster has a topology similar to that of the first cluster, with vertebrate, fungal, plant, insect, and two ciliate branches, the first one containing the sequences TTHERM_00077800 and TTHERM_00348230 and two P. tetraurelia hypothetical proteins (C1), and the second branch containing TTHERM_00876970 and three hypothetical proteins from P. tetraurelia (C2).
FIG. 6.
Phylogenetic analysis of C-5(6) sterol desaturases, C-4 sphingolipid hydroxylases, sterol C-4 methyl oxidases, and putative proteins of Tetrahymena thermophila. The phylogenetic tree was created using the neighbor-joining method, with 10,000 replicates, in MEGA-4 (24). The accession numbers of the amino acid sequences used for the analysis were the following: 1, NP_766357; 2, NP_008849; 3, NP_001133588; 4, ACO51759; 5, NP_013157; 6, XP_962923; 7, AAN27998; 8, CAL53849; 9, NP_186907; 10, XP_001023372 (TTHERM_00446080); 11, XP_001029976 (TTHERM_01194720); 12, XP_001440490; 13, XP_001017777 (TTHERM_00438800); 14, ZP_01908611; 15, ZP_01017596; 16, NP_420481; 17, ZP_02181983; 18, YP_798571; 19, ZP_01689977; 20, XP_001441873; 21, XP_001453241; 22, XP_001441700; 23, XP_001450297; 24, XP_001453136; 25, XP_001016978 (TTHERM_00758950); 26, XP_001426093; 27, XP_001459383; 28, AAN77731; 29, EDN60625; 30, EEF39915; 31, NP_001149259; 32, XP_001015720 (TTHERM_00077800); 33, XP_001449651; 34, XP_001455915; 35, XP_001022978 (TTHERM_00348230); 36, XP_001016047 (TTHERM_00876970); 37, XP_001448034; 38, XP_001460322; 39, XP_001459715; 40, XP_001861819; 41, XP_001657694; 42, NP_850133; 43, EEF41918; 44, ACG34890; 45, CAL54207; 46, NP_011574; 47, XP_713456; 48, XP_002174187; 49, XP_569526; 50, NP_006736; 51, NP_079712; 52, XP_420391; 53, NP_001072809; and 54, NP_998518. Specific branches are indicated as V (vertebrates), F (fungi), P (plant), B (bacteria), I (insect), and C1 and C2 (ciliates). The bar shows the percentages of substitutions.
DISCUSSION
Previous characterization of C-7(8) and C-22(23) sterol-desaturating activities in T. thermophila microsomal fractions revealed their dependence on Cyt b5 for the transfer of electrons from the reduced cofactor NAD(P)H (17). As other C-5(6) desaturases studied so far show similar Cyt b5 dependence and subcellular localization, we speculated that T. thermophila C-5(6) desaturase has similar requirements (15, 19).
Based on the consensus sequence of known Cyt b5-dependent C-5(6) sterol desaturases, particularly those from H. sapiens and S. cerevisiae, we used the complete sequence of these proteins as queries for BLAST searches in the Tetrahymena database (14). Eight sequences were retrieved, all of which contained the three conserved histidine boxes that are characteristic of this kind of enzyme. TTHERM_00487050 was not taken into consideration, as it encodes a very large protein with an N-terminal domain that is highly similar to that of protein kinases. Only a small C-terminal domain showed similarity to sterol C-4 methyl oxidases and, more distantly, to C-5 desaturases. TTHERM_01194720, which we named DES5A, showed the highest similarity to C-5 desaturases, and it was selected for further analysis.
In this study, we report the identification, by gene knockout and sequence analysis, of the first sterol desaturase from T. thermophila. The mutation introduced in the DES5A gene by targeted knockout generated cells with either the complete or nearly complete replacement of the copies of the WT gene. As is described for biolistic transformation, the knockout most probably targeted a macronuclear gene, and therefore the presence in the genome of at least one copy of a WT gene (Fig. 3B) could be explained by the germ line micronucleus. In accordance with this observation, no RNA transcripts from this gene could be detected at 6 or 24 h of culturing in the presence of cholestanol.
Nevertheless, a minor enzymatic activity (6%) remains in the mutant. This activity could be explained by the presence of another desaturase with low specifity, such as a C-7 sterol desaturase, which has not been characterized so far, or by the existence of more than one gene encoding a C-5(6) sterol desaturase, like the putative gene TTHERM_OO446080, which is grouped very closely to the DES5A gene in the phylogenetic tree. The presence of more than one C-5(6) sterol desaturase gene was reported previously for Aspergillus fumigatus (1).
As part of a genetic and functional characterization of the mutant, growth and morphological parameters were tested. They did not reveal significant differences between the WT and knockout mutant. In principle, these data, as well as its capacity to grow at high concentrations of paromomycin (75 mg/ml), suggest that the DES5A gene is nonessential.
We used the mutant KO270 to show that the DES5A gene product supports the specific C-5(6) desaturation of sterols using substrates such as lathosterol and cholestanol. Significantly, only C-5(6) desaturase activity was strongly diminished in the KO270 mutant, while C-7(8) and C-22(23) desaturase activities were not affected.
The DES5A expression analysis also revealed that the gene is transcribed in the absence of sterols, and that this process is not suppressed by the external addition of cholestanol. Actually, the expression appears to be stimulated by the sterol (Fig. 3C). This response seems different, in principle, from those of mammals and yeast, where sterol addition inhibits the expression of C-5(6) desaturase as well as the expression of other enzymes of the sterol biosynthetic pathway. This difference could be explained by the fact that this protozoon does not synthesize sterols, rather it modifies them to more unsaturated species of unknown function in the ciliate.
Acknowledgments
We acknowledge with thanks the advice of Martin Gorovsky and Jody Bowen in the frame of the ASM International Fellowship for Latin America (2005), granted to A. Nusblat, and the critical reading of the manuscript by Klaas Hellingwerf (University of Amsterdam, The Netherlands). A.D.N., A.D.U., and C.B.N. are members of Carrera del Investigador Científico, CONICET, Argentina.
The work was supported by grants ANPCYT 8301 (Argentinean S&T Council) and UBACYT B108 (University of Buenos Aires).
Footnotes
Published ahead of print on 12 June 2009.
REFERENCES
- 1.Alcazar-Fuoli, L., E. Mellado, G. Garcia-Effron, M. J. Buitrago, J. F. Lopez, J. O. Grimalt, J. M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2006. Aspergillus fumigatus C-5 sterol desaturases Erg3A and Erg3B: role in sterol biosynthesis and antifungal drug susceptibility. Antimicrob. Agents Chemother. 50453-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackburn, E. H., and J. G. Gall. 1978. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J. Mol. Biol. 12033-53. [DOI] [PubMed] [Google Scholar]
- 3.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37911-917. [DOI] [PubMed] [Google Scholar]
- 4.Cassidy-Hanley, D., J. Bowen, J. H. Lee, E. Cole, L. A. VerPlank, J. Gaertig, M. A. Gorovsky, and P. J. Bruns. 1997. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146135-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conner, R. L., F. B. Mallory, J. R. Landrey, and C. W. Iyengar. 1969. The conversion of cholesterol to delta-5,7,22-cholestatrien-3-beta-ol by Tetrahymena pyriformis. J. Biol. Chem. 2442325-2333. [PubMed] [Google Scholar]
- 6.Conner, R. L., J. R. Landrey, C. H. Burns, and F. B. Mallory. 1968. Cholesterol inhibition of pentacyclic triterpenoid biosynthesis in Tetrahymena pyriformis. J. Protozool. 15600-605. [DOI] [PubMed] [Google Scholar]
- 7.Eisen, J. A., R. S. Coyne, M. Wu, D. Wu, M. Thiagarajan, J. R. Wortman, J. H. Badger, Q. Ren, P. Amedeo, K. M. Jones, L. J. Tallon, A. L. Delcher, S. L. Salzberg, J. C. Silva, B. J. Haas, W. H. Majoros, M. Farzad, J. M. Carlton, R. K. Smith, Jr., J. Garg, R. E. Pearlman, K. M. Karrer, L. Sun, G. Manning, N. C. Elde, A. P. Turkewitz, D. J. Asai, D. E. Wilkes, Y. Wang, H. Cai, K. Collins, B. A. Stewart, S. R. Lee, K. Wilamowska, Z. Weinberg, W. L. Ruzzo, D. Wloga, J. Gaertig, J. Frankel, C. C. Tsao, M. A. Gorovsky, P. J. Keeling, R. F. Waller, N. J. Patron, J. M. Cherry, N. A. Stover, C. J. Krieger, C. del Toro, H. F. Ryder, S. C. Williamson, R. A. Barbeau, E. P. Hamilton, and E. Orias. 2006. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frankel, J. 2000. Cell biology of Tetrahymena thermophila. Methods Cell Biol. 6227-125. [DOI] [PubMed] [Google Scholar]
- 9.Fukushima, H., T. Takeda, N. Sasaki, T. Watanabe, and Y. Nozawa. 1983. Purification and characterization of microsomal cytochrome b560ms from a unicellular eukaryote Tetrahymena pyriformis. J. Biol. Chem. 25811991-11996. [PubMed] [Google Scholar]
- 10.Gaertig, J., L. Gu, B. Hai, and M. A. Gorovsky. 1994. High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 225391-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43405-413. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann, K., and W. Stoffel. 1993. TMBASE—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374166. [Google Scholar]
- 13.Honjo, K., T. Ishibashi, and Y. Imai. 1985. Partial purification and characterization of lathosterol 5-desaturase from rat liver microsomes. J. Biochem. (Tokyo) 97955-959. [DOI] [PubMed] [Google Scholar]
- 14.Husselstein, T., H. Schaller, D. Gachotte, and P. Benveniste. 1999. Delta7-sterol-C-5-desaturase: molecular characterization and functional expression of wild-type and mutant alleles. Plant Mol. Biol. 39891-906. [DOI] [PubMed] [Google Scholar]
- 15.Kawata, S., J. M. Trzaskos, and J. L. Gaylor. 1985. Microsomal enzymes of cholesterol biosynthesis from lanosterol. Purification and characterization of delta 7-sterol 5-desaturase of rat liver microsomes. J. Biol. Chem. 2606609-6617. [PubMed] [Google Scholar]
- 16.Mallory, F. B., and R. L. Conner. 1971. Dehydrogenation and dealkylation of various sterols by Tetrahymena pyriformis. Lipids 6149-153. [DOI] [PubMed] [Google Scholar]
- 17.Nusblat, A. D., L. Muñoz, G. A. Valcarce, and C. B. Nudel. 2005. Characterization and properties of cholesterol desaturases from the ciliate Tetrahymena thermophila. J. Eukaryot. Microbiol. 5261-67. [DOI] [PubMed] [Google Scholar]
- 18.Osumi, T., T. Nishino, and H. Katsuki. 1979. Studies on the delta 5-desaturation in ergosterol biosynthesis in yeast. J. Biochem. 85819-826. [PubMed] [Google Scholar]
- 19.Rahier, A., M. Smith, and M. Taton. 1997. The role of cytochrome b5 in 4alpha-methyl-oxidation and C5(6) desaturation of plant sterol precursors. Biochem. Biophys. Res. Commun. 236434-437. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki, N., H. Fukushima, T. Watanabe, and Y. Nozawa. 1984. Studies on Tetrahymena microsomal electron transport systems: solubilization of microsomal electron transport enzymes involved in fatty acid desaturation. Comp. Biochem. Physiol. B 79219-223. [DOI] [PubMed] [Google Scholar]
- 21.Shang, Y., X. Song, J. Bowen, R. Corstanje, Y. Gao, J. Gaertig, and M. A. Gorovsky. 2002. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 993734-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shanklin, J., E. Whittle, and B. G. Fox. 1994. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 3312787-12794. [DOI] [PubMed] [Google Scholar]
- 23.Shevchuk, N. A., A. V. Bryksin, Y. A. Nusinovich, F. C. Cabello, M. Sutherland, and S. Ladisch. 2004. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 32e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 25.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turkewitz, A. P., E. Orias, and G. Kapler. 2002. Functional genomics: the coming of age for Tetrahymena thermophila. Trends Genet. 1835-40. [DOI] [PubMed] [Google Scholar]
- 27.Valcarce, G. May 2002. Methods for treating foodstuffs using cell free extracts from ciliates. U.S. patent 6,391,351.
- 28.Valcarce, G., A. Nusblat, J. Florin-Christensen, and B. C. Nudel. 2002. Bioconversion of egg cholesterol to pro-vitamin D sterols with Tetrahymena thermophila. J. Food Sci. 672405-2409. [Google Scholar]
- 29.Valcarce, G., L. Munoz, A. Nusblat, C. Nudel, and J. Florin-Christensen. 2001. The improvement of milk by cultivation with ciliates. J. Dairy Sci. 842136-2143. [DOI] [PubMed] [Google Scholar]
- 30.Zaug, A. J., and T. R. Cech. 1986. The intervening sequence RNA of Tetrahymena is an enzyme. Science 231470-475. [DOI] [PubMed] [Google Scholar]