Abstract
The processing of polycistronic pre-mRNAs in trypanosomes requires the spliceosomal small ribonucleoprotein complexes (snRNPs) U1, U2, U4/U6, U5, and SL, each of which contains a core of seven Sm proteins. Recently we reported the first evidence for a core variation in spliceosomal snRNPs; specifically, in the trypanosome U2 snRNP, two of the canonical Sm proteins, SmB and SmD3, are replaced by two U2-specific Sm proteins, Sm15K and Sm16.5K. Here we identify the U2-specific, nuclear-localized U2B″ protein from Trypanosoma brucei. U2B″ interacts with a second U2 snRNP protein, U2-40K (U2A′), which in turn contacts the U2-specific Sm16.5K/15K subcomplex. Together they form a high-affinity, U2-specific binding complex. This trypanosome-specific assembly differs from the mammalian system and provides a functional role for the Sm core variation found in the trypanosomal U2 snRNP.
In trypanosomes, trans-splicing is an essential step in the expression of all protein-coding genes. The resulting mRNAs always carry a noncoding spliced leader (SL) sequence of 39 nucleotides at their 5′ ends, which is derived from the SL RNA. In addition to the SL RNA, the small nuclear RNAs (snRNAs) U2, U4, U5, and U6 are essential cofactors during trans-splicing (reviewed in reference 14).
In previous studies, we characterized some of the protein components of the spliceosomal small nuclear ribonucleoproteins (snRNPs) from Trypanosoma brucei. All snRNPs contain a core of seven Sm polypeptides (18). Recently, we reported that the identity of the Sm proteins varies among spliceosomal snRNPs; specifically, two of the canonical Sm proteins, SmB and SmD3, are replaced in the U2 snRNP by two novel, U2 snRNP-specific Sm proteins, Sm15K and Sm16.5K (34). There is a similar case of Sm core variation in the U4 snRNP, where a single Sm protein, SmD3, is replaced by a U4-specific LSm protein (32; N. Jaé and A. Bindereif, unpublished data). Trypanosomal snRNAs also differ significantly from what we know in other systems, reflecting the large evolutionary distance and trypanosome-specific properties. For example, both the U1 and U5 snRNAs from trypanosomes represent the shortest known orthologues (6, 19).
In addition to the Sm proteins, some snRNP-specific protein factors were found in trypanosomes. Sequence comparisons identified the U2-40K protein as the trypanosomal homologue of the human U2A′ protein (5), a finding that was unexpected, since no immunological relationship could be detected between these proteins (17). As characterized in other systems, including those of humans, yeasts, and plants (9, 29, 31), the U2 snRNP contains a second specific protein, U2B″, a protein closely related to the U1 snRNP protein U1A. Except for the Saccharomyces cerevisiae orthologue, the known U2B″ proteins are built of two RNA recognition motifs (RRMs), with the N-terminal RRM being responsible for snRNA binding specificity (25). The close relatedness of these two proteins is also reflected in Drosophila melanogaster, where a single protein, SNF/D25, combines the functions of both individual proteins (11, 22). Furthermore, genetic and functional redundancy was demonstrated for the two proteins in Caenorhabditis elegans (24).
From previous studies on the mammalian U2 snRNP, we know that the U2- specific proteins U2A′ and U2B″ interact with each other, independently of U2 snRNA; moreover, U2B″ binds directly to loop nucleotides of stem-loop IV, but only with the assistance of interacting U2A′ (23, 25, 26). Analogous to the cis-splicing mechanism, the U2 snRNP is likely to play an important role in early trans-spliceosome assembly. Compared with the other snRNAs, the trypanosomatid U2 snRNA differs in several important aspects from its highly conserved counterparts in other species. First, stem-loop III is precisely deleted. Second, the branch point recognition region located between stem-loops I and IIa is missing; in parallel, there is no stringent consensus of branch points in the 3′-splice site region of the polycistronic pre-mRNA. Third, the Sm protein binding site does not follow the general consensus. Finally, only some of the otherwise highly conserved loop IV nucleotides occur in the trypanosomatid U2 snRNAs (7, 12, 15, 33).
Here we report the identification and characterization of the U2-specific protein U2B″ of T. brucei. Sequence analysis revealed that the trypanosomal orthologue contains only a single RRM, in contrast to the mammalian, two-RRM domain structure, and that the homology is restricted to this single RRM. We show that U2-40K (U2A′) binds very efficiently to U2B″ in the absence of U2 snRNA and increases the binding affinity of U2B″ to U2 snRNA. Furthermore U2-40K (U2A′) contacts the two specific components of the U2 Sm core, Sm16.5K and Sm 15K, forming together a high-affinity, U2-specific binding complex. This establishes a specific function of the U2 Sm core variation in mediating U2-specific protein-protein interactions.
MATERIALS AND METHODS
Growth of trypanosomes and preparation of cell extracts.
Cell culture of the procyclic form of T. brucei strain 427 and of stably transfected cell lines was done as described previously (5, 27). Cell extracts were prepared in IPP-150 buffer (150 mM KCl, 20 mM Tris-Cl [pH 7.7], 3 mM MgCl2, 0.5 mM dithiothreitol [DTT]) containing a Complete mini-, EDTA-free protease inhibitor cocktail tablet (Roche) by using a Polytron PT 3100 cell homogenizer (Kinematica AG, Switzerland). Cell lysates were supplemented with 0.1% Tween 20 (Sigma) and centrifuged twice at 14,000 rpm for 15 min to remove aggregates.
Database analysis.
Protein sequence alignments were performed by the ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and T-Coffee (http://www.ebi.ac.uk/t-coffee/) programs. Pattern and profile searches were done by SMART (http://smart.embl-heidelberg.de/). Protein structure prediction was carried out via the GeneSilico metaserver (13).
Recombinant proteins.
All recombinant plasmids were transformed into Escherichia coli BL21(DE3)pLysS, and proteins were expressed as described previously (30). The open reading frames (ORFs) of T. brucei U2-40K and U2B″ were PCR amplified from genomic DNA and cloned into pGEX-2TK. For the His-tag derivatives U2B″ and U2B″-RRM, the ORFs were cloned into pQE30 or pET151/D/lacZ vectors. Recombinant proteins were purified by glutathione S-transferase (GST) or Ni-nitrilotriacetic acid affinity chromatography on an ÄKTApurifier high-pressure liquid chromatography system (GE Healthcare).
Protein-protein interaction assays by GST pulldown.
For assaying protein-protein interactions, 1 μg of GST-U2-40K protein (or GST alone as a negative control) was immobilized on 25 μl packed glutathione-Sepharose 4B beads (GE Health care) and incubated with ∼500 ng of His-tagged Sm subcomplexes (SmD1/D2, SmE/F/G, SmD3/B, or Sm16.5K/15K; His-tagged proteins are underlined) in 900 μl binding buffer (50 mM Tris-Cl [pH 7.5], 300 mM KCl, 5 mM MgCl2, 0.05% NP-40, 0.5 mM DTT). After a 4-h incubation at 4°C, beads were washed with the same buffer, which contained 500 mM KCl. Bound proteins were released from the beads and analyzed by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting with anti-His6 monoclonal antibody (Sigma). To detect the interaction between U2-40K and U2B″ proteins, the same GST pulldown assay was performed. Eight hundred nanograms of N-terminally His-FLAG-tagged U2B″ was added to 2 μg of GST-U2-40K or GST protein in 500 μl binding buffer, incubated for 1 h at 25°C, and then washed with the same buffer, which contained 300 mM KCl.
Western blot analysis.
Protein samples were separated by SDS-15% PAGE, blotted, and reacted with anti-His6 (Sigma) or anti-40K antibodies (17) at a dilution of 1:5,000. Western blots were developed by the ECL chemiluminescent detection system.
TAP tag construct of U2B″, pulldown assays, and Northern analysis.
The complete ORF and an upstream region (from position −591 relative to the translational initiation codon) of T. brucei U2B″ were cloned into a tandem affinity purification (TAP) tag vector (pD11) containing a duplicate protein A epitope, a tobacco etch virus (TEV) cleavage site, a calmodulin binding peptide, and a neomycin resistance marker (28). The ORF was PCR amplified from T. brucei genomic DNA and cloned upstream of the TAP tag sequence, between the ApaI and NotI sites of the pD11 vector. For genomic integration, 10 μg of the TAP-U2B″ construct was linearized by NsiI (upstream of the U2B″ ORF) and transfected into procyclic T. brucei 427 cells by electroporation, replacing one allele of U2B″ by homologous recombination. Stably transfected cells were selected with G418 (40 μg/ml Geneticin; Gibco-BRL).
For pulldown assays via TAP tag, cell extracts from stably transfected cell lines were incubated at 4°C with 25 μl of packed immunoglobulin G (IgG) Sepharose 6 fast flow beads (Invitrogen) equilibrated in IPP-150 buffer. After the beads were washed with the same buffer (or with IPP-500, which contains 500 mM KCl), coselected RNAs were released by proteinase K buffer treatment and analyzed by denaturing PAGE, followed by Northern blotting, using digoxigenin-labeled probes (1).
Oligonucleotides, mutagenesis, and in vitro transcription.
The sequences of DNA oligonucleotides are available upon request. The [α-32P]UTP-labeled, uncapped TbU2-3′half wild-type transcript, which contains nucleotides 67 through 148 of the T. brucei U2 snRNA with the Sm site sequence, was transcribed by T7 RNA polymerase, using as a template two overlapping DNA oligonucleotides filled in by Taq DNA polymerase. In the mutant derivative TbU2-3′half ΔG, nucleotide G94 was deleted (34). In the TbU2-3′half hul4 construct (8), the trypanosome loop IV sequence (nucleotides 118 through 129) was replaced by nucleotides 159 through 171 of the human U2 snRNA.
Reconstitution of recombinant Sm cores and His-tag pulldown assays.
First, 100 pmol of purified His-tagged Sm subcomplexes (for the canonical Sm core, SmD3/B, SmD1/D2, and SmE/F/G; for the U2 Sm core, Sm16.5K/15K, SmD1/D2, and SmE/F/G; His-tagged proteins are underlined) were mixed in equimolar amounts in 10 μl of 5× reconstitution buffer (100 mM Tris-Cl [pH 7.5], 1 M NaCl, 25 mM MgCl2, 5 mM DTT). Second, ∼30 ng (1.5 × 106 cpm) of wild-type or ΔG mutant U2-3′half transcripts were added to the reaction to a final volume of 50 μl. The reconstitution reactions were incubated at 30°C for 30 min and then at 37°C for 15 more minutes. In the following procedures, either GST-U2-40K, GST-U2B″, GST-U2-40K/His-FLAG-U2B″, or GST alone was added and incubated for 30 min at 30°C. For the pulldown assays, the reconstituted complexes were incubated with 25 μl of packed glutathione-Sepharose 4B beads (Amersham) in 1× reconstitution buffer (20 mM Tris-Cl [pH 7.5], 200 mM of NaCl, 5 mM MgCl2, 1 mM DTT, 0.05% NP-40) at room temperature. After the beads were washed with the same buffer, bound RNAs were released from the beads and analyzed by denaturing PAGE. Signals were quantitated using Tina version 2.07d software.
Bandshift assays.
RNA binding reactions were performed in 10 μl 1× reconstitution buffer (20 mM Tris-Cl [pH 7.5], 200 mM NaCl, 5 mM MgCl2, 1 mM DTT) containing the purified proteins and 32P-labeled RNA (5 nM). The effects of U2-40K and U2B″ on RNA binding was assessed by adding the protein at concentrations of 50 nM to 2 μM. In addition, the affinity of U2B″ for U2-3′half RNA was measured at a U2-40K concentration of 300 nM (see Fig. 3A to C). For competition experiments (see Fig. 3D), 32P-labeled RNA (2 nM) was incubated with GST-U2-40K (300 nM), His-FLAG-U2B″ (2,000 nM), and increasing amounts of unlabeled competitor RNA [TbU2-3′half wild type or TbU2-3′half hul4 RNA at a 1- to 10-fold molar excess]. Binding reaction mixtures were incubated for 30 min at room temperature, combined with 30% glycerol, and loaded immediately onto cold, nondenaturing Tris-glycine-10% polyacrylamide gels that had been prerun for 30 min. After 2 h of electrophoresis at 180 V, complexed and free RNAs were visualized by a Phosphorimager system (Bio-Rad).
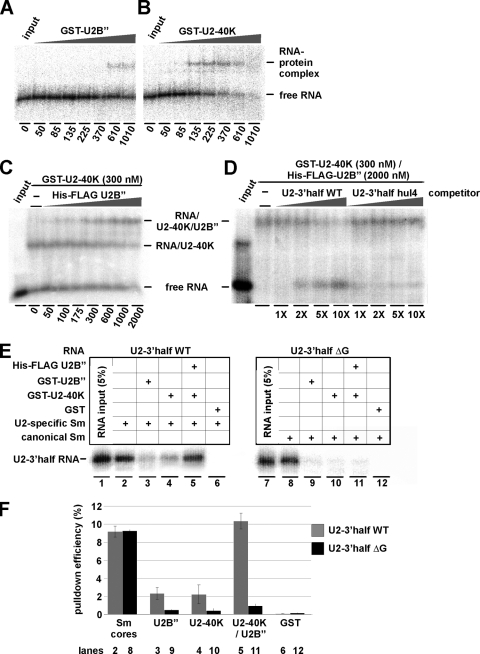
FIG. 3.
U2 snRNA interactions of U2-40K, U2B″, and the U2-specific Sm proteins. (A through D) Analysis of U2B″ and U2-40K protein binding to U2-3′half RNA. Increasing amounts of GST-U2B″ (50 to 1,010 nM, as indicated below) (A), or GST-U2-40K protein (50 to 1,010 nM) (B), or His-FLAG U2B″ (50 to 2,000 nM) (C) were combined with 32P-labeled U2-3′half snRNA (5 nM). The reactions shown in panel C also contained a constant amount of GST-U2-40K (300 nM). (D) In addition, to test for the role of the 3′-terminal loop of U2 snRNA, the formation of the RNP complex of GST-U2-40K (300 nM), His-FLAG-U2B″ (2,000 nM), and 32P-labeled TbU2-3′half wild-type RNA (2 nM) was analyzed in the presence of unlabeled competitor RNA [TbU2-3′half wild type or TbU2-3′half hul4 RNA at a 1- to 10-fold molar excess, as indicated below]. RNP complexes were separated from free RNA by non-denaturing gel electrophoresis. In each panel, free RNA was analyzed as well (input lanes). (E and F) snRNA binding specificity of the U2-40K/B″ complex. 32P-labeled wild-type and ΔG94 mutant (ΔG) T. brucei U2-3′half RNAs (nucleotides 67 through 148 of U2 snRNA) were reconstituted in vitro with recombinant canonical (lanes 8 through 12) or U2-specific (lanes 2 through 6) Sm core proteins, followed by the addition of GST-U2B″, GST-U2-40K, His-FLAG-U2B″/GST-U2-40K, or GST alone (as indicated) and further incubation for 30 min at 30°C. The respective RNA inputs are analyzed in lanes 1 and 7 (5% of each) (E). To control for efficient Sm core assembly, His-tag pulldown assays were performed (lanes 2 and 8). For the other reconstitutions, coprecipitated RNAs were recovered by GST pulldown and analyzed by denaturing gel electrophoresis (E). (F) The efficiencies of the reconstitutions were quantitated and diagrammed, using as a measure the percentages of precipitated RNAs to their respective inputs with standard deviations (n = 3). Gray bars, wild-type U2-3′half RNA; black bars, ΔG mutant RNA. The lane numbers refer to the gel shown in panel E.
Immunofluorescence.
T. brucei cells expressing TAP-U2B″ were harvested and fixed on coverslips with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min at 4°C. The cells were blocked with 1% cold water fish gelatin (Sigma-Aldrich) in PBS and incubated with a 1:40,000 dilution of anti-protein A primary antibody (Sigma-Aldrich). Following washing with PBS containing 0.05% Tween 20, a solution containing 1 μg/ml DAPI (4′,6-diamidino-2-phenylindole) and Alexa 594 secondary antibody (Invitrogen) at a 1:400 dilution was added. The coverslips were mounted in Vectashield (Vector Laboratories). For imaging, a Zeiss Axio Scope 20 microscope and AxioVision software were used.
RESULTS AND DISCUSSION
Identification of the trypanosome orthologue of the human U2B″ protein.
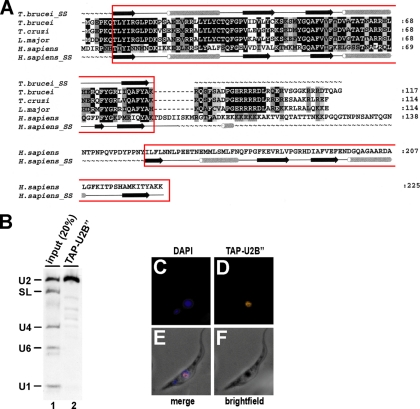
Based on sequence homology and the T. brucei genome project (3), we were able to identify a potential orthologue of the human U2B″ (GeneDB accession number Tb927.3.3480; 13.6 kDa; 117 amino acids) (an alignment of the three trypanosomatid and the human U2B″ protein sequences is shown in Fig. 1A). After the introduction of a gap in the trypanosomatid U2B″ sequences, the human and T. brucei U2B″ sequences are 32% identical. Strikingly, homology is restricted to the N-terminal region (amino acids 1 through 84 of trypanosomal U2B″ and amino acids 1 through 85 of human U2B″; 42% identity). In contrast to the human U2B″ protein with its two RRMs, the trypanosomatid U2B″ proteins contain only a single, N-terminal RRM, which in the human system is sufficient for U2 snRNA binding specificity (2).
FIG. 1.
Sequence alignment, U2 snRNA binding specificity, and nuclear localization of T. brucei U2B″. (A) Multiple sequence alignment of trypanosomatid and human U2B″ sequences. ClustalW alignments of the U2B″ protein sequences from T. brucei, Trypanosoma cruzi, and Leishmania major and human U2B″ are shown, with the RRMs boxed. Residues that are conserved or whose physicochemical character is conserved are highlighted in black and gray, respectively. The secondary structures of U2B″ from T. brucei and Homo sapiens are shown above and below the alignment, respectively. α-Helices are represented as tubes and ß strands as arrows, while disordered regions are shown as tildes (∼). The GeneDB or NCBI accession numbers of U2B″ sequences are as follows: T. brucei, Tb927.3.3480; T. cruzi, Tc00.1047053507951.140; L. major, LmjF29.0865; and H. sapiens, NP_003083. The total numbers of amino acids are given on the right. (B) U2 snRNA specificity of T. brucei U2B″. Extract was prepared from a T. brucei cell line stably expressing TAP-tagged U2B″, followed by IgG pulldown of TAP-tagged complexes and Northern blot analysis of copurifying RNAs, detecting U2, SL, U4, U6, and U1 snRNAs (as indicated on the left; lane 2). For comparison, 20% of the total input was analyzed (lane 1). (C through F) Nuclear localization of T. brucei U2B″. TAP-U2B″-expressing cells were fixed (F) and stained with DAPI (C). In parallel, TAP-tagged U2B″ was detected by anti-protA primary antibody and Alexa 594 secondary antibody (D). (E) Merged view of DAPI and TAP-U2B″ staining.
To further confirm the identity of the trypanosome U2B″, we used the first step of the TAP procedure. A stable T. brucei cell line expressing C-terminally TAP-tagged U2B″ was generated (see Materials and Methods). Cell lysate was prepared, tagged complexes were purified via IgG Sepharose, and copurifying snRNAs were detected by Northern blotting (Fig. 1B). We concluded that T. brucei U2B″ is specifically associated with U2 snRNA.
By indirect immunofluorescence, we also investigated the subcellular localization of U2B″ in the procyclic form of T. brucei, using the same cell line and immunostaining with anti-protein A antibodies (Fig. 1C through F). Clearly, TAP-tagged U2B″ is nuclear localized, and no cytoplasmic staining was detectable (Fig. 1C through E). Controls with wild-type cells showed no nuclear staining (data not shown). Note that the nuclear localization of trypanosome U2B″ is consistent with our previous finding that the other U2 snRNP protein, U2-40K (U2A′), resides predominantly in the nucleus as well (17).
Protein-protein interactions in the U2 snRNP: U2B″ associates with U2-40K, and U2-40K contacts the U2-specific Sm core.
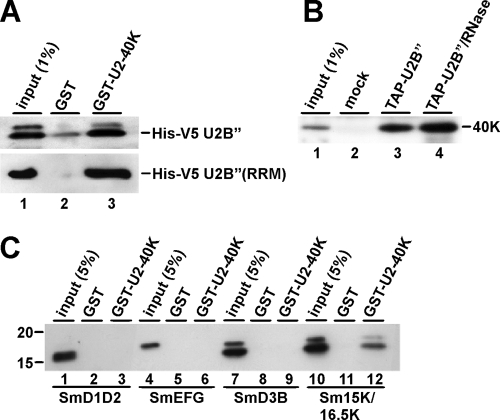
In the human system, in the absence of RNA, U2B″ stably interacts with another U2-specific component, U2A′ (4, 26). Since the trypanosome U2B″ differs in its domain structure from its mammalian counterpart, we tested for this protein-protein interaction in T. brucei, using GST pulldown assays (Fig. 2A). Note that the trypanosome orthologue to U2A′ is called U2-40K (5). E. coli lysates expressing His-V5-U2B″ and purified recombinant GST-tagged U2-40K protein (or GST as a control) were incubated together, followed by GST pulldown. After washing, bound proteins were eluted and analyzed by SDS-PAGE and Western blotting, using anti-V5 antibodies. As a result, U2B″ could be efficiently recovered (Fig. 2A, top panel) only with GST-40K, but not with GST alone, demonstrating that trypanosome U2B″ and U2A′ interact in vitro in the absence of RNA. For this interaction, the single RRM domain of the trypanosome U2B″ is sufficient (Fig. 2A, bottom panel), similar to previous findings for the mammalian proteins (26).
FIG. 2.
Protein-protein interactions of U2-40K, U2B″, and the U2-specific Sm proteins. (A) U2-40K and U2B″ interact in vitro. Immobilized GST-40K protein (lane 3) or GST as a control (lane 2) were incubated with lysate from E. coli cells expressing His-V5-tagged U2B″ protein (top panel) or the RRM domain only of His-V5 U2B″ (bottom panel). After washing, bound proteins were eluted and analyzed by SDS-PAGE and Western blotting, using anti-V5 antibodies. (B) U2-40K and U2B″ interact in vivo. Lysates were prepared from a T. brucei cell line stably expressing TAP-tagged U2B″ protein without (lane 3) or with (lane 4) an additional RNase treatment or from the wild-type strain (mock; lane 2), followed by precipitation by IgG Sepharose. TAP-U2B″-associated proteins were analyzed by SDS-PAGE and Western blotting with anti-40K antibodies. (C) U2-40K interacts specifically with the U2-specific Sm proteins. Immobilized GST-40K protein (lanes 3, 6, 9, and 12) or GST as a control (lanes 2, 5, 8, and 11) were incubated with purified His-tagged SmD1/D2, SmE/F/G, SmD3/B, or Sm16.5K/15K proteins (His-tagged proteins are underlined). After washing, bound proteins were eluted and analyzed by SDS-PAGE and Western blotting, using anti-His6 antibodies.
In addition, we assayed for a U2-40K/U2B″ interaction in vivo, using the T. brucei cell line stably expressing TAP-tagged U2B″ (Fig. 2B; see also Fig. S1 in the supplemental material for a specificity control). Tagged proteins were purified from lysate on IgG Sepharose, and copurifying U2-40K was analyzed by Western blotting (Fig. 2B, lanes 1 through 3). Since both proteins are present in vivo within the U2 snRNP, we also assayed for an interaction in the absence of RNA by incubating the lysate with RNase A, followed by IgG Sepharose purification (Fig. 2B, lane 4). In both cases, TAP-tagged U2B″ efficiently interacted with U2-40K, in contrast to the mock control with lysate from wild-type cells (Fig. 2B, compare lanes 2 through 4). We concluded that trypanosome U2B″ and U2-40K stably associate with each other independently of RNA. In summary, it appears that the trypanosomal U2B″ protein represents a minimal version of this protein, with its single RRM being sufficient for U2-40K interaction.
The U2 snRNP also carries a special Sm core, in which the canonical SmB and SmD3 proteins are replaced by two U2-specific Sm proteins, Sm15K and Sm16.5K, respectively (34). Therefore, the latter two proteins are good candidates to play a role in the specific recruitment of further U2 snRNP-specific proteins. To investigate the potential role of the Sm core proteins in U2 snRNP assembly, we assayed both U2-40K and U2B″ for interactions with the purified His-tagged Sm subcomplexes SmD1/D2, SmD3/B, SmE/F/G, and Sm16.5K/15K (His-tagged proteins are underlined) (for an analysis of the purified Sm subcomplexes used, see Fig. S2 in the supplemental material). First, GST-40K bound proteins were analyzed by SDS-PAGE and Western blotting, using anti-His6 antibodies. GST-tagged U2-40K associated efficiently only with the U2-specific Sm16.5K/15K subcomplex but not detectably with the canonical SmD1/D2, SmE/F/G, and SmD3/B proteins (Fig. 2C). Second, we tested U2B″ binding to the Sm subcomplexes, but here no interaction could be observed (data not shown). We concluded that U2-40K is the U2 specificity determinant of the U2-specific heterodimer U2A′/U2B″. This finding is the first evidence for a functional and specific protein-protein interaction mediated by the Sm core variation in the U2 snRNP.
Interaction of U2B″ and U2-40K proteins with U2 snRNA.
After characterizing protein-protein interactions in the U2 snRNP, we next asked whether the U2-specific Sm core in the 3′ domain of the U2 snRNP might be involved in the binding of U2-40K and U2B″. When identifying the U2-40K protein, we had initially shown that U2-40K by itself binds RNA nonspecifically but that the addition of extract confers U2 snRNA specificity (5), suggesting that other protein components cooperate with the U2-40K protein in U2 snRNP assembly. Therefore, we assayed T. brucei U2-40K and U2B″, both separately and in combination, to determine their U2 snRNA binding (Fig. 3A through D). Based on our initial study (5), we focused on the 3′ terminal domain of U2 snRNA, including stem-loops IIb and IV (nucleotides 67 through 148; called U2-3′half).
32P-labeled U2-3′half RNA was incubated in different molar ratios with recombinant U2B″ (Fig. 3A) or U2-40K (Fig. 3B) or with both proteins in combination (Fig. 3C), followed by analysis of complex formation by native gel electrophoresis. U2-40K exhibited a higher U2 RNA binding affinity than U2B″ (Fig. 3, compare panels A and B). We next investigated whether U2-40K can enhance the low affinity of U2B″ for U2-3′half RNA and whether a ternary complex can form (Fig. 3C). When His-tagged U2B″ was combined with U2-3′half RNA in the presence of U2-40K in a concentration which is sufficient for RNA binding (300 nM), we observed a ternary complex that migrated more slowly than the binary complex formed by U2-40K and U2-3′half RNA in the absence of U2B″ (Fig. 3C).
To test for the sequence specificity of this ternary complex, we also did competition experiments (Fig. 3D). The ternary RNP complex of GST-U2-40K (300 nM), His-FLAG-U2B″ (2,000 nM), and 32P-labeled TbU2-3′half wild-type RNA (2 nM) (Fig. 3C, last lane) was formed in the presence of increasing amounts of unlabeled competitor RNA, TbU2-3′half wild-type RNA, or TbU2-3′half hul4 RNA, in which the trypanosome 3′-terminal loop sequence of 12 nucleotides had been replaced by the corresponding human loop sequence. Since only the wild type, but not the loop mutant of U2-3′half RNA was able to compete, we concluded that the 3′-terminal loop of the trypanosome U2 snRNA plays an essential role in U2 snRNP assembly.
U2-specific Sm proteins are necessary for assembly of the U2-40K/U2B″ dimer on U2 snRNA.
After characterizing the specific interaction between U2-40K and the U2-specific Sm proteins, we addressed the question of whether the U2-specific Sm core is required for specific assembly of U2-40K/U2B″ onto the U2 snRNA. Therefore, we established an in vitro reconstitution assay, using recombinant canonical (SmD3/B, SmD1/D2, and SmE/F/G) or U2-specific (Sm16.5K/15K, SmD1/D2, and SmE/F/G) Sm subcomplexes.
To examine the binding specificity, in parallel with the wild-type U2 snRNA sequence, we assayed a mutant derivative in which G94 is deleted, both as short U2 derivatives containing nucleotides 67 through 148 (U2-3′half wild type and ΔG). Note that this single nucleotide deletion, ΔG94, makes the U2 Sm site identical to a canonical Sm site. As we had shown previously, the unusual guanosine at position 94 in the U2 Sm site is sufficient for discriminating between the U2-specific Sm core and the canonical Sm core. U2-3′half wild-type RNA specifically associates with the U2-specific Sm proteins, and U2-3′half ΔG RNA with the canonical Sm proteins (34).
32P-labeled U2 snRNA derivatives were reconstituted with the U2-specific Sm core (U2-3′half wild type) (Fig. 3E, lanes 2 through 6) or the canonical Sm core (U2-3′half ΔG) (Fig. 3E, lanes 8 through 12). The efficiency of these reconstitutions was controlled by His-tag pulldown, using Ni-nitrilotriacetic acid agarose beads, and by analyzing coprecipitated RNAs by denaturing PAGE (Fig. 3E, lanes 2 and 8). Subsequently, the recombinant trypanosomal U2 snRNP-specific proteins GST-U2-40K or GST-U2B″ or the GST-U2-40K/His-FLAG-U2B″ heterodimer were incubated together with the reconstituted Sm cores. Finally, after GST pulldown, associated RNAs were analyzed by denaturing PAGE and quantitation (Fig. 3E and F).
As a result, U2-40K and U2B″ alone associated weakly with the reconstituted U2-specific Sm core (U2-40K, 2.0%; U2B″, 2.7% [Fig. 3E and F, compare lanes 3 and 4]), but even less efficiently with the canonical Sm core (U2-40K, 0.6%; U2B″, 0.5% [Fig. 3E and F, compare lanes 9 and 10]). In contrast, the U2-40K/U2B″ heterodimer associated efficiently with U2-3′half wild-type RNA and the U2-specific Sm core, but not or only very weakly with the U2-3′half ΔG RNA and the canonical Sm core (10.5% versus 1% [Fig. 3E and F, compare lanes 5 and 11]).
In summary, this finding underlines the importance of the U2 Sm site with its unusual guanosine at position 94, which not only specifies assembly of the U2-specific Sm core proteins but also directs the recruitment of the U2-specific 40K/U2B″ heterodimer. Thereby the unusual U2 Sm site is indirectly responsible for the accurate assembly of further U2 snRNP-specific proteins.
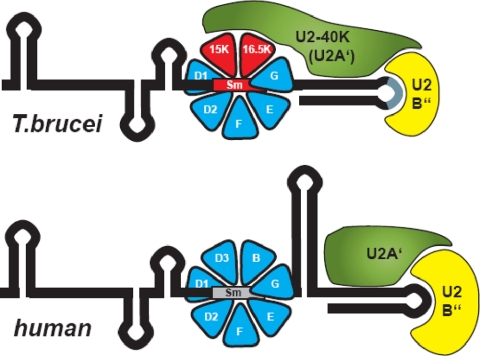
Our data suggest a model for the assembly of the trypanosome U2 snRNP and its RNA-protein interactions (Fig. 4). The initial assembly of the U2-specific Sm core on the special Sm site of the U2 snRNA directs the association of the U2-40K/U2B″ heterodimer, involving contacts between U2-40K and the U2-specific Sm proteins. This trypanosome-specific U2-40K/Sm protein interaction correlates with the lack of stem-loop III.
FIG. 4.
Model of the interactions in the core of the trypanosomal U2 snRNP (top), in comparison with the human counterpart (bottom). RNA secondary structures are indicated by black lines; boxed regions represent the Sm sites (the U2-specific Sm site is in red). Protein homologies are represented by the same colors. Direct binding of the trypanosome U2B″ to U2 snRNA hairpin loop IV is drawn in analogy to the human RNP structure, consistent with the competition experiment shown in Fig. 3D. The interactions between trypanosome U2-40K (U2A′) and U2B″ (Fig. 2A and B), as well as between U2-40K (U2A′) and the U2-specific Sm16.5K/15K subcomplex (Fig. 2C) are schematically represented. It is not known which of the two U2-specific Sm proteins is contacted by U2-40K (U2A′). Note the trypanosome-specific characteristics, the U2-40K interaction with the U2-Sm proteins, the lack of stem-loop III, several deviations of the loop IV nucleotides conserved in other species (gray line) (discussed in reference 7), and the unusually small, single-RRM U2B″. This model addresses only the 3′-terminal domain of the U2 snRNP, including the U2-specific Sm site region.
In two other cases, the Sm core components of the U1 snRNP and the LSm proteins of the U7 snRNP Sm proteins participate in the RNP function. First, in the mammalian U7 snRNP, the U7 Sm binding site not only ensures the assembly of the U7-specific Sm core but also plays an important functional role during histone mRNA processing (20, 21). A second example comes from the spliceosomal snRNPs. The Sm proteins of the human U1 snRNP stabilize binding of the U1 snRNP-specific proteins U1-70K and U1-C (10, 16).
As we have shown here, the trypanosome U2 snRNP represents a third example of Sm proteins playing an active role in RNP function. The U2-specific Sm core variation not only mediates specific recognition of the special Sm site in the U2 snRNA (34) but also engages in a network of multiple protein-protein interactions that result in a stable 3′-terminal RNP complex.
Supplementary Material
Acknowledgments
We thank members of our laboratory for critical comments and advice and Arthur Günzl for the TAP-tag vector.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 535 and IRTG 1384).
Footnotes
Published ahead of print on 19 June 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Bell, M., and A. Bindereif. 1999. Cloning and mutational analysis of the Leptomonas seymouri U5 snRNA gene: function of the Sm site in core RNP formation and nuclear localization. Nucleic Acids Res. 273986-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley, R. C., and J. D. Keene. 1991. Recognition of U1 and U2 small nuclear RNAs can be altered by a 5-amino-acid segment in the U2 small nuclear ribonucleoprotein (snRNP) B″ protein and through interactions with U2 snRNP-A′ protein. Mol. Cell. Biol. 111829-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berriman, M., et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309416-422. [DOI] [PubMed] [Google Scholar]
- 4.Boelens, W., D. Scherly, R. P. Beijer, E. J. Jansen, N. A. Dathan, I. W. Mattaj, and W. J. van Venrooij. 1991. A weak interaction between the U2A′ protein and U2 snRNA helps to stabilize their complex with the U2B″ protein. Nucleic Acids Res. 19455-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross, M., B. Wieland, Z. Palfi, A. Günzl, U. Röthlisberger, H. W. Lahm, and A. Bindereif. 1993. The trans-spliceosomal U2 snRNP protein 40K of Trypanosoma brucei: cloning and analysis of function domains reveals homology to a mammalian snRNP protein. EMBO J. 121239-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dungan, J. M., K. P. Watkins, and N. Agabian. 1996. Evidence for the presence of a small U5-like RNA in active trans-spliceosomes of Trypanosoma brucei. EMBO J. 154016-4029. [PMC free article] [PubMed] [Google Scholar]
- 7.Günzl, A., M. Cross, and A. Bindereif. 1992. Domain structure of U2 and U4/U6 small nuclear ribonucleoprotein particles from Trypanosoma brucei: identification of trans-spliceosomal specific RNA-protein interactions. Mol. Cell. Biol. 12468-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Günzl, A., M. Cross, Z. Palfi, and A. Bindereif. 1993. Assembly of the U2 small nuclear ribonucleoprotein from Trypanosoma brucei. A mutational analysis. J. Biol. Chem. 26813336-13343. [PubMed] [Google Scholar]
- 9.Habets, W. J., P. T. Sillekens, M. H. Hoet, J. A. Schalken, A. J. Roebroek, J. A. Leunissen, W. J. van den Ven, and W. Jvan Venrooij. 1987. Analysis of a cDNA clone expressing a human autoimmune antigen: full-length sequence of the U2 small nuclear RNA-associated B″ antigen. Proc. Natl. Acad. Sci. USA 842421-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamm, J., M. Kazmaier, and I. W. Mattaj. 1987. In vitro assembly of U1 snRNPs. EMBO J. 63479-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harper, D. S., L. D. Fresco, and J. D. Keene. 1992. RNA binding specificity of a Drosophila snRNP protein that shares sequence homology with mammalian U1A and U2B″proteins. Nucleic Acids Res. 203645-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartshorne, T., and N. Agabian. 1990. A new U2 RNA secondary structure provided by phylogenetic analysis of trypanosomatid U2 RNAs. Genes Dev. 42121-2131. [DOI] [PubMed] [Google Scholar]
- 13.Kurowski, M. A., and J. M. Bujnicki. 2003. GeneSilico protein structure prediction metaserver. Nucleic Acids Res. 313305-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang, X. H., A. Haritan, S. Uliel, and S. Michaeli. 2003. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lücke, S., K. Jürchott, L. H. Hung, and A. Bindereif. 2005. mRNA splicing in Trypanosoma brucei: branch-point mapping reveals differences from the canonical U2 snRNA-mediated recognition. Mol. Biochem. Parasitol. 142248-251. [DOI] [PubMed] [Google Scholar]
- 16.Nelissen, R. L., C. L. Will, W. J. van Venrooij, and R. Lührmann. 1994. The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 134113-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palfi, Z., and A. Bindereif. 1992. Immunological characterization and intracellular localization of trans-spliceosomal small nuclear ribonucleoproteins in Trypanosoma brucei. J. Biol. Chem. 26720159-20163. [PubMed] [Google Scholar]
- 18.Palfi, Z., S. Lücke, H. W. Lahm, W. S. Lane, V. Kruft, E. Bragado-Nilsson, B. Seraphin, and A. Bindereif. 2000. The spliceosomal snRNP core complex of Trypanosoma brucei: cloning and functional analysis reveals seven Sm protein constituents. Proc. Natl. Acad. Sci. USA 978967-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palfi, Z., W. S. Lane, and A. Bindereif. 2002. Biochemical and functional characterization of the cis-spliceosomal U1 small nuclear RNP from Trypanosoma brucei. Mol. Biochem. Parasitol. 121233-243. [DOI] [PubMed] [Google Scholar]
- 20.Pillai, R. S., M. Grimmler, G. Meister, C. L. Will, R. Lührmann, U. Fischer, and D. Schümperli. 2003. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 172321-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pillai, R. S., C. Will, R. Lührmann, D. Schümperli, and B. Müller. 2001. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain LSm10, a new 14 kDa Sm D1-like protein. EMBO J. 205470-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polycarpou-Schwarz, M., S. I. Gunderson, S. Kandels-Lewis, B. Seraphin, and I. W. Mattaj. 1996. Drosophila SNF/D25 combines the functions of the two snRNP proteins U1A and U2B′ that are encoded separately in human, potato, and yeast. RNA 211-23. [PMC free article] [PubMed] [Google Scholar]
- 23.Price, S. R., P. R. Evans, and K. Nagai. 1998. Crystal structure of the spliceosomal U2B″-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature 394645-650. [DOI] [PubMed] [Google Scholar]
- 24.Saldi, T., C. Wilusz, M. MacMorris, and T. Blumenthal. 2007. Functional redundancy of worm spliceosomal proteins U1A and U2B″. Proc. Natl. Acad. Sci. USA 1049753-9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scherly, D., W. Boelens, N. A. Dathan, W. J. van Venrooij, and I. W. Mattaj. 1990. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B″ and their cognate RNAs. Nature 345502-506. [DOI] [PubMed] [Google Scholar]
- 26.Scherly, D., N. Dathan, W. Boelens, W. van Venrooij, and I. W. Mattaj. 1990b. The U2B″ RNP motif as a site of protein-protein interaction. EMBO J. 93675-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schimanski, B., G. Laufer, L. Gontcharova, and A. Günzl. 2004. The Trypanosoma brucei spliced leader RNA and rRNA gene promoters have interchangeable TbSNAP50-binding elements. Nucleic Acids Res. 32700-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schimanski, B., T. N. Nguyen, and A. Günzl. 2005. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot. Cell 41942-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson, G. G., P. Vaux, G. P. Clark, R. Waugh, J. D. Beggs, and J. W. Brown. 1991. Evolutionary conservation of the spliceosomal protein, U2B″. Nucleic Acids Res. 195213-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 18560-89. [DOI] [PubMed] [Google Scholar]
- 31.Tang, J., N. Abovich, and M. Rosbash. 1996. Identification and characterization of a yeast gene encoding the U2 small nuclear ribonucleoprotein particle B″ protein. Mol. Cell. Biol. 162787-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tkacz, I. D., Y. Lustig, M. Z. Stern, M. Biton, M. Salmon-Divon, A. Das, V. Bellofatto, and S. Michaeli. 2007. Identification of novel snRNA-specific Sm proteins that bind selectively to U2 and U4 snRNAs in Trypanosoma brucei. RNA 1330-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tschudi, C., S. P. Williams, and E. Ullu. 1990. Conserved sequences in the U2 snRNA-encoding genes of Kinetoplastida do not include the putative branchpoint recognition region. Gene 9171-77. [DOI] [PubMed] [Google Scholar]
- 34.Wang, P., Z. Palfi, C. Preusser, S. Lücke, W. S. Lane, C. Kambach, and A. Bindereif. 2006. Sm core variation in spliceosomal small nuclear ribonucleoproteins from Trypanosoma brucei. EMBO J. 254513-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.