Abstract
Treatment of systemic fungal infections is difficult because of the limited number of antimycotic drugs available. Thus, there is an immediate need for simple and innovative systems to assay the contribution of individual genes to fungal pathogenesis. We have developed a pathogenesis assay using Caenorhabditis elegans, an established model host, with Saccharomyces cerevisiae as the invading fungus. We have found that yeast infects nematodes, causing disease and death. Our data indicate that the host produces reactive oxygen species (ROS) in response to fungal infection. Yeast mutants sod1Δ and yap1Δ, which cannot withstand ROS, fail to cause disease, except in bli-3 worms, which carry a mutation in a dual oxidase gene. Chemical inhibition of the NADPH oxidase activity abolishes ROS production in worms exposed to yeast. This pathogenesis assay is useful for conducting systematic, whole-genome screens to identify fungal virulence factors as alternative targets for drug development and exploration of host responses to fungal infections.
Nosocomial microbial infections are a growing health problem. Among these, fungal infections are especially threatening, with an estimated mortality rate of 40% (47). The key reason for this alarming mortality rate is the limited range of antifungal agents. Identification of new drug targets requires high-throughput infection assays that are complicated by the very fact that they involve two organisms: a host and a pathogen.
We have taken a reductionistic approach to studying host-pathogen interactions and have developed a Saccharomyces cerevisiae-based assay to understand the genetic and molecular mechanisms of fungal pathogenesis. Using Caenorhabditis elegans as a model host, we have found that S. cerevisiae infects the worm, producing visible disease phenotypes. The two organisms used in our study are specifically suited for host-pathogen infection studies because both genomic sequences have been completely determined and mutants are readily available. A complete genome knockout collection is available for S. cerevisiae, a resource that does not exist for any fungal pathogen. Likewise an RNA interference (RNAi)-mediated knockdown genomic library is available for C. elegans. These unique tools are key in the context of a genetic screen and allow us to systematically scan the entire genomes to identify fungal virulence factors and modulators of host immunity that combat a fungal pathogen.
The budding yeast S. cerevisiae has recently been described as an emerging pathogen and has been isolated from human patients (34, 35). It is routinely used as a model for pathogenic fungi because a large proportion of its genes are conserved in pathogenic fungi (for a review, see reference 32). Homologs of genes and pathways identified in S. cerevisiae have been shown to be important in bona fide pathogens. It has also been used for the identification of gene products important for fungal survival in the mammalian host environment (21, 46). For example, the SSD1 allele type affects pathogenicity of yeast, indicating that allelic variation at the SSD1 locus may be important for survival under various conditions (46). This has allowed investigators to use reverse genetic approaches to study contributions of genes whose importance has been established in S. cerevisiae.
Caenorhabditis elegans has emerged as a valuable model host in which to study pathogenesis and innate immunity (for a review, see reference 22). Microbial genes essential for virulence in mammalian models have been shown to be required for pathogenicity in nematodes (43). These studies have primarily explored bacterial species and have tested only a few fungi, such as Cryptococcus neoformans and Candida albicans, to explore virulence strategies. These studies focus on a killing assay using C. elegans and have identified several virulence factors with homologs in S. cerevisiae (4, 37), suggesting that genes and pathways we have identified in S. cerevisiae are likely to be found in pathogens. Moreover, other pathogenic fungi tested are limited in the repertoire of laboratory tools available for their study, making them recalcitrant to genetic manipulation and inappropriate for whole-genome high-throughput approaches to studying fungal virulence. Recently, Breger et al. described the application of a C. elegans-based infection assay as a tool to screen a chemical library for candidate antifungal compounds (9). Our investigation complements these studies in two significant ways. First, it allows us to identify genes that exacerbate as well as attenuate the pathogenic process, because we use an intermediate disease phenotype, while most other studies have used death as an end point phenotype. This aspect, taken together with the fact that S. cerevisiae shares significant genetic identity with pathogenic fungi, suggests that our study will yield a basic understanding of fungal pathogenesis. Second, it allows us to conduct a systematic, unbiased, whole-genome screen, which is currently not available for pathogenic fungi. Furthermore, genes and pathways identified may be targeted for antimycotic drug development.
Facets of innate immunity are evolutionarily conserved from nematodes to mammals. For example, a common defense strategy of mammals (phagocytes), (14), plants (3), and insects (23) is to produce reactive oxygen species (ROS), which directly damage pathogens. In human phagocytes, an NADPH oxidase enzyme complex produces ROS in host defense (19, 41). In Drosophila melanogaster, ROS are generated in the intestine by a NADPH oxidase to combat ingested bacteria (23). Loss of NADPH oxidase activity makes the fly susceptible to the bacterial infection (23, 24). Likewise, C. elegans has also been shown to produce ROS, such as superoxide and/or hydrogen peroxide, when it ingests bacterial pathogens (12). In each case, pathogen death can be abrogated by the addition of enzymes such as catalase that break down ROS (8, 27, 36), suggesting that ROS production plays a key role in a variety of pathogenic interactions.
We have found that S. cerevisiae can cause infection and death in C. elegans. Our data indicate that the nematode host produces ROS in response to fungal infection. We demonstrate that mutant yeast carrying deletions of genes that mediate oxidative stress responses fail to induce the Dar disease phenotype except in mutant worms with an altered dual oxidase gene, suggesting that the generation of ROS is a part of the defense strategy for the host and the neutralization of ROS is needed for persistent fungal infection.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Strains used for the study, for both S. cerevisiae and C. elegans, are listed in Table 1. Deletions in the yeast deletion set strains were confirmed by PCR or recreated using a PCR-mediated gene disruption cassette (45).
TABLE 1.
Strains used in this study
| Strain | Description | Sourcea |
|---|---|---|
| S. cerevisiae strains | ||
| BY4741 | MATahis3Δ1leu2Δ0met15Δ0ura3Δ0 | IDL |
| Pdc1::mcherry | PDC1::mcherry in BY4741 | G. Fink, MIT |
| yap1Δ | yap1Δ in BY4741 | IDL |
| yap2Δ | yap2Δ in BY4741 | IDL |
| yap4Δ | yap4Δ in BY4741 | IDL |
| sod1Δ | sod1Δ in BY4741 | IDL |
| pdr1Δ | pdr1Δ in BY4741 | IDL |
| pdr15Δ | pdr15Δ in BY4741 | IDL |
| C. elegans strains | ||
| N2 Bristol | Wild type | CGC |
| bli-3(e767)I | Dual oxidase | CGC |
| jnk-1(gk7)IV | MAPK of JNK MAP kinase pathway | CGC |
| mek-2(n1989)I | MAPK kinase of ERK MAP kinase pathway | CGC |
IDL, Invitrogen Deletion Library; CGC, Caenorhabditis Genetics Center.
The C. elegans wild-type strain was variant Bristol, strain N2. Mutant and wild-type C. elegans strains were obtained from the Caenorhabditis Genetics Center (MN). C. elegans stocks used for the study were grown on nematode growth agar medium (NGM) on Escherichia coli OP50 and maintained as described previously (10). E. coli OP50 was grown overnight in Luria broth at 37°C. Yeast growth medium was prepared as described by Sherman et al. (42) and strains were grown overnight at 30°C.
Egg preparation.
Worms were grown for 4 to 5 days on NGM agar containing E. coli OP50 at 20°C. Eggs and worms were washed off four plates with M9 buffer (10) and centrifuged at 900 × g for 2 min. The pellet was resuspended and washed twice with M9. This was then resuspended in a 1:4 dilution of commercial bleach (5.25%) containing 0.25 M sodium hydroxide solution, mixed gently by inversion for 3 min, and centrifuged for 2 min at 2,000 × g. The pellet was washed and centrifuged twice with M9 buffer at 2,000 × g for 2 min each and then finally resuspended in M9 buffer. The egg suspension was diluted or concentrated with M9 as required to obtain approximately 5 to 6 eggs/μl.
Pathogenesis assay.
E. coli and yeast strains were grown overnight at 37°C and 30°C, respectively. Culture aliquots were centrifuged at full speed in a microcentrifuge, washed twice in sterile water, and finally resuspended to a final concentration of 200 mg/ml and 20 mg/ml, respectively. A 3:1 E. coli/yeast ratio (wt/wt) (an estimated 30:1 ratio by number of organisms) mixture was prepared. A mixture of 10 μl of a 50-mg/ml streptomycin sulfate stock and 10 μl of E. coli:yeast mix (2.5 μl:7.5 μl) was spotted on each NGM plate and 5 μl of C. elegans egg suspension was transferred to each plate. Plates were observed for 4 to 5 days unless specified otherwise. Analysis of variance (ANOVA) was used to check the statistical significance of the differences observed between mutants and wild-type yeast strains.
Test of Koch's postulates and measurement of CFU.
Twenty worms each from S. cerevisiae test plates and E. coli control plates were picked and washed four times with sterile water. Worms were then crushed using a freeze fracture technique in individual microcentrifuge tubes and resuspended in 100 μl of sterile water. Appropriate dilutions of the mixture were transferred to yeast extract-peptone-dextrose (YPD) plates and incubated overnight. Colonies were counted to estimate CFU. Resulting colonies were replica plated to selective medium to test for auxotrophic markers. Two of these colonies were retested using the pathogenesis assay described above.
Microscopic analysis of C. elegans.
A 2% agarose pad containing 0.01 M sodium azide as anesthetic was prepared on a slide. A 3-μl drop of M9 buffer was added to the pad. Worms were picked and transferred to the drop on the slide. Mounted worms were then covered with a coverslip and observed at 40× and 20× magnification using an Axiovision Zeiss microscope under differential interference contrast (Nomarski) and epifluorescence optics. An ApoTome attachment was used to enhance fluorescence images.
Amplex Red hydrogen peroxide assay.
NGM plates were spotted with 20 μl of a 1:1 (vol/vol) mixture of streptomycin (50 mg/ml) and overnight yeast culture. These plates were then kept overnight at 30°C. Approximately L3- to L4-stage worms were washed off stock plates with M9 and then transferred to these plates and kept at 20°C for 10 h. The Amplex Red assay kit (Molecular Probes) was used to detect hydrogen peroxide. Postincubation the worms were washed four times with 1 ml of reaction buffer and resuspended to a final volume of 100 worms/50 μl. Fifty microliters of this suspension was added to the wells of a 96-well polystyrene plate. Diphenyleneiodonium (DPI) was added to some samples to make a final concentration of 100 μM and allowed to stand for 10 min. Then, 50 μl of Amplex Red reaction buffer was added to each well and color change was observed over 3 to 5 h.
C. elegans survival analysis.
For survival analysis, test plates and C. elegans eggs were prepared as described under “Pathogenesis assay,” above. Each plate was started with 30 ± 5 eggs; each experiment included three yeast and three E. coli plates per strain. Beginning on the second day after plating eggs, the numbers of dead and live worms on each plate were recorded daily. Live worms were transferred to new plates as necessary to avoid confusing the original worms with their offspring. All plates were of the same composition as the original test plates.
SigmaStat 3.5 (Systat Software, Inc.) was used to analyze the survival curve data. Significance, defined as a P value of <0.05, was assessed using the Gehan-Breslow test. In our experiments, worms that left the plates in the first several days were “censored,” i.e., removed from the counts of subsequent days. The Gehan-Breslow test assumes that the data from early survival times are more accurate than later times and weights the data accordingly. The number of censored worms was taken as the total number of worms (dead plus live worms) on that day minus total worms on the previous day. All of the data were collected from two independent experiments.
RESULTS
Development of an assay for fungal pathogenesis.
Study of host-pathogen relationships could be greatly simplified by the use of two well-characterized model systems with readily available mutants. This would allow simultaneous study of the genetics of both host and pathogen. To this end, we developed a pathogenesis assay using C. elegans as the model host and S. cerevisiae as the invading fungus. This system will allow us to use systematic whole-genome approaches to study fungal pathogenesis as well as host responses to fungal infections.
In order to study the interaction of S. cerevisiae with C. elegans, the organisms were cocultured on NGM plates. L3- to L4-stage larvae or adult hermaphrodites that were offered only S. cerevisiae as food successfully laid eggs but their progeny arrested at an early (L1 to L2 larval) stage of development. One possible explanation for the larval-stage growth arrest phenotype is that the yeast cells were too large for young larvae to ingest. To test this we cocultured hermaphrodite worms on lawns of red fluorescent protein (RFP)-labeled yeast. Within a few hours, labeled yeast (or yeast particles) were visible in the intestinal lumen of adult and late-stage larvae (data not shown) but not in the early larval stages. Therefore, in subsequent assays E. coli was added as a nutritional supply along with small amounts of S. cerevisiae. In this way small larvae could overcome the growth arrest while being exposed to yeast.
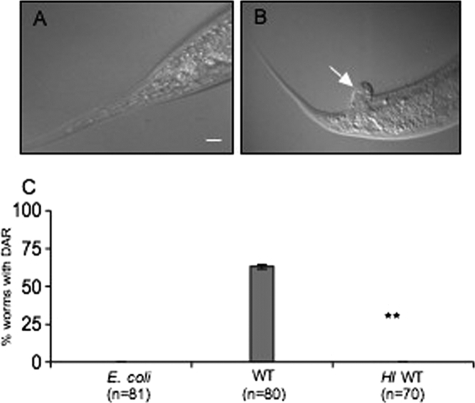
C. elegans grown on E. coli as a control (Fig. 1A) and mixed lawns of E. coli strain OP50 and small amounts of S. cerevisiae strain S288c (BY4741) exhibited the deformed anal region (Dar) phenotype (Fig. 1B). This phenotype was never seen in worms grown on E. coli OP50 alone (Fig. 1A). The Dar phenotype, which has been established as a disease symptom of C. elegans infected with the bacterium Microbacterium nematophilum (22, 26, 38), is characterized as a distinctive swelling in the postanal region. It has been suggested that the anal region of the worm is enlarged in response to infection, as worm mutants that do not show this phenotype are much more adversely affected by M. nematophilum infection. This deformity has been genetically characterized as a part of a defense reaction of the worm and can be used as a marker for infection (22). An intermediate phenotype allows us to identify factors that exacerbate (for example, Dar visible earlier or death) as well as attenuate the pathogenic response. Late-stage larvae or adult worms grown on yeast alone also display the Dar response. In this study we used Dar as a phenotypic marker and previously characterized host genes, involved in the Dar response, as genotypic markers of the disease condition.
FIG. 1.
S. cerevisiae causes a deformity in the postanal region (Dar) in C. elegans. (A) C. elegans exposed to E. coli as a control. (B) Worms exposed to S. cerevisiae show the Dar phenotype (arrow). Bar, 20 μm. (C) The Dar phenotype was scored on day 4 for nematodes exposed to E. coli, S. cerevisiae (WT), and heat-inactivated (HI) wild-type S. cerevisiae. The difference between WT and HI was statistically significant (P < 0.01, ANOVA).
Worms were monitored twice daily from the time the eggs were added to the microbial cultures up to day 4, when the Dar disease was clearly visible. At day 4 the Dar phenotype was scored for worms exposed to yeast or heat-inactivated yeast and compared to worms reared on E. coli (Fig. 1C). These results clearly indicated that the Dar phenotype affected worms exposed to yeast and that only metabolically active yeast cells were capable of eliciting Dar. To test whether the Dar phenotype is reversible, we transferred the affected worms to plates containing E. coli. The worms were “cured” of the Dar phenotype within 2 days of transfer. This supports the notion that yeast causes the deformity in the anal region and that worms can recover by clearing the yeast when they are no longer exposed to it.
The nematode's response to yeast is unlikely to be due to starvation because microscopic observation of the pathogenesis assay plates indicated that at the time when the Dar phenotype was visible, ample food was present. Furthermore, the inability of heat-inactivated yeast and mutant yeast (described below) to evoke the Dar phenotype strongly supports the notion that this manifestation of infection is a response to the pathogen rather than a response to starvation. We also ruled out the possibility that the worms are unable to digest the yeast because L3- to L4-stage larvae that are reared on yeast develop into fertile adults, suggesting that they are able to meet their nutritional requirements from yeast.
As a test of Koch's postulates, we reisolated and cloned microbes from an infected worm after extensive washing to remove externally associated yeast cells (see Materials and Methods). Genotypic characterization of the reisolated yeast cells demonstrated that they harbored the same mutant markers as the original yeast strain. Furthermore, yeast cells from two independent colonies isolated from the worm were used to reinfect wild-type worms. These worms showed the same extent of Dar disease progression observed with the original yeast strain. These results indicate that the etiological agent for the Dar disease was the one that we introduced, not a spurious contaminant.
Progressive distension and accumulation of yeast in the intestinal lumen.
To visualize yeast present in the intestine and clearly differentiate them from yeast sticking to the outside of the worm, we used a fluorescent-tagged housekeeping protein to follow progression of disease and specifically test the hypothesis that the intestinal distension is due to the accumulation of yeast. We chose an in-frame fusion of the yeast protein pyruvate decarboxylase (Pdc1) with RFP because its RFP fluorescence was clearly visible and maintained over the 5-day period of our assay. Moreover, it was indistinguishable from a yeast strain containing the native untagged version of PDC1 in all phenotypic tests (G. Fink, MIT, personal communication).
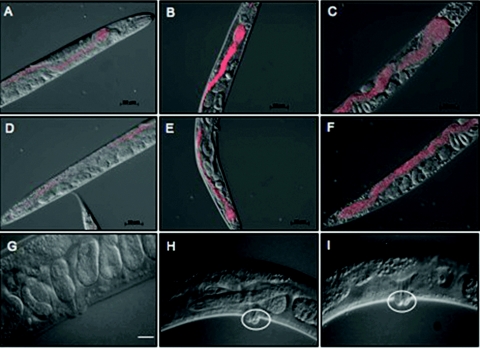
A time course of microscopic evaluation of infection using RFP-labeled yeast revealed that by day 3, RFP-labeled yeast cells had started to accumulate in the pharynx and the intestine (Fig. 2A and D). By day 4 (Fig. 2B and E) and day 5 (Fig. 2C and F), progressively more yeast cells had accumulated in the pharynx and intestine, causing the lumen to be severely distended compared to uninfected worms, in which the intestinal lumen is a narrow tube. Intestinal distension is also observed in M. nematophilum infection, but bacteria accumulate at the anus and in the rectum near the Dar tail swelling and do not extend into the intestine (38). This is in contrast to what we observed: 4 days postexposure, the intestinal lumen appeared to be packed with yeast cells, and the yeast were not observed to accumulate in the rectum. This striking difference may be indicative of the primary route of entry for the fungal pathogen, which appears to be oral ingestion. Distension of the intestinal lumen was also visible by differential interference contrast (Nomarski) micrographs of infected worms (Fig. 1B).
FIG. 2.
Time course of intestinal distention in C. elegans worms exposed to S. cerevisiae. Worms were exposed to RFP-marked, wild-type yeast from hatching and photographed on day 3 (A and D), day 4 (B and E), and day 5 (C and F). The experiment was done three times, and 60 to 75 worms were observed over a 3-day period. (A to C) Anterior region of the worm; (D to F) posterior region of the worm. Accumulation of yeast began in the pharynx region (compare panels A and D) and proceeded to the posterior. (G to I) S. cerevisiae also induces vulval swelling in the worms. Worms exposed to S. cerevisiae (H to I) show abnormal vulval swelling (white circles) compared to the control sample grown on E. coli (G). Bars: A to G, 50 μm; H and I, 20 μm.
Consistent with these microscopic observations, numbers of intact yeast cells released from infected worms increased during the course of infection. CFU were determined for yeast recovered from the worm intestine on days 2, 3, and 5. A 4-fold increase in the number of yeast CFU was observed between day 2 and day 3 and a10-fold increase between day 2 and day 5.
Between days 5 and 7, a 5 to 17% decrease in viability was observed on yeast test plates. The progeny of some dead worms appeared to have hatched inside the animal. Upon closer inspection on days 4 and 5, we noted a pronounced swelling in the vulval region of these worms (Fig. 2H and I). It is possible that this swelling impeded eggs from being laid and caused them to hatch within the worm, resulting in matricidal death. These qualitative assessments suggest that viable S. cerevisiae causes visible lesions near two of the three body openings of C. elegans. Coincidentally, such body openings are common sites of fungal infection in mammals.
Having established phenotypic characteristics of the diseased condition in the worm host, we wanted to assay the contribution of known molecular markers of bacterial disease in our S. cerevisiae-C. elegans-based pathogenesis assay.
Requirements for C. elegans ERK MAPK pathway in yeast infection: verification of molecular markers of host response.
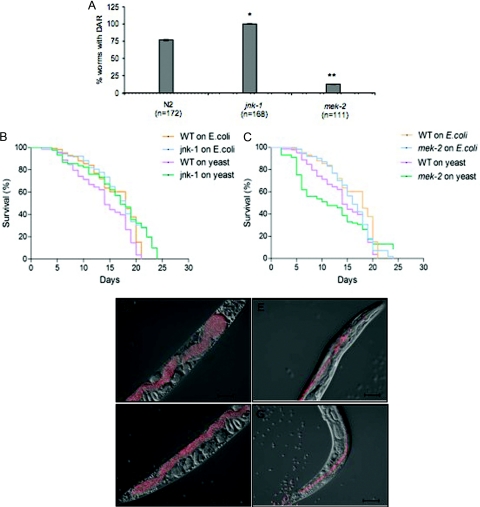
The C. elegans response to pathogens involves the mitogen-activated protein kinase (MAPK) pathways. An active extracellular signal-regulated kinase (ERK) pathway has been shown to be required for the Dar response of worms exposed to M. nematophilum (38), while the jnk-1 gene, encoding a JNK-like MAP kinase, is not required. The JNK pathway has been implicated in immunity in other organisms (16). We tested representative loss-of-function mutants in the ERK and JNK MAPK pathways for effects on S. cerevisiae pathogenesis in C. elegans (Table 1). The mek-2 gene encodes a MAPK kinase in the C. elegans ERK pathway (51). The loss-of-function mutant mek-2(n1989) showed a very low percentage of tail deformity when exposed to yeast compared to N2 wild-type worms (Fig. 3A). However, a loss-of-function mutation in the JNK-like MAP kinase, jnk-1(gk7), exhibited the Dar phenotype in response to S. cerevisiae. These effects are analogous to the Dar response of nematodes with M. nematophilum infections, suggesting that the ERK MAPK pathway is required for the tail deformities while the JNK MAPK pathway is not.
FIG. 3.
An active ERK MAPK pathway is required for the DAR phenotype and survival of worms upon yeast infection. (A) Worm mutants jnk-1(gk7) of the JNK MAPK pathway and mek-2(n1989) of the ERK MAPK pathway were exposed to wild-type yeast, and the DAR phenotype was scored on day 4. The mek-2(n1989) mutants, but not jnk-1(gk7) mutants, showed decreased Dar compared to the wild-type N2 worms. Differences between mek-2(n1989) or jnk-1(gk7) and wild type were statistically significant. Values for mek-2(n1989) and jnk-1(gk7) were significantly different from wild-type values (*, P < 0.01; **, P < 0.001; ANOVA). (B and C) Survival curves for the mek-2(n1989) and jnk-1(gk7) mutants indicate that mek-2(n1989) mutant worms were more susceptible to yeast than wild-type worms (P < 0.001, Gehan-Breslow test). The jnk-1(gk7) mutants survived just as long as their wild-type counterparts. (D to G) Worms showing Dar accumulated less yeast, at both the anterior and posterior regions compared to worms without Dar. Wild-type nematodes were exposed to RFP-marked, wild-type yeast and photographed on day 5. The experiment was done three times, and 20 to 25 worms were observed in each experiment. (D to E) Anterior region of the worm; (F and G) posterior end of the worm. Panels E and G show worms with Dar.
To study the contribution of these MAPK pathways to the survival of the host worm upon exposure to yeast, we conducted survival assays. This assay measured the survival of mutant worms compared to wild-type worms when exposed to S. cerevisiae. Survival of wild-type worms exposed to yeast versus E. coli was also monitored. Worm mutants were grown on E. coli to evaluate their general health. Our data indicated that the mek-2 mutant that was unable to exhibit the Dar response also showed enhanced susceptibility to yeast killing compared to wild-type C. elegans on yeast (P < 0.001, Gehan-Breslow test) (Fig. 3B). By contrast, the jnk-1 mutant that showed no defect in its Dar phenotype also showed no difference in survival compared to wild-type C. elegans on yeast and in fact appeared to be less susceptible than wild type (P = 0.055, Gehan-Breslow test) (Fig. 3C). Wild-type C. elegans showed a slight yet significant reduction in survival on S. cerevisiae compared to E. coli (P < 0.008, Gehan-Breslow test). The direct correlation between survival of the worm mutants and the severity of their Dar response (Fig. 3A, compared to B and C) suggests that the Dar response might be a protective phenotype. To address this we observed the extent of intestinal distention (or constipation) in worms exhibiting the Dar phenotype compared to worms that did not show the Dar response. We used the RFP-marked yeast strain for visualization (Table 1). We found that wild-type worms exhibiting the Dar phenotype accumulated less yeast in their intestine, thus enhancing survival (Fig. 3D to G). Together these results suggest that the ERK MAPK pathway is important for the Dar response and protecting the worms from S. cerevisiae infection. Although the exact defensive mechanism is not clearly understood, a similar observation was noted for M. nematophilum infection of C. elegans (22, 38).
Fungal resistance to oxidative stress is required to establish disease.
In order to use this pathogenesis assay to identify fungal virulence factors, we used a candidate gene approach in which yeast gene-specific deletion mutants were tested for their ability to induce the Dar phenotype in C. elegans.
We chose Yap1 and its paralog, Yap2, for this study because these genes have been shown to be important in host-pathogen interactions of plant and human fungal pathogens (2, 31, 40). It has been shown that Yap1 is activated in conidial germ tubes of Cochliobolus heterostrophus, a fungal pathogen of maize, at the earliest stage of plant infection and persists during infection (31). Furthermore, overexpression of CAP1 (the C. albicans ortholog of YAP1) confers resistance to the popular clinical antifungal fluconazole, an azole derivative (1). However, until this study no one had demonstrated that Yap1, or its orthologs, is required for pathogenesis. Yap1 has been recognized as a likely candidate for antimycotic drug development because it is a fungus-specific transcription factor of the AP-1 family that is involved in multidrug resistance (2, 31, 40). It also regulates oxidative stress responses in fungi, as the mutants show lack of growth in the presence of reactive oxygen species (13, 52). Otherwise there are no apparent growth defects in yap1Δ and yap2Δ mutants. Therefore, we chose to test yap1Δ and its paralog yap2Δ in our pathogenesis assay.
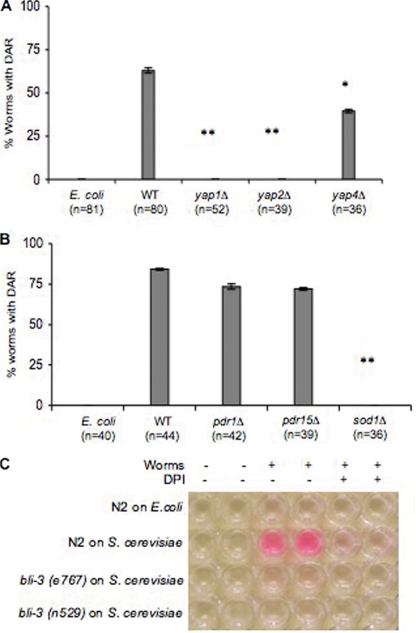
C. elegans exposed to either yap1Δ or yap2Δ mutants failed to exhibit the Dar phenotype (Fig. 4A). As a control we showed that a mutant with a mutation in a related transcription factor, Yap4, did exhibit the Dar phenotype (Fig. 4A). These results strongly support previous observations in bona fide fungal pathogens that Yap1 plays a key role in the infection process. These studies suggest that Yap1 and its paralog, Yap2, may be important virulence factors.
FIG. 4.
Yap1, Yap2, and Sod1 are required for fungal virulence. The Dar phenotype was scored on day 4 in nematodes exposed to yap1Δ, yap2Δ, and yap4Δ mutants (A) or sod1Δ, pdr1Δ, or pdr15Δ mutants (B) and compared to E. coli or the isogenic wild-type yeast strains. Values for yap1Δ, yap2Δ, yap4Δ, or sod1Δ were significantly different from wild-type values (*, P < 0.01; **, P < 0.001; ANOVA). The pdr1Δ and pdr15Δ mutants were not significantly different from the wild type (P > 0.01, ANOVA). (C) Production of ROS was monitored using Amplex Red reagent in wild-type and bli-3 (alleles e767 and n529) mutant worms exposed to S. cerevisiae in the presence or absence of DPI. Each experiment was performed in duplicate and repeated three times. Worms treated similarly with E. coli were used as controls.
In addition to the regulation of oxidative stress responses, Yap1 plays a role in regulating multidrug resistance genes (1, 2). To define which aspect of Yap1 is required to evoke the Dar response we tested mutants that were defective in one or the other response but not both. The specific mutants tested were superoxide dismutase (sod1Δ) and pleiotropic drug resistance (pdr1Δ and pdr15Δ) genes using the same pathogenesis assay. The physiological role for Sod1 is to guard cells against oxidative damage by neutralizing reactive oxygen species, and the mutants show normal growth except when in the presence of reactive oxygen species (6, 11, 33). Pdr1, like Yap1, is a transcription factor and regulates resistance to a variety of drugs, while PDR15 encodes an ATP binding cassette transporter of the plasma membrane implicated in general cellular detoxification (15, 25, 48-50). Like yap1Δ and yap2Δ mutants, sod1Δ mutants failed to elicit the Dar response, while pdr1Δ and pdr15Δ mutants were indistinguishable from their wild-type counterparts (Fig. 4B).
These experiments demonstrate that the fungus-specific transcription factors Yap1 and Yap2 are required to elicit the Dar response. These proteins mediate oxidative stress responses and drug resistance. Using mutants defective in only one of these two pathways made it unlikely that multidrug resistance is required for virulence; we believe that the inability of Yap mutants to tolerate oxidative stress compromises their ability to elicit Dar. Furthermore, these results suggest that fungi must be able to neutralize ROS in order to cause disease. This presents a testable hypothesis that this requirement stems from the fact that C. elegans produces ROS when it is infected with S. cerevisiae.
Worms produce reactive oxygen species in response to a fungal infection.
ROS have been shown to play an important role in pathogenesis and defense. Human phagocytes are known to produce ROS when fighting infections (5). It has recently been shown that C. elegans produces ROS when infected with bacteria (12, 36), presumably as a part of its defense mechanisms. Worms also produce lipofuscin, a marker of oxidative stress, in intestinal cells as they age (20), and lipofuscin is produced earlier in development when young worms are infected with a pathogen that elicits ROS production (12). We tested the hypothesis that young worms exposed to yeast produce more ROS by measuring relative amounts of ROS produced in worms exposed to yeast compared to worms reared on bacteria. We used a commercially available biochemical assay (Amplex Red peroxidase kit; Molecular Probes) to test ROS produced by the worms in response to pathogenic attack (12). Hydrogen peroxide produced by the host oxidizes the substrate, Amplex Red, to form a red product. Here we show that worms exposed to yeast produced more ROS than the control population grown on E. coli (Fig. 4C). Furthermore, we showed that ROS production can be chemically compromised using DPI, a specific inhibitor of NADPH oxidase activity that does not inhibit peroxidase (7). ROS production in phagocytes is catalyzed by an NADPH oxidase (5), and DPI has recently been shown to inhibit production of ROS in nematodes in response to pathogenic attack (12). Worms exposed to yeast that are treated with DPI oxidize Amplex Red only as well as unexposed worms (Fig. 4C), suggesting that DPI-mediated inhibition of the NADPH oxidase activity impedes the ability of the worm to produce ROS. These results directly implicate NADPH oxidase activity in the host response to fungal infection and suggest that such an activity might constitute a protective response against yeast infection. These observations prompted us to look for C. elegans proteins that contain a domain similar to the mammalian NADPH oxidase domain, gp91phox.
Homology searches revealed that an NADPH oxidase motif is present in the coding sequence of the bli-3 (blister-3) locus. In the encoded protein, the gp91phox motif is juxtaposed with a peroxidase domain in a single polypeptide; hence, it is referred to as a dual oxidase (CeDuox1) (17, 29, 44). Therefore we tested the hypothesis that CeDuox1 is responsible for generating an environment of elevated ROS.
Mutant worms that are unable to produce ROS are susceptible to S. cerevisiae.
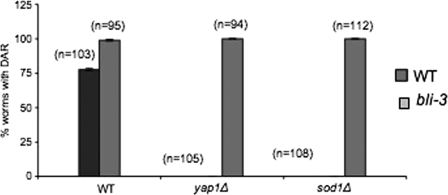
Two mutations, bli-3(e767) and bli-3(n529) have been identified in the gene encoding CeDuox1 (44). The mutant phenotypes of these strains recapitulate those produced by RNAi of CeDuox1 (17). Using the Amplex Red assay described above we showed that both mutant alleles of bli-3 produced less ROS when exposed to yeast (Fig. 4C). We hypothesized that mutant yeast that are sensitive to ROS, and hence unable to induce Dar in a wild-type nematode host, would be able to do so in a bli-3 mutant worm. To address this we tested the bli-3(e767) mutant allele in our pathogenesis assay. The bli-3 gene encodes the dual oxidase (CeDuox1) that we believe is involved in creating an environment of elevated ROS. Yeast yap1Δ and sod1Δ mutants that are unable to evoke the Dar response in wild-type worms are competent to induce Dar in bli-3 mutant worms (Fig. 5). Furthermore, the bli-3 mutant displays the Dar disease earlier, after 3 days, in contrast to the wild type, which typically shows Dar only after 4 days. No Dar phenotype was observed when wild-type N2 and the bli-3 mutant were grown on E. coli as a control (data not shown). These results strongly support the interpretation that worms exposed to yeast generate a burst of ROS via the action of the bli-3 gene product, CeDuox1, that inhibits the ability of yeast to induce Dar. In the absence of this defensive response, even the yap1Δ and sod1Δyeast mutants can induce Dar.
FIG. 5.
The yeast yap1Δ and sod1Δ mutants are able to cause Dar disease in the C. elegans bli-3(e767) mutant. The bli-3(e767) gene encodes a dual oxidase (CeDuox1) which contains an NADPH oxidase motif that has been implicated in ROS production in phagocytes. C. elegans bli-3(e767) mutants were exposed to yap1Δ and sod1Δ mutants, and the isogenic wild type was included as a control. The Dar response was scored on day 4 and compared to that in wild-type worms that were exposed to the yeast yap1Δ and sod1Δ mutants and wild-type counterparts.
We have used genetic and biochemical approaches on both sides of the host-pathogen equation in a pathogenesis assay to demonstrate a role for ROS in the host defense against fungal pathogenesis. We have identified Yap1 and its homolog, Yap2, as factors which may be used as targets for antifungal drug development. We have also identified CeDuox1 as a modulator of host responses to yeast infections. Studies such as the current one have the potential of conducting unbiased, whole-genome screens to identify novel fungal virulence factors and will greatly enhance our knowledge of host defense mechanisms in response to fungi.
DISCUSSION
Our studies have demonstrated that ROS play a central role in mediating host-pathogen interactions in our model. Using our novel pathogenesis assay with S. cerevisiae as a model pathogen and C. elegans as the model host, we found that yeast can infect worms, resulting in intestinal distension, a deformity in the postanal region (DAR phenotype), and death of the host nematode. In this study we show that ROS play an important role in fungal pathogenesis and host defense. The C. elegans bli-3 gene encodes a dual oxidase known to have mammalian homologs capable of producing ROS. Our results show that two bli-3 mutants with point mutations in the peroxidase domain (44) do not produce hydrogen peroxide in the Amplex Red assay. This may seem surprising, because classically, superoxide produced by NADPH oxidase is converted to hydrogen peroxide by dismutation and then to antimicrobial oxidants by peroxidase. According to this model, a mutation in the peroxidase domain would not be expected to alter production of the hydrogen peroxide. However, it has been suggested that the peroxidase domain of the dual oxidases catalyzes dismutation as well as production of antimicrobial oxidants (30). In that case, a mutation in the peroxidase domain might well cause a decrease in hydrogen peroxide production. Alternatively, a mutation in the peroxidase domain may alter the topology of the oxidase domain and decrease its activity. Further studies of the dual oxidases will be required to understand the details of their catalytic mechanisms.
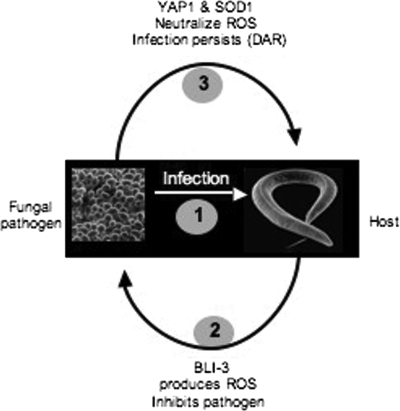
Our working model (Fig. 6) is based on the following results. In C. elegans the dual oxidase, CeDuox1, encoded by bli-3 is responsible for creating an environment of elevated ROS in response to the presence of yeast. Yeast mutants such as yap1Δ and sod1Δ that are sensitive to oxidizing environments are unable to evoke the Dar response in wild-type worms but are able to induce it in bli-3 mutants. Based on our data we predict a mechanism in which the nematode produces ROS in response to yeast. The Yap1 and Sod1 gene products of the invading fungus must neutralize these ROS before they can cause Dar. CeDuox1 is expressed in the cuticle (17), and our data predict that it should also be active in the intestinal lumen of the worms. In the future, in situ localization of CeDuox1 can be used to further test our hypothesis. Mammals also express the Nox family oxidases, which include the Duox enzymes, in epithelial tissues, including the intestine (18, 28), and evidence is mounting that these enzymes are involved in host defense (30).
FIG. 6.
Working model for the role of ROS in early events in host defense and fungal pathogenesis.
C. elegans has successfully been used as a model host for several microbial pathogens. Likewise, S. cerevisiae has proved its worth as a prototype for fungal pathogens. This is the first study that demonstrates a pathogenic interaction between these two powerful genetic model organisms. The advantages of being able to study both the host and pathogen genetically have been noted previously (39). The present study demonstrates how insights from one side of the host-pathogen interaction can be used to suggest experiments that can inform what is going on on the other side of the interaction.
This is exciting because in principle, such a study can be extended to the whole genome using existing methodologies. S. cerevisiae is the only fungal species with a systematic single-gene deletion library. This assay will enable us to conduct a high-throughput, unbiased, whole-genome screen to identify fungal virulence factors. Promising genes and pathways identified may be targeted for antifungal drug development. On the host side, there are plasmid libraries available containing RNAi constructs that downregulate single transcripts in C. elegans. Our pathogenesis assay will allow us to screen such a library and identify host factors that may be different from those identified in bacterial interactions.
Fungal infections are hard to treat because chemicals that are toxic to fungi are also often harmful to human patients. Thus, genes that are unique to fungi are potential targets for antifungal agents. It has previously been suggested that Yap1, a fungus-specific transcription factor, is involved in pathogenesis (2, 31, 40); however, it has not been shown to be required for pathogenesis. We use this pathogenesis assay to demonstrate that Yap1 and Yap2 are required to elicit a potentially protective host response, the Dar phenotype. This key observation, along with the finding that Yap1 is a fungus-specific transcription factor, makes it a likely target for broad-spectrum antimycotic drug development.
Acknowledgments
We thank F. Winston and J. Argüello for critical reading of the manuscript. We acknowledge S. Chan for the RFP yeast strain used in this study. C. elegans strains were provided by the Caenorhabditis Genetics Center, which is supported by the NIH National Center for Research Resources. We also thank G. Fink, M. Lorenz, D. Garsin, L. Ryder, and J. Duffy for useful discussions. Experiments were designed by C.J., S.M.P., and R.P.R. M.Y. performed experiments and analyzed data shown in Fig. 3B and C. C.J. performed all other experiments. R.P.R. wrote the paper.
This work was performed at and supported by Worcester Polytechnic Institute.
Footnotes
Published ahead of print on 5 June 2009.
REFERENCES
- 1.Alarco, A. M., I. Balan, D. Talibi, N. Mainville, and M. Raymond. 1997. AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J. Biol. Chem. 27219304-19313. [DOI] [PubMed] [Google Scholar]
- 2.Alarco, A. M., and M. Raymond. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apel, K., and H. Hirt. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55373-399. [DOI] [PubMed] [Google Scholar]
- 4.Apidianakis, Y., L. G. Rahme, J. Heitman, F. M. Ausubel, S. B. Calderwood, and E. Mylonakis. 2004. Challenge of Drosophila melanogaster with Cryptococcus neoformans and role of the innate immune response. Eukaryot. Cell 3413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babior, B. M., J. T. Curnutte, and B. J. McMurrich. 1976. The particulate superoxide-forming system from human neutrophils. Properties of the system and further evidence supporting its participation in the respiratory burst. J. Clin. Investig. 58989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bermingham-McDonogh, O., E. B. Gralla, and J. S. Valentine. 1988. The copper, zinc-superoxide dismutase gene of Saccharomyces cerevisiae: cloning, sequencing, and biological activity. Proc. Natl. Acad. Sci. USA 854789-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bindschedler, L. V., J. Dewdney, K. A. Blee, J. M. Stone, T. Asai, J. Plotnikov, C. Denoux, T. Hayes, C. Gerrish, D. R. Davies, F. M. Ausubel, and G. Paul Bolwell. 2006. Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 47851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolm, M., W. T. Jansen, R. Schnabel, and G. S. Chhatwal. 2004. Hydrogen peroxide-mediated killing of Caenorhabditis elegans: a common feature of different streptococcal species. Infect. Immun. 721192-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breger, J., B. B. Fuchs, G. Aperis, T. I. Moy, F. M. Ausubel, and E. Mylonakis. 2007. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 3e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 7771-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, E. C., B. F. Crawford, Z. Hong, T. Bilinski, and D. J. Kosman. 1991. Genetic and biochemical characterization of Cu,Zn superoxide dismutase mutants in Saccharomyces cerevisiae. J. Biol. Chem. 2664417-4424. [PubMed] [Google Scholar]
- 12.Chavez, V., A. Mohri-Shiomi, A. Maadani, L. A. Vega, and D. A. Garsin. 2007. Oxidative stress enzymes are required for DAF-16 mediated immunity due to generation of reactive oxygen species by C. elegans. Genetics 1761567-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coleman, S. T., E. A. Epping, S. M. Steggerda, and W. S. Moye-Rowley. 1999. Yap1p activates gene transcription in an oxidant-specific fashion. Mol. Cell. Biol. 198302-8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross, A. R., and A. W. Segal. 2004. The NADPH oxidase of professional phagocytes: prototype of the NOX electron transport chain systems. Biochim. Biophys. Acta 16571-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decottignies, A., A. M. Grant, J. W. Nichols, H. de Wet, D. B. McIntosh, and A. Goffeau. 1998. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 27312612-12622. [DOI] [PubMed] [Google Scholar]
- 16.Dong, C., R. J. Davis, and R. A. Flavell. 2002. MAP kinases in the immune response. Annu. Rev. Immunol. 2055-72. [DOI] [PubMed] [Google Scholar]
- 17.Edens, W. A., L. Sharling, G. Cheng, R. Shapira, J. M. Kinkade, T. Lee, H. A. Edens, X. Tang, C. Sullards, D. B. Flaherty, G. M. Benian, and J. D. Lambeth. 2001. Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91phox. J. Cell Biol. 154879-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El Hassani, R. A., N. Benfares, B. Caillou, M. Talbot, J. C. Sabourin, V. Belotte, S. Morand, S. Gnidehou, D. Agnandji, R. Ohayon, J. Kaniewski, M. S. Noel-Hudson, J. M. Bidart, M. Schlumberger, A. Virion, and C. Dupuy. 2005. Dual oxidase 2 is expressed all along the digestive tract. Am. J. Physiol. Gastrointest. Liver Physiol. 288G933-G942. [DOI] [PubMed] [Google Scholar]
- 19.Gauss, K. A., L. K. Nelson-Overton, D. W. Siemsen, Y. Gao, F. R. DeLeo, and M. T. Quinn. 2007. Role of NF-κB in transcriptional regulation of the phagocyte NADPH oxidase by tumor necrosis factor-alpha. J. Leukoc. Biol. 82729-741. [DOI] [PubMed] [Google Scholar]
- 20.Gerstbrein, B., G. Stamatas, N. Kollias, and M. Driscoll. 2005. In vivo spectrofluorimetry reveals endogenous biomarkers that report healthspan and dietary restriction in Caenorhabditis elegans. Aging Cell 4127-137. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein, A. L., and J. H. McCusker. 2001. Development of Saccharomyces cerevisiae as a model pathogen. A system for the genetic identification of gene products required for survival in the mammalian host environment. Genetics 159499-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gravato-Nobre, M. J., H. R. Nicholas, R. Nijland, D. O'Rourke, D. E. Whittington, K. J. Yook, and J. Hodgkin. 2005. Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics 1711033-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ha, E. M., C. T. Oh, Y. S. Bae, and W. J. Lee. 2005. A direct role for dual oxidase in Drosophila gut immunity. Science 310847-850. [DOI] [PubMed] [Google Scholar]
- 24.Ha, E. M., C. T. Oh, J. H. Ryu, Y. S. Bae, S. W. Kang, I. H. Jang, P. T. Brey, and W. J. Lee. 2005. An antioxidant system required for host protection against gut infection in Drosophila. Dev. Cell 8125-132. [DOI] [PubMed] [Google Scholar]
- 25.Hikkel, I., A. Lucau-Danila, T. Delaveau, P. Marc, F. Devaux, and C. Jacq. 2003. A general strategy to uncover transcription factor properties identifies a new regulator of drug resistance in yeast. J. Biol. Chem. 27811427-11432. [DOI] [PubMed] [Google Scholar]
- 26.Hodgkin, J., P. E. Kuwabara, and B. Corneliussen. 2000. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr. Biol. 101615-1618. [DOI] [PubMed] [Google Scholar]
- 27.Jansen, W. T., M. Bolm, R. Balling, G. S. Chhatwal, and R. Schnabel. 2002. Hydrogen peroxide-mediated killing of Caenorhabditis elegans by Streptococcus pyogenes. Infect. Immun. 705202-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawahara, T., Y. Kuwano, S. Teshima-Kondo, R. Takeya, H. Sumimoto, K. Kishi, S. Tsunawaki, T. Hirayama, and K. Rokutan. 2004. Role of nicotinamide adenine dinucleotide phosphate oxidase 1 in oxidative burst response to Toll-like receptor 5 signaling in large intestinal epithelial cells. J. Immunol. 1723051-3058. [DOI] [PubMed] [Google Scholar]
- 29.Lambeth, J. D., G. Cheng, R. S. Arnold, and W. A. Edens. 2000. Novel homologs of gp91phox. Trends Biochem. Sci. 25459-461. [DOI] [PubMed] [Google Scholar]
- 30.Leto, T. L., and M. Geiszt. 2006. Role of Nox family NADPH oxidases in host defense. Antioxid. Redox. Signal. 81549-1561. [DOI] [PubMed] [Google Scholar]
- 31.Lev, S., R. Hadar, P. Amedeo, S. E. Baker, O. C. Yoder, and B. A. Horwitz. 2005. Activation of an AP1-like transcription factor of the maize pathogen Cochliobolus heterostrophus in response to oxidative stress and plant signals. Eukaryot. Cell 4443-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, H. 2001. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4728-735. [DOI] [PubMed] [Google Scholar]
- 33.Longo, V. D., L. L. Liou, J. S. Valentine, and E. B. Gralla. 1999. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch. Biochem. Biophys. 365131-142. [DOI] [PubMed] [Google Scholar]
- 34.McCusker, J. H. 2006. Saccharomyces cerevisiae: an emerging and model pathogenic fungus. ASM Press, Washington, DC.
- 35.McCusker, J. H., K. V. Clemons, D. A. Stevens, and R. W. Davis. 1994. Genetic characterization of pathogenic Saccharomyces cerevisiae isolates. Genetics 1361261-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moy, T. I., E. Mylonakis, S. B. Calderwood, and F. M. Ausubel. 2004. Cytotoxicity of hydrogen peroxide produced by Enterococcus faecium. Infect. Immun. 724512-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mylonakis, E., F. M. Ausubel, J. R. Perfect, J. Heitman, and S. B. Calderwood. 2002. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc. Natl. Acad. Sci. USA 9915675-15680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholas, H. R., and J. Hodgkin. 2004. The ERK MAP kinase cascade mediates tail swelling and a protective response to rectal infection in C. elegans. Curr. Biol. 141256-1261. [DOI] [PubMed] [Google Scholar]
- 39.Persson, J., and R. E. Vance. 2007. Genetics-squared: combining host and pathogen genetics in the analysis of innate immunity and bacterial virulence. Immunogenetics 59761-778. [DOI] [PubMed] [Google Scholar]
- 40.Prusty, R., P. Grisafi, and G. R. Fink. 2004. The plant hormone indoleacetic acid induces invasive growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1014153-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinn, M. T., and K. A. Gauss. 2004. Structure and regulation of the neutrophil respiratory burst oxidase: comparison with nonphagocyte oxidases. J. Leukoc. Biol. 76760-781. [DOI] [PubMed] [Google Scholar]
- 42.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 43.Sifri, C. D., J. Begun, and F. M. Ausubel. 2005. The worm has turned: microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 13119-127. [DOI] [PubMed] [Google Scholar]
- 44.Simmer, F., C. Moorman, A. M. van der Linden, E. Kuijk, P. V. van den Berghe, R. S. Kamath, A. G. Fraser, J. Ahringer, and R. H. Plasterk. 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 101793-1808. [DOI] [PubMed] [Google Scholar]
- 46.Wheeler, R. T., M. Kupiec, P. Magnelli, C. Abeijon, and G. R. Fink. 2003. A Saccharomyces cerevisiae mutant with increased virulence. Proc. Natl. Acad. Sci. USA 1002766-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wisplinghoff, H., H. Seifert, S. M. Tallent, T. Bischoff, R. P. Wenzel, and M. B. Edmond. 2003. Nosocomial bloodstream infections in pediatric patients in United States hospitals: epidemiology, clinical features and susceptibilities. Pediatr. Infect. Dis. J. 22686-691. [DOI] [PubMed] [Google Scholar]
- 48.Wolfger, H., Y. Mahe, A. Parle-McDermott, A. Delahodde, and K. Kuchler. 1997. The yeast ATP binding cassette (ABC) protein genes PDR10 and PDR15 are novel targets for the Pdr1 and Pdr3 transcriptional regulators. FEBS Lett. 418269-274. [DOI] [PubMed] [Google Scholar]
- 49.Wolfger, H., Y. M. Mamnun, and K. Kuchler. 2001. Fungal ABC proteins: pleiotropic drug resistance, stress response and cellular detoxification. Res. Microbiol. 152375-389. [DOI] [PubMed] [Google Scholar]
- 50.Wolfger, H., Y. M. Mamnun, and K. Kuchler. 2004. The yeast Pdr15p ATP-binding cassette (ABC) protein is a general stress response factor implicated in cellular detoxification. J. Biol. Chem. 27911593-11599. [DOI] [PubMed] [Google Scholar]
- 51.Wu, Y., M. Han, and K. L. Guan. 1995. MEK-2, a Caenorhabditis elegans MAP kinase kinase, functions in Ras-mediated vulval induction and other developmental events. Genes Dev. 9742-755. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, X., M. De Micheli, S. T. Coleman, D. Sanglard, and W. S. Moye-Rowley. 2000. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol. Microbiol. 36618-629. [DOI] [PubMed] [Google Scholar]